The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
108
|
sequence length |
327
|
structure length |
327
|
Chain Sequence |
SKARVYADVNVLRPKEYWDYEALTVQWGEQDDYEVVRKVGRGKYSEVFEGINVNNNEKCIIKILKPVKKKKIKREIKILQNLCGGPNIVKLLDIVRDQHSKTPSLIFEYVNNTDFKVLYPTLTDYDIRYYIYELLKALDYCHSQGIMHRDVKPHNVMIDHELRKLRLIDWGLAEFYHPGKEYNVRVASRYFKGPELLVDLQDYDYSLDMWSLGCMFAGMIFRKEPFFYGHDNHDQLVKIAKVLGTDGLNVYLNKYRIELDPQLEALVGRHSRKPWLKFMNADNQHLVSPEAIDFLDKLLRYDHQERLTALEAMTHPYFQQVRAAENS
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
pubmed doi rcsb |
| molecule keywords |
Protein kinase CK2
|
| molecule tags |
Transferase
|
| source organism |
Zea mays
|
| total genus |
108
|
| structure length |
327
|
| sequence length |
327
|
| ec nomenclature |
ec
2.7.11.1: Non-specific serine/threonine protein kinase. |
| pdb deposition date | 2002-05-07 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00069 | Pkinase | Protein kinase domain |
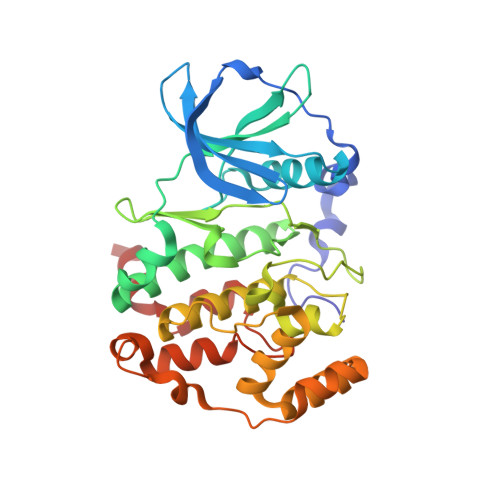
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Alpha | Orthogonal Bundle | Transferase(Phosphotransferase); domain 1 | Transferase(Phosphotransferase) domain 1 | ||
| Alpha Beta | 2-Layer Sandwich | Phosphorylase Kinase; domain 1 | Phosphorylase Kinase; domain 1 |
#chains in the Genus database with same CATH superfamily 1A06 A; 5BVW A; 1H1Q A; 2EB3 A; 4UMQ A; 2VAG A; 3BE9 A; 3QWJ A; 3SVV A; 2O8Y A; 5AP7 A; 3C4E A; 5DRB A; 5ITA A; 2GQR A; 4ERW A; 1U5Q A; 4XMO A; 5EOB A; 3VS3 A; 5CAV A; 2PUL A; 3U8W A; 3HV6 A; 3FML A; 3QLG A; 1YHV A; 3GCQ A; 1ZAR A; 3CQW A; 1B6C B; 2H6D A; 2J0K A; 4MXX A; 2JDS A; 5D41 A; 5AUV A; 2QC6 A; 5CU6 A; 4EHG A; 1OUY A; 3D5W A; 2YWV A; 5KMN A; 2FSM X; 3FC1 X; 4BZO A; 3RWP A; 3BKB A; 3KK9 A; 1E1V A; 3JUH A; 5J8I A; 3FDN A; 5HD7 A; 4DCE A; 2XEZ A; 4NUS A; 1UU7 A; 2QOL A; 1Q61 A; 3UNJ A; 1W0X C; 3BEA A; 2BIL B; 2C1A A; 2YIT A; 3EKK A; 4G6N A; 2ZM4 A; 1XH7 A; 5IDN A; 3TDV A; 3WF7 A; 3PIZ A; 3LLT A; 3PZE A; 3QQK A; 2HW7 A; 4XHK B; 4D2T A; 1ZYJ A; 4DCA A; 4J8M A; 3DAJ A; 4IZA A; 1R3C A; 2BDF A; 1GNG A; 5C8K A; 4A4O A; 3WIL A; 4BTM A; 4UYA A; 3FPM A; 4W4Y A; 3TKH A; 4TWP A; 1XWS A; 3CD3 A; 4RIW A; 4UV0 A; 3ZUT A; 2J2I B; 1H1S A; 4JRV A; 3DJ7 A; 3QRI A; 3FE3 A; 2BIY A; 3IW7 A; 3Q4C A; 3ZYA A; 2KUL A; 3F3W A; 5KE0 A; 5A6N A; 5JFV A; 5CEM A; 3IDB A; 4CMT A; 4NM3 A; 1WVW A; 5CI7 A; 5CSW A; 5HBH A; 4MED A; 4BIE A; 3OWP A; 5FP5 A; 3VBQ A; 5LWN A; 4B9D A; 3EQI A; 4BB4 A; 5I4N A; 3EL8 A; 5LVL A; 1J3H A; 1KV1 A; 3BLR A; 3PVU A; 4AGC A; 4DHF A; 2W0J A; 5HVK A; 3V01 A; 2F57 A; 3HVC A; 3RM6 A; 4HVG A; 4LI5 A; 2ACX A; 5EQY A; 2V5Q A; 3QRJ A; 4EZ5 A; 3V8S A; 3UTO A; 1XR1 A; 3ZUV A; 3DLS A; 2X8E A; 5FM2 A; 4C61 A; 2PVY A; 4D5H A; 1GAG A; 4NTT A; 1PXO A; 4II5 A; 1PKG A; 2VTI A; 3HV3 A; 4JI9 A; 4I0S A; 1Y8Y A; 5AA8 A; 3SQQ A; 3CC6 A; 3E3B X; 4I92 A; 4QNY A; 4ZLO A; 3K5V A; 2IVV A; 3QRK A; 4FC0 A; 2JKO A; 2A27 A; 3GU7 A; 5EDQ A; 3FMD A; 3HMO A; 3S95 A; 5M09 A; 5JGA A; 4L44 A; 4UZD A; 5D12 A; 3BBW A; 3OOM A; 5BVK A; 4FYN A; 2OI4 X; 3I6W A; 2W4K A; 2C68 A; 1WFC A; 4F9A A; 4DFU A; 1F0Q A; 