The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
14
|
sequence length |
89
|
structure length |
89
|
Chain Sequence |
VHFCDKCGLPIKVYGRMIPCKHVFCYDCAILHEKKGDKMCPGCSDPVQRIEQCTRGSLFMCSIVQGCKRTYLSQRDLQAHINHRHMRAG
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
Dimeric switch of Hakai-truncated monomers during substrate recognition: insights from solution studies and NMR Structure
doi rcsb |
| molecule tags |
Ligase
|
| source organism |
Mus musculus
|
| molecule keywords |
E3 ubiquitin-protein ligase Hakai
|
| total genus |
14
|
| structure length |
89
|
| sequence length |
89
|
| ec nomenclature |
ec
2.3.2.27: RING-type E3 ubiquitin transferase. |
| pdb deposition date | 2014-06-11 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF18408 | zf_Hakai | C2H2 Hakai zinc finger domain |
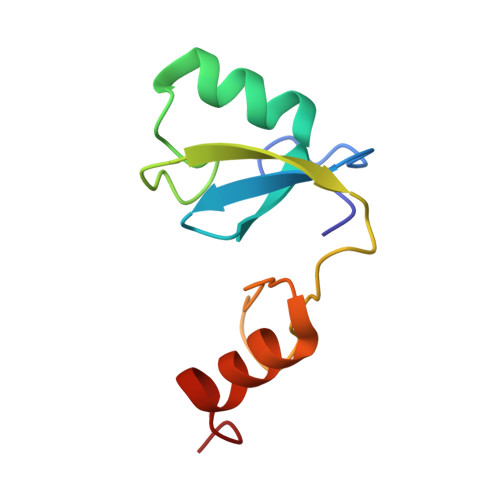
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...