4IDV A; 3PSD A; 4FSN A; 3PY0 A; 4X3F C; 5IEY A; 3TM0 A; 4CFN A; 3ENH A; 3JPV A; 4RC3 A; 2CCI A; 3BBT B; 3UJG A; 1SVH A; 5J7B A; 2OXX A; 2OF2 A; 3UQF A; 1UKI A; 2VGO A; 4NU1 A; 5HE3 A; 4QYY A; 4GK4 A; 4U41 A; 1PME A; 3UOH A; 2XNG A; 5EI6 A; 4PMT A; 3GCV A; 3ZZW A; 2VX1 A; 2WIH A; 4OTW A; 1QPC A; 1U4D A; 5E90 A; 5K5N A; 4FBX A; 3OW4 A; 4R3P A; 2YM5 A; 2JAM A; 3U9N A; 4NZW B; 5EQP A; 3EN9 A; 3QD3 A; 5FRI A; 4X3F A; 4DH7 A; 3R28 A; 3TTJ A; 5IA2 A; 1FGI A; 1RW8 A; 5L2I A; 2OH4 A; 4FIF A; 1FMO E; 4LGG A; 4Q2A A; 4IJP A; 2XJ2 A; 5HTC A; 4G1W A; 4IZY A; 3TAC A; 2V22 A; 3EN5 A; 4KA3 A; 4AWI A; 1S9I A; 4DEA A; 5I5Z A; 3R1S A; 1FMK A; 4QMV A; 4B0G A; 4HPU E; 3R9R A; 2R3P A; 1XHA A; 4D4Y A; 4U97 A; 1PJK A; 2OJJ A; 4CKI A; 4OTD A; 3OWJ A; 2JDO A; 3DV3 A; 4QQT A; 4ZMF A; 2QOB A; 1R0E A; 4FVP A; 3W16 A; 2G9X A; 2WTI A; 4QMY A; 1BMK A; 5D11 A; 3FZO A; 2UW8 A; 4CCU A; 5LQF A; 4DG0 E; 4IW0 A; 2XP2 A; 5LMK C; 2J6M A; 5E91 A; 2XRW A; 5AP1 A; 2J51 A; 2C69 A; 3AD6 A; 3QUE A; 2ITW A; 2Y4I C; 3H10 A; 3QTS A; 4RWK A; 3EQG A; 4U80 A; 5B2M A; 3CLY A; 2JFM A; 3DK6 A; 4LOO A; 3FI8 A; 3O7L D; 4GK2 A; 1UVR A; 2BKZ A; 2C47 A; 2PZP A; 2PVK A; 3VUL A; 3DCV A; 4W9W A; 4NJD A; 4H3B A; 3LXL A; 1OMW A; 4QMP A; 4JG6 A; 4ASZ A; 3JY0 A; 5KMO A; 1YOL A; 4J96 A; 5E8Y A; 3R02 A; 5EYD A; 3EB0 A; 4JBQ A; 2PVM A; 3R2Y A; 3FQE A; 5LMK A; 5EM7 A; 3JR1 A; 2ETM A; 5CVG A; 5DMZ A; 5AAF A; 5DR2 A; 2E9V A; 4XCU A; 4CFW A; 3QCY A; 2GNL A; 4YJN A; 3P7A A; 4C4J A; 2NP8 A; 4IDT A; 5LVM A; 4I0R A; 3DBE A; 3LFQ A; 4IFG A; 4DGM A; 2XIX A; 3F3U A; 2Z02 A; 2YAC A; 2PE1 A; 4XNE A; 1BKX A; 5KNJ A; 1RJB A; 4FSR A; 4ARK A; 1UKH A; 2PZY A; 3POO A; 3VC4 A; 2HW6 A; 4M15 A; 1F5Q A; 2ESM A; 3FV8 A; 4ZSL A; 1J91 A; 4XYF A; 5HX8 A; 5AWM A; 3FC2 A; 3FY0 A; 4LG4 A; 4XG7 A; 4YTI A; 3D14 A; 5IME A; 5AV2 A; 3MGY A; 4F70 A; 4XX9 A; 4HPT E; 4EK8 A; 3AQV A; 4IWO A; 4ZOG A; 4PL4 A; 4YO4 A; 1JKT A; 2VRX A; 3VS5 A; 2ERK A; 3K3J A; 3QZG A; 4F0F A; 5TE0 A; 3F2R A; 4O7T A; 2ITO A; 3EID A; 2OFV A; 5DLS A; 4JAJ A; 3NWW A; 5DYJ A; 4Q5E A; 1MQ4 A; 3P4K A; 5L8K A; 4ERK A; 1A9U A; 2XEY A; 4ZAU A; 5AAD A; 3QCQ A; 3F3V A; 3PJ2 A; 4U3Z A; 2QOF A; 3PVW A; 3Q9X A; 3NIZ A; 3QRT A; 4AAC A; 4FTL A; 1ZYS A; 5JN2 A; 4XBR A; 4QD6 A; 4IC8 A; 4QTE A; 3V04 A; 3LFD A; 4CXA A; 2BAK A; 4AU8 A; 3AMB A; 4S32 A; 4C62 A; 4BDD A; 3QZH A; 3ORP A; 4UWC A; 3E92 A; 3ZLY A; 2HZ4 A; 3ODZ X; 3UO4 A; 5HLN A; 4H05 A; 2X7G A; 3DKC A; 4AT4 A; 4DED A; 3L8X A; 3VS4 A; 1Q99 A; 1IAS A; 2WFY A; 3R7Y A; 4ASE A; 4E5A X; 5G6V A; 3V8T A; 3E8C A; 3LFF A; 2CNQ A; 5BVN A; 2XNP A; 3VBY A; 4AZS A; 4L7F A; 5TF9 A; 2F7X E; 3S1H A; 2C5X A; 3H9R A; 4C4H A; 5JFS A; 4FOD A; 2QOC A; 2P2I A; 4BCI A; 3PXF A; 2R4B A; 1KOB A; 2XUU A; 2PE0 A; 4N6Y A; 1PMU A; 2UZT A; 3ZDU A; 5T6K A; 2VTH A; 2YJR A; 4WNM A; 4BTF A; 1H08 A; 4P90 A; 4O2Z A; 1AQ1 A; 3QUP A; 3OFM A; 4M13 A; 4M97 A; 5EI8 A; 5AAE A; 4I22 A; 4CVA A; 2G2H A; 3P23 A; 4DEI A; 3GVU A; 4J99 A; 3DLZ A; 5HNG A; 5BVE A; 4EOI A; 5M4F A; 4DEH A; 1AGW A; 2R9S A; 4ZK5 A; 3RKB A; 4PDY A; 3EKN A; 3D15 A; 2CGU A; 4J53 A; 5AUY A; 2WNT A; 2YIW A; 3L8V A; 3LM5 A; 4TYH B; 3R7V A; 5IPJ A; 3LVP A; 4ZTQ A; 3FZR A; 4GVJ A; 1CTP E; 5E8W A; 4C0T A; 4U9A A; 2B9I A; 2B55 A; 2YHV A; 4K1B A; 3PVG A; 2C0I A; 4L3P A; 4XLI A; 1HCK A; 5J87 A; 5DNR A; 2GQS A; 5L2T A; 3AC4 A; 4UX9 A; 4M3Q A; 5L2W A; 3REP A; 2C5V A; 4O0W A; 2OSC A; 4J97 A; 4CTB A; 1U46 A; 5J5S A; 4IWP A; 4IAA A; 3LFB A; 4IB5 A; 4OTP A; 3R9H A; 4QQ5 A; 3ZRL A; 4RLL A; 3FME A; 1NW1 A; 3O2M A; 4U8Z A; 4DGN A; 5KUP A; 2B9F A; 3MVJ A; 4GEO A; 2A0C X; 4CTC A; 3CKW A; 1VYZ A; 5M08 A; 2BDJ A; 5FDX A; 2GCD A; 1XO2 B; 4FNY A; 3KF4 A; 1PXK A; 4RFY A; 4BCN A; 3F6X A; 4FL1 A; 1SNU A; 4GYG A; 5AFV A; 3QAL E; 4FTT A; 4RX8 A; 3R9D A; 3UOL A; 1IA9 A; 5L2Q A; 3SD0 A; 1KE9 A; 3QX4 A; 5CS6 A; 4FSQ A; 4WT6 A; 4QP3 A; 3LPB A; 2Q0N A; 3UZT A; 4MNE B; 5EYC A; 3CIK A; 4C7T A; 3PJC A; 4O0U A; 4MK0 A; 4XV9 A; 2Z7S A; 3PXY A; 3PG1 A; 3Q32 A; 1WBN A; 2C3I B; 3SKC A; 3BZ3 A; 1LEZ A; 2C6M A; 5CXH A; 4ZSG A; 2CN8 A; 4A9S A; 3QGW A; 2P3G X; 4G9R A; 2Z8C A; 3T9I A; 3AC8 A; 4F08 A; 2PY3 A; 3MSS A; 4R5S A; 3W2Q A; 2R3M A; 4BI1 A; 2I6L A; 2QHN A; 1ZOG A; 4H3P A; 4CKR A; 3V3V A; 2YCS A; 2VX3 A; 3A8W A; 3NUN A; 4Z16 A; 5TT7 A; 3CGO A; 5H8B A; 4OUC A; 5CEO A; 4U7Z A; 5A14 A; 3HMI A; 3FLZ A; 4A9U A; 4RA5 A; 3B2W A; 3R7I A; 3ANR A; 2I0Y A; 4UWB A; 3V6R A; 1WCC A; 3V8W A; 2WMS A; 4ZS4 A; 3UQG A; 3FPQ A; 1JKS A; 4RJ5 A; 3BGZ A; 4Y83 A; 4ALW A; 2GNI A; 4MD7 E; 2WQB A; 1XH5 A; 4WUA A; 5M07 A; 4MYG A; 2EWA A; 2B7A A; 3OWK A; 4UMU A; 3I5Z A; 5CU3 A; 4BDA A; 4FG9 A; 4C36 A; 3FHR A; 3CGF A; 3SOA A; 4C4I A; 3BGQ A; 1J1B A; 2R3O A; 3KUL A; 4APC A; 2FH9 A; 5EFQ A; 2G2I A; 3RPY A; 3FXZ A; 1RDQ E; 2C6E A; 2YFX A; 5CX9 A; 5ACB C; 4AFJ A; 5E1E A; 4USF A; 4R7H A; 3Q5Z A; 2Q0B A; 2VNY A; 5AR5 A; 3TXO A; 1OZ1 A; 4FOC A; 1M2R A; 1FVT A; 2E9U A; 4WD5 A; 3HP2 A; 3NEW A; 4EOO A; 4RX7 A; 3UMW A; 2Y8O A; 2ZM3 A; 3GBZ A; 3SGC A; 2WGJ A; 4FV8 A; 2NRU A; 2V55 A; 4BN1 A; 3U51 A; 2YZL A; 1YXU A; 2ETR A; 1MRY A; 3COI A; 3DKO A; 4W4X A; 4C4G A; 2FA2 A; 4LOQ A; 4KIO A; 3CCN A; 3HRC A; 4U79 A; 3V51 A; 4LGH A; 2YLE A; 1JWH A; 3HLL A; 4A4X A; 3N4T A; 1FPU A; 4QYF A; 3BV2 A; 3V5L A; 3UPZ A; 3FQS A; 3DAK A; 2BHH A; 5CZH A; 2XBA A; 3NAY A; 4LFI A; 3RPV A; 2B54 A; 3LAU A; 5FUT A; 5JK3 A; 3EWH A; 4IAF A; 3F5G A; 1I09 A; 3QYW A; 4A9R A; 5KKR B; 5B0X A; 2GDO A; 2Z2W A; 3F66 A; 4BGH A; 3FZP A; 4KIQ A; 4ITJ A; 4WBO A; 2CKO A; 4NIF B; 4IX5 A; 2GTN A; 2C5N A; 4RFZ A; 2X6E A; 2UW3 A; 2GSF A; 4EWQ A; 5KMJ A; 4G31 A; 4EH4 A; 2KTY A; 2WU7 A; 2AC5 A; 1IAH A; 1PXP A; 4U40 A; 4ONA A; 2JBO A; 2VWZ A; 4FEU A; 4A9Y A; 3R04 A; 4DLJ A; 2CN5 A; 2HY0 A; 1V0O A; 5EHO A; 2Z60 A; 4S34 A; 4DTA A; 2QOI A; 2WMV A; 4ZTS A; 4YZ9 A; 4NH1 A; 1PXJ A; 3KXN A; 2HXQ A; 4XP2 A; 4FKP A; 4JT3 A; 4IM0 A; 3ZXZ A; 5JH6 A; 2UZO A; 2W5H A; 3Q2M A; 4NW6 A; 4TY1 A; 3RAL A; 3RIN A; 3OCT A; 3W10 A; 4M84 A; 4IC7 A; 4DH8 A; 3UGC A; 5GIO E; 5JZJ A; 1ZZL A; 4UB7 A; 2ZYB A; 2UW4 A; 4FZ7 A; 3PWD A; 3ZHP C; 4KRD A; 4AN2 A; 2XNM A; 4AS0 A; 3V6S A; 1OUK A; 1GZ8 A; 3HV5 A; 2R3G A; 4PUZ A; 4U0I A; 5FP6 A; 3ORX A; 1FIN A; 2YLF A; 4FNW A; 4Q9Z A; 1OL2 A; 5A6O A; 3ODY X; 3QL8 A; 3QZF A; 3PFQ A; 2YN8 A; 2BFX A; 4MBL A; 3SHE A; 3D5X A; 1FGK A; 3PLS A; 3TUC A; 2GFC A; 4BDB A; 3NR9 A; 3L9L A; 5DHJ A; 4GL9 A; 3DNE A; 4GV1 A; 4NKA A; 5EKO A; 4L9I A; 5AUU A; 5HG9 A; 1NT2 A; 3UP2 A; 2XK3 A; 4LMN A; 5BMM A; 3SLS A; 1UWJ A; 3G0E A; 3LFA A; 3OXI A; 4Y5H A; 5A4C A; 4FV7 A; 2WXV A; 4LL0 A; 3KRR A; 3ZFX A; 1YOJ A; 2QOH A; 4OBO A; 2UVX A; 3KRJ A; 4ANO A; 4BBE A; 5IA3 A; 4RC4 A; 2GS2 A; 3OCB A; 4ITH A; 2JDR A; 4MXA A; 4USD A; 3VBT A; 2FVD A; 1WBV A; 2X2M A; 3NYO A; 4EOJ A; 1J7L A; 5FEE A; 3H9F A; 5IA0 A; 2HAK A; 2BCJ A; 3MVM A; 3VS1 A; 5LWM A; 5C1Q B; 4I94 A; 3EJ1 A; 2VNW A; 3RPO A; 1BUH A; 3NAX A; 3AG9 A; 3F61 A; 4O1O A; 4YJO A; 3R8V A; 2R3J A; 1YKR A; 3HL7 A; 5B2K A; 4AOJ A; 5AP0 A; 3BHY A; 3P78 A; 4KIP A; 4BTK A; 3IPH A; 4D58 A; 2XKD A; 3F9N A; 4LRK A; 4OLI A; 4ZY6 A; 4JRN A; 2X39 A; 1J7U A; 4BDG A; 4D9T A; 1BYG A; 2GFS A; 2YDK A; 3F2A A; 2UW7 A; 4JXF A; 4MF1 A; 2DYL A; 2NPQ A; 4QOX A; 4Y8D A; 5M44 A; 4MQ1 A; 3Q9W A; 2OF4 A; 2RGP A; 2C6T A; 3ZRM A; 1Q8Z A; 3CY2 A; 3QKM A; 4O7Z A; 5HI2 A; 4R5Y A; 4FL2 A; 5CEI A; 4N57 A; 2C6I A; 2QLQ A; 5FTQ A; 1PKD A; 2ZMC A; 3WIG A; 4X3J A; 1APM E; 5I3O A; 4NM5 A; 2OWB A; 4ZLY A; 3UO5 A; 2X81 A; 3F3Z A; 4PMP A; 5CQU A; 4P5Q A; 3OG7 A; 4FK3 A; 2YIY A; 3E8N A; 3HRF A; 4H58 A; 4RIW B; 3MFT A; 4NKS A; 3F5U A; 5ANK A; 3IW5 A; 3OWL A; 4HYS A; 2JKM A; 4W9X A; 3OVV A; 1ZZ2 A; 4QQC A; 3QC4 A; 2Y7J A; 3ROC A; 1GZO A; 3F7W A; 3BHU A; 3IW8 A; 1OPJ A; 2W9Z B; 2R3N A; 3NIE A; 3OBG A; 3C4Z A; 5AV1 A; 4E73 A; 5A54 A; 2X0G A; 4XEY A; 5A4T A; 4I5M A; 3G9L X; 4MD9 E; 4YJP A; 4YMJ A; 4EI4 A; 1H24 A; 3N4V A; 3PRF A; 4Y46 A; 4U6R A; 3G51 A; 2VTA A; 1K3A A; 3RCJ A; 3FZS A; 1F3M C; 4DG3 E; 2YM3 A; 2YDJ A; 2OGV A; 4GT5 A; 5BVO A; 5GZ9 A; 2UV2 A; 3NCZ A; 3Q96 A; 4BIC A; 5IU2 A; 4HGT A; 3CKX A; 4F4P A; 3E7O A; 3ZFM A; 4DIT A; 3L1S A; 4WG5 A; 3Q3B A; 2EXC X; 5K5X A; 5IQE A; 3HV7 A; 4O82 A; 3P08 A; 3R21 A; 5L3A A; 3SX9 A; 5HEZ A; 4OTG A; 2C5Y A; 2W1F A; 2WEL A; 3DBF A; 3EFW A; 3MB6 A; 5AR3 A; 4AOI A; 3O7L B; 4DH3 A; 4KB8 A; 2PL0 A; 3JXW A; 3MPT A; 4P7E A; 4BI2 A; 3DDP A; 4MAO A; 5GIN E; 3TNQ B; 3L9N A; 4M67 A; 4DG2 E; 3F88 A; 3QQG A; 1SVE A; 2HXL A; 3CS9 A; 4K11 A; 4PY1 A; 4FTI A; 2PZR A; 3ZMM A; 3R1Y A; 4I6B A; 1JNK A; 1JSV A; 3VJN A; 1YDR E; 5EI2 A; 4N4S A; 3UVQ A; 3B8R A; 4C57 A; 4RPV A; 5H3Q A; 2RSV A; 5E1S A; 3DFC B; 4MD8 E; 4S2Z A; 5G1X A; 1Q8T A; 4QMM A; 3SV0 A; 4A06 A; 2PWL A; 2UZB A; 2VX0 A; 3NCG A; 4QP2 A; 5EOL A; 5HCY A; 4X3F B; 1S9J A; 2QOD A; 3KRE A; 4BID A; 4AZV A; 3NLB A; 2DUV A; 5E8T A; 4GS6 A; 4QYE A; 4UAL A; 4YLL A; 3CQE A; 4NW5 A; 5EW8 A; 4X2K A; 4XG9 A; 2PVH A; 3K5U A; 4DTB A; 4FVR A; 4PDO A; 3LXN A; 5HCZ A; 5IIS A; 2ERZ E; 3NNW A; 3RAH A; 4XUF A; 3E5A A; 3ZCL A; 4YBJ A; 4MCV A; 4KBA A; 3NX8 A; 1HOW A; 5BPY A; 5GJD A; 1X8B A; 5KCV A; 3IGO A; 4EH2 A; 4IH8 A; 3RCD A; 1TQP A; 4YR8 A; 4CD0 A; 5J9Z A; 5CU4 A; 1BLX A; 3SXR A; 4QMQ A; 3WF8 A; 3QX2 A; 5AV3 A; 3PDT A; 1W8C A; 4AN3 A; 4RJ6 A; 3SAY A; 5CF6 A; 4Z9L A; 4ACG A; 1V0P A; 4U5J A; 4KS8 A; 3R8P A; 1H26 A; 3TJD A; 3BHT A; 3KXG A; 1KE5 A; 1ZXE A; 5CWZ A; 1JVP P; 3P9J A; 3PPK A; 4FIC A; 4KRC A; 4QTB A; 3GOP A; 4YGA A; 4ZS0 A; 2BRH A; 3OCG A; 4ZTL A; 4YZM A; 2X6D A; 5ETC A; 4PTE A; 4GG5 A; 4YNO A; 4NFN A; 5CEQ A; 1P4O A; 2Y4I B; 4HYH A; 4RX9 A; 4XV3 A; 4BR3 A; 1XJD A; 3OTU A; 2LGC A; 3SA0 A; 3ZBX A; 1Y6B A; 1REK A; 1ZYC A; 4BDF A; 3A61 A; 5CI6 A; 4AX8 A; 1NVQ A; 4BCP A; 5IA1 A; 3QD0 A; 4WOV A; 3KMU A; 5B2L A; 4DA5 A; 2HY8 1; 3EQF A; 1I44 A; 2BTR A; 4Z7G A; 1PXI A; 3EN6 A; 3TNP C; 3ZLW A; 3TUD A; 3O8T A; 2JII A; 1PF8 A; 3QBN A; 3R7G A; 4EH7 A; 5AND A; 3ALN A; 2VTT A; 5CQW A; 3LFE A; 5T68 A; 2R3H A; 4JIK A; 4JJR A; 5AP4 A; 3OS3 A; 4BDC A; 1OM1 A; 3SG8 A; 2G1T A; 4E4N A; 3TZ7 A; 2I0H A; 2R5T A; 4GUE A; 4C34 A; 4E1Z A; 3GXL A; 2W1D A; 2I7Q A; 2EB2 A; 2WU6 A; 4AA4 A; 3VQH A; 4J71 A; 5CF5 A; 4BWX A; 3PWY A; 4BDK A; 4L43 A; 4RVK A; 5HGI A; 4W4W A; 3RP9 A; 2F2U A; 3DBD A; 4X2N A; 4L53 A; 3WE4 A; 3E7V A; 1TQI A; 3O23 A; 4RMZ A; 4C4F A; 1JSU A; 3AT2 A; 2RF9 A; 5AM7 A; 5BYL A; 2BRG A; 3IEC A; 3ZLX A; 4BKZ A; 2RFN A; 3MY5 A; 4I0T A; 3FMN A; 3EFL A; 3WIK A; 4EOM A; 5IQA A; 4JLC A; 5IUI A; 4R78 A; 4BCO A; 2GQG A; 2OXD A; 3A7G A; 2IVS A; 5IEV A; 3GC0 A; 3G0F A; 4WG3 A; 4NL0 A; 2B9H A; 3RAK A; 1ZMU A; 3PUP A; 2O65 A; 2IZR A; 5ANI A; 4D2S A; 4I6F A; 2PMO X; 2UZV A; 4I5H A; 1V0B A; 1XKK A; 3ZFY A; 4RX5 A; 3B2T A; 3DGK A; 3G5D A; 2P0C A; 3QKL A; 3KCK A; 4I5P A; 4HVH A; 1IAJ A; 3UC4 A; 3HA6 A; 4BC6 A; 3DS6 A; 4NFM A; 1H1R A; 1W82 A; 3GB2 A; 3NUX A; 4O83 A; 4V0G B; 3GC8 A; 5IQH A; 4YTH A; 3SW7 A; 3CEK A; 2F49 A; 4FG7 A; 4KIK B; 3O71 A; 2ITQ A; 4G3E A; 2JAV A; 4O7N A; 5IQD A; 4EKL A; 2WIP A; 1PW2 A; 4X2G A; 4JG7 A; 4M12 A; 4OTI A; 2WTD A; 5CU0 A; 4M8T A; 3I60 A; 1YXS A; 2BMC A; 3F2N A; 5I3R A; 3E64 A; 1PYE A; 2XJ0 A; 4FKR A; 4YLK A; 4EWH A; 1L8T A; 1OIR A; 2XS0 A; 4EK3 A; 4D4R A; 3D0E A; 4AQC A; 4FKV A; 4RT7 A; 3C4F A; 4IAC A; 4BFM A; 1KE7 A; 4TND A; 3R2B A; 3JS2 A; 4FUY A; 3R8U A; 3R30 A; 4NCT C; 4TYG A; 2XIR A; 4KNB A; 4TKS A; 3VUK A; 3KMW A; 3N51 A; 4RFM A; 1Y6A A; 1FBN A; 3CSV A; 2F2C B; 2GNJ A; 3SW4 A; 2B4S B; 5IQF A; 3LFC A; 3LHJ A; 4U45 A; 2CSN A; 4ZJJ A; 4B8M A; 2QG7 A; 3F5X A; 5CNN A; 5JSM A; 4O84 A; 3LJ2 A; 4G5P A; 5HOA A; 2WTK B; 2XKF A; 1Q41 A; 1FVV A; 4FGB A; 4PF4 A; 2C1B A; 4X6Q C; 3ZO4 A; 3DA6 A; 3F69 A; 3L9P A; 4NCT A; 2WQE A; 2PUI A; 5AX3 A; 4EQC A; 4MKC A; 3CP9 A; 4YU2 A; 3UZR A; 3POZ A; 4PNI A; 3D9V A; 3DXP A; 3Q9Y A; 1KE6 A; 3MV5 A; 4QNA A; 4XG4 A; 3MH2 A; 1BI8 A; 3U87 A; 4BWK A; 4FT7 A; 5E8S A; 4CZY B; 3ERK A; 2R7B A; 4PMM A; 4QP4 A; 4ENY A; 4B4L A; 1YW2 A; 3KK8 A; 3AGM A; 3D4Q A; 4GJ2 A; 2CHL A; 4Q5H A; 5D1J A; 4EZ3 A; 5GJG A; 4F7L A; 3RTP A; 3ET7 A; 4BTJ A; 3PRI A; 4RIX A; 2YER A; 3UMX A; 2QQ7 A; 2QVS E; 4PRJ A; 2OJI A; 1UNH A; 3EMG A; 5AIK A; 3RNI A; 2ZM1 A; 4X0M A; 3SRV B; 3TKI A; 1PHK A; 4EYJ A; 4TTH B; 4FGR A; 3A4P A; 2NNW B; 3Q04 A; 1VR2 A; 4ZSJ A; 2F4J A; 4UMT A; 1J7I A; 1WBP A; 3QWK A; 3O8P A; 3K3I A; 4AT5 A; 4O22 A; 4RLK A; 2YWP A; 5IF1 A; 2BRB A; 1GY3 A; 3R00 A; 5HG8 A; 3CTQ A; 4Y5Q A; 1FQ1 B; 5CSX A; 4EJN A; 3HEG A; 3ID6 C; 4GII A; 3TV6 A; 2CKE A; 3NRM A; 3QQF A; 2WQN A; 4QMZ A; 3MFS A; 2WMR A; 3QQJ A; 4CQE A; 5IQC A; 5FLF A; 2ONL C; 3PXK A; 4IB3 A; 3RZF A; 3THB A; 3UE4 A; 3BI6 A; 3KU2 A; 3OHT A; 4KAB A; 3LW0 A; 4NWM A; 4ZTN A; 5C26 A; 1U54 A; 2W1Z A; 5HZN A; 2LAV A; 3DZQ A; 4RZW A; 4CG9 A; 4DN5 A; 2W4O A; 3ORL A; 5LMA A; 3WE8 A; 3M1S A; 5A3X A; 3EHA A; 4BVU A; 2B53 A; 5A4L A; 3SDJ A; 3Q52 A; 2VTQ A; 3DZO A; 5FDP A; 5AX9 A; 1KV2 A; 1YI3 A; 4IMY A; 3MA3 A; 4WBB B; 4ZY4 A; 5KMK A; 4XP0 A; 5LCJ A; 1EH4 A; 2RL5 A; 2ONL A; 3PA3 A; 4FTA A; 4S33 A; 3RHX A; 4D0W A; 2GNH A; 5KML A; 4M66 A; 1XH8 A; 4GT4 A; 4X2J A; 3P7C A; 3IW6 A; 4GKI A; 3C51 A; 3BIZ A; 3ORN A; 4QMT A; 3NUS A; 4GVA A; 4K77 A; 5C8M A; 1G8A A; 4D5K A; 4PPB A; 2ZV7 A; 3LXP A; 2QON A; 2P4I A; 3KUL B; 4K6Z A; 4CKJ A; 2E9O A; 2JDT A; 4BZN A; 2JKK A; 5E8U A; 3A4O X; 1YRP A; 3W2P A; 4D28 A; 4I4E A; 4F1O A; 3PXZ A; 3VHK A; 2OKR A; 1UNG A; 4KKG A; 5IA5 A; 2I40 A; 3GC7 A; 4IAD A; 5E8X A; 3W2R A; 4PX6 A; 4USE A; 3G6H A; 3QTX A; 5TKD A; 4KAO A; 4EC9 A; 4QMN A; 4JQ7 A; 2EUF B; 4CRL A; 1R78 A; 5HES A; 4ANM A; 1PXL A; 1Q3W A; 2B1P A; 4I5C A; 4G3G A; 1SZM A; 3VUH A; 4ENX A; 5IEX A; 2VTS A; 5JEB A; 3UIM A; 4G3F A; 3W8L A; 4ANS A; 1OIQ A; 4IFC A; 3I0Q A; 4I21 A; 4JG8 A; 2YJS A; 4KS7 A; 4E7W A; 4DLI A; 5F95 A; 4UYN A; 4YC6 A; 4WA9 A; 2VWX A; 4FIJ A; 4L46 A; 4PMS A; 3TIZ A; 3BLH A; 3TZM A; 4GK3 A; 2ZOQ A; 4AZE A; 3EFJ A; 5AUW A; 2OIQ A; 3RK5 A; 2OO8 X; 1U5R A; 3PP0 A; 3V5P A; 2GHM A; 4R1Y A; 3RK7 A; 5I9X A; 3WZD A; 4ZJV A; 1P14 A; 5EM6 A; 1YHW A; 2JBP A; 1YI6 A; 1OKZ A; 2REI A; 3R9O A; 2J0J A; 4K2R A; 1PXN A; 4AE6 A; 3Q2J A; 4EL9 A; 3DTC A; 3QCS A; 4TZR A; 5TOZ A; 3Q9Z A; 4RVT A; 1WBO A; 3ANQ A; 2GMX A; 2WTW A; 3HP5 A; 4GYI A; 3Q4Z A; 2DWB A; 2FSL X; 3A7F A; 2YDI A; 4F63 A; 3VUI A; 2C30 A; 4K18 A; 5ANO A; 1H00 A; 5HX6 A; 1UU8 A; 4LUD A; 5IUH A; 5KX8 A; 2Z7R A; 3CPC A; 4ACD A; 2WD1 A; 2W05 A; 2YAK A; 2ZV2 A; 4B6L A; 3VBV A; 3MFR A; 4BYI A; 4L3L A; 2XIZ A; 3FMM A; 1LP4 A; 4FKG A; 4GCJ A; 4DE4 A; 2Y9Q A; 3ZU7 A; 2QUR A; 4DC2 A; 5L2S A; 2VV9 A; 4FTR A; 4O27 B; 2XK8 A; 2VTR A; 3OEZ A; 5GJF A; 2C5O A; 4KD1 A; 4DGL C; 5HO8 A; 3RMF A; 2XB7 A; 3AC2 A; 4F64 A; 4AW1 A; 4E20 A; 3EQB A; 3UZP A; 3FLN C; 5JFX A; 1H0V A; 4GH2 A; 4MBI A; 3ZLS A; 2WOT A; 4FTN A; 5LMB A; 5CZI A; 2UZD A; 2CPK E; 2WPA A; 5F94 A; 2C6D A; 5HCX A; 2C3K A; 5AMN A; 2E9P A; 2VZ6 A; 4GSB A; 3S3I A; 3VS0 A; 2PVF A; 2VWW A; 2WMW A; 2OZA A; 3AD4 A; 3C1X A; 4EHZ A; 3OUK A; 2ZVA A; 5CEK A; 5DE2 A; 1OEC A; 5EW3 A; 5L6W L; 1W7H A; 1Y57 A; 1K9A A; 2HWO A; 4FT9 A; 4FTK A; 5FG8 A; 4F6U A; 4ZTR A; 4Z3V A; 3RVG A; 4IJ9 A; 3QQL A; 2V7O A; 5D9H A; 2BUJ A; 5LCK A; 3U55 A; 4FKO A; 3ORT A; 4E6A A; 4GMY A; 4HNF A; 5DBX A; 3VUM A; 4RA4 A; 2PML X; 3I0O A; 4R6V A; 2WMU A; 3IDP A; 4KWP A; 2JLD A; 3EQC A; 4I20 A; 1JKL A; 2XNO A; 3MVL A; 4HYI A; 1URW A; 3FSF A; 4YSJ A; 4YNE A; 2UW9 A; 5JZN A; 1VZO A; 2WAJ A; 3PIY A; 3OT3 A; 4HVI A; 2FO0 A; 4JBP A; 4P5Z A; 4WNK A; 3R8M A; 1OKW A; 4XG3 A; 3GC9 A; 3NUP A; 3Q4T A; 4AA5 A; 4BBF A; 4DEG A; 5HQ0 A; 4IZ7 A; 3BYO A; 4IX6 A; 3ZM4 A; 2HCK A; 4IWQ A; 2PVR A; 1WAK A; 5AP6 A; 4GJ3 A; 2EXE A; 4FMQ A; 2A19 B; 2B9J A; 4RIO A; 1T45 A; 4WSY A; 3FHI A; 3LMG A; 4Z55 A; 2IW6 A; 4YJQ A; 2GU8 A; 5HG7 A; 1IEP A; 4H39 A; 3MBL A; 4FZF B; 2C6O A; 3BPR A; 1WZY A; 1LR4 A; 2RFS A; 2RFE A; 2BTS A; 3ENM A; 4W8D A; 5HIB A; 4IXP A; 4I23 A; 3DJ5 A; 3R6X A; 5DEW A; 5AP3 A; 1VEB A; 1H1P A; 1YQJ A; 1H07 A; 4LL5 A; 5KHW A; 3D7T B; 3GFE A; 2SRC A; 4JNW A; 3QTZ A; 2W96 B; 5HVJ A; 4YO6 A; 4BCH A; 3D7Z A; 1JKK A; 2GNF A; 1G5S A; 3RPR A; 1OL6 A; 3V5W A; 3RI1 A; 2J50 A; 5HKM A; 5CAP A; 1W84 A; 2QHM A; 3KXM A; 4WSQ A; 3CD8 A; 4O96 A; 3B8Q A; 4QP7 A; 4FEV A; 2YM7 A; 4BDI A; 3LIJ A; 5ANJ A; 3PXQ A; 2J5E A; 5HE2 A; 3BLQ A; 2YIX A; 3KY2 A; 4Y95 A; 4G16 A; 5CSV A; 5I9V A; 4MXZ A; 1U59 A; 4NTS A; 4FIG A; 3GEN A; 2XNE A; 5IUG A; 3F5P A; 3FYJ X; 4ZTM A; 4RWJ A; 4QML A; 2BIK B; 4FZ6 A; 2OJG A; 2V0D A; 3D2K A; 4HCV A; 5CSH A; 4HGS A; 2UZE A; 3IOK A; 5J79 A; 3NS9 A; 4HNI A; 1M7Q A; 4U44 A; 4O21 A; 4AZF A; 1Q3D A; 2CNU A; 3ORI A; 2QU5 A; 3MES A; 2P2H A; 5GHV A; 5HE1 A; 3U6H A; 2YCR A; 4L6Q A; 4TL0 A; 2V7A A; 3KXX A; 4FE2 A; 2PUP A; 5BUI A; 3GU8 A; 3BWF A; 5D7A A; 1KOA A; 3BYU A; 4ZME A; 3NGA A; 5CT0 A; 3A7I A; 3II5 A; 5D7V A; 4BKY A; 4DFZ E; 4E93 A; 3MTL A; 3Q53 A; 4G2F A; 3KVW A; 4BHN A; 3WBL A; 2JGZ A; 2GS6 A; 3FLW A; 5FBN C; 2QOQ A; 4QMW A; 2ETK A; 3UIB A; 3G6G A; 3CXW A; 3HMM A; 5DR9 A; 3W8Q A; 3BU6 A; 3FAA A; 1QPD A; 2WMA A; 2OIB A; 5DT4 A; 3SXS A; 3KQ7 A; 4C2W A; 4CV8 A; 3TNI A; 4MVF A; 5IG1 A; 3TUB A; 1G8S A; 5DG5 A; 1JLU E; 2H96 A; 2BPM A; 2VWU A; 3O0G A; 4N6Z A; 3FLQ A; 3EQR A; 4WIH A; 3VNT A; 3HDN A; 4JDK A; 4E5B A; 3PG3 A; 4DFN A; 3C4X A; 3W55 A; 3QHW A; 4C38 A; 3HRB A; 4GFG A; 4D1S A; 2VO7 A; 3A7H A; 4FKU A; 3I1A A; 3W2C A; 4B7T A; 4J1R A; 4DTK A; 4EOS A; 4RIY B; 4F7S A; 4MWH A; 3MJ1 A; 3NUY A; 3KGA A; 2BZI B; 1UV5 A; 1CMK E; 4ORK A; 3QQH A; 3PE2 A; 5CF8 A; 4FV1 A; 3DND A; 2OLC A; 3WYY A; 3HZT A; 4AA0 A; 3T8O A; 3KMM A; 3ZC5 A; 4ZLZ A; 3CJG A; 3H0Y A; 4M14 A; 5D10 A; 4FV9 A; 4RRV A; 5FM3 A; 5A9U A; 2C4G A; 1ZYD A; 4XRL A; 4F9W A; 3T3V A; 3BGP A; 1TVO A; 4NEU A; 4KKH A; 4A7C A; 5BYY A; 2RKU A; 2CJM A; 3KN6 A; 5CT7 A; 1L3R E; 4MWI A; 3HAV A; 4BGG A; 4FA2 A; 2QO2 A; 4KKE A; 4R7I A; 4WHZ A; 3P86 A; 3QXP A; 2W4J A; 3W33 A; 2W5B A; 5EG3 A; 3OMV A; 3PJ8 A; 4RJ4 A; 4J7B A; 2PVJ A; 4M7I A; 2V62 A; 3DQW A; 3LE6 A; 4DGO A; 2FB8 A; 1R0P A; 4O86 A; 3SC1 A; 5M4C A; 4LGD A; 2O3P A; 3ZEP A; 3NVK I; 2WMX A; 3R63 A; 5HOR A; 3RAW A; 1KUT A; 2HZI A; 4C8B A; 4EUT A; 4HOK A; 4JQE A; 3QUD A; 4FV6 A; 4CFX A; 4DFX E; 3PPZ A; 3O17 A; 5GNK A; 3R0T A; 4JDI A; 4PTC A; 1GIJ A; 4ANL A; 4F09 A; 2O64 A; 2W1C A; 3VF9 A; 4CFU A; 4YJT A; 5CAN A; 3KB7 A; 2JIT A; 4TYH A; 4DBN A; 2CLQ A; 4RWI A; 3MY0 A; 2VWI A; 4QTC A; 1R1W A; 4FTJ A; 4AT3 A; 4UY9 A; 4WUN A; 4CLI A; 1Y91 A; 4RQK A; 2CGW A; 4HVD A; 5ETI A; 2GHG A; 2OU7 A; 2CMW A; 4O0Y A; 4TNB A; 4UZH A; 3C0I A; 2WTV A; 1VYW A; 3DTW A; 5DT0 A; 3VVH A; 4CMO A; 1TQM A; 3NW6 A; 4CT1 A; 3PA4 A; 1QCF A; 3COM A; 3PJ3 A; 3TG1 A; 3RNY A; 3AMA A; 4BCQ A; 4OAV B; 5IZF A; 1BL6 A; 4E26 A; 4EOQ A; 4FKQ A; 2YM4 A; 2JD5 A; 4LRM A; 3GCS A; 3DJ6 A; 3OXT A; 4EH3 A; 3U4W A; 4E5W A; 3S0O A; 2ZMD A; 5EH0 A; 5M56 A; 5AP5 A; 1A48 A; 4AOT A; 1DS5 A; 3R01 A; 4IEB A; 3ZSI A; 3DB6 A; 4EKK A; 2VO0 A; 4FSY A; 4FZD B; 4IVD A; 3BEG A; 3OEF X; 2IZS A; 3R1Q A; 2IW8 A; 4WG4 A; 3AC5 A; 5FTO A; 5HG5 A; 1OBG A; 2QO7 A; 4I6H A; 5EHY A; 4GFM A; 5BUJ A; 4ANB A; 2XBJ A; 3AXW A; 3ETA A; 2A1A B; 2X2L A; 1ATP E; 4FTM A; 4A9T A; 5L6P A; 3CE3 A; 4TT7 A; 2CNV A; 1BX6 A; 5M55 A; 1OI9 A; 2JFL A; 4FZA B; 5J95 A; 3TT0 A; 4D2W A; 2PK9 A; 2QG5 A; 2XKC A; 4HJO A; 5TO8 A; 5JKG A; 4CSV A; 4DT9 A; 3PYY A; 1OL7 A; 4DF3 A; 1Q4L A; 1XBA A; 4EC8 A; 2DS1 A; 3D5U A; 5BVD A; 3GU5 A; 5DVR A; 2C0T A; 3HYH A; 4FKW A; 4HGL A; 4AGU A; 4E4L A; 4X7Q A; 1CDK A; 2PUN A; 2R3Q A; 4EH8 A; 5J1V A; 2F9G A; 3MH0 A; 2IW9 A; 2PU8 A; 3U4U A; 2IWI A; 3OZ6 A; 3ZO2 A; 2XK7 A; 1Q24 A; 2QR8 A; 4HCU A; 3KRX A; 4L3J A; 3ORZ A; 4YP8 A; 5A4Q A; 3DK3 A; 3WF9 A; 3TZ9 A; 1YWV A; 3EFK A; 4ZTH A; 2X4Z A; 2PVN A; 3ORK A; 3IDC A; 4FV4 A; 5HID A; 3TTI A; 5IQG A; 5KBQ A; 1M52 A; 3DDQ A; 3KXZ A; 1W83 A; 3ZSH A; 4E3C A; 3IKA A; 3MIY A; 3H4J A; 5AIR A; 3C4W A; 3HA8 A; 4GT3 A; 2OXY A; 2FYS A; 3I5N A; 3NNV A; 4QMO A; 1OB3 A; 4G3C A; 3Q6W A; 3MJ2 A; 2ITN A; 5HHW A; 3IO7 A; 1ZOE A; 4DH1 A; 2YAA A; 3N9X A; 4IAK A; 1DI8 A; 3A99 A; 3QFV A; 2HYY A; 1OL5 A; 4C37 A; 3EH9 A; 4F9Y A; 4GIH A; 3RBW A; 3T9T A; 3CBL A; 4CEG A; 2YM8 A; 1YDT E; 2OK1 A; 4RJ3 A; 3UOD A; 4JQ8 A; 4PQN A; 4DAW A; 5JQ5 A; 2BDW A; 3FI4 A; 4H1J A; 4F9C A; 4O1P A; 3KL8 A; 1B38 A; 3K54 A; 4FV5 A; 5BVF A; 5KKR C; 3FMJ A; 2Y4P A; 3ZZE A; 3VAP A; 5HIC A; 4L42 A; 4H36 A; 4H3Q A; 1JAM A; 1ZAO A; 5ICP A; 1FOT A; 1GII A; 4IIR A; 5ANG A; 4ZXT A; 3QD2 B; 3PY1 A; 4FG8 A; 4JX7 A; 4FT5 A; 2P55 A; 5L6O A; 2XYU A; 5GZA A; 1PMV A; 2J0L A; 4YJV A; 4FEX A; 2J7T A; 4OT6 A; 5K4I A; 1NA7 A; 2OID A; 1H8F A; 4G3D A; 1Z5M A; 3E62 A; 4QPA A; 4TWC A; 4ANQ A; 5M4I A; 4JS8 A; 3PVB A; 2QNJ A; 4FR4 A; 1GZK A; 3Q6U A; 1B39 A; 4OBP A; 5CVH A; 4E6Q A; 5K0K A; 4DT8 A; 4DFY A; 1ZY5 A; 4ZZM A; 3IG7 A; 3LJ0 A; 4NIF A; 1ZMW A; 5HLW A; 1M2Q A; 4QTD A; 2YEX A; 1WVX A; 2XA4 A; 3IOP A; 3VBW A; 3GEQ A; 4AZW A; 1STC E; 2QO9 A; 4EOL A; 2X2K A; 1WVY A; 4EHE A; 1NVR A; 3VS2 A; 5C27 A; 2X8I A; 3MIA A; 3OP5 A; 2X7F A; 3PJ1 A; 3TCP A; 4G5J A; 2C6K A; 4GFO A; 3EQH A; 3MB7 A; 4CT2 A; 4HGE A; 2QOK A; 3WOW A; 3DKG A; 4HZR A; 2ITP A; 2UVZ A; 3OBJ A; 4FKT A; 4N0S A; 3EQD A; 4PTG A; 3VF8 A; 3ZO3 A; 2QU6 A; 4GUB A; 4IQ6 A; 4YPD A; 4O0X A; 4EBV A; 2W06 A; 5AUX A; 3UIU A; 2WTC A; 1ZOH A; 2P33 A; 4ACC A; 4DEE A; 4Q9S A; 2RFD A; 2XZS A; 3BEL A; 4TXC A; 4RWL A; 1IR3 A; 4TYE A; 4G17 A; 4L8M A; 4OTR A; 2B52 A; 5JFW A; 2VTP A; 4XLV A; 1ZMV A; 5EYK A; 3M11 A; 3GOK A; 3R7O A; 2ZJW A; 3QF9 A; 3UIX A; 4D4V A; 1OKY A; 5BQ0 A; 4YZC A; 5H8E A; 4F0G A; 4IHP A; 3O96 A; 3ZOS A; 3EYG A; 2W17 A; 3NW5 A; 2HZ0 A; 3DU8 A; 1ZYL A; 3QRU A; 2J0M B; 2ZV8 A; 5CAO A; 3OCS A; 3HX4 A; 5EHL A; 3ZO1 A; 3I7C A; 4O0T A; 2A4L A; 4B99 A; 4BYJ A; 1YXX A; 3MDY A; 1K2P A; 2GHL A; 2BAL A; 2UW5 A; 4RZV A; 5HTB A; 4M69 A; 1ZLT A; 2QD9 A; 2UZW E; 3W32 A; 3FJQ E; 3PXR A; 4NST A; 3BE2 A; 3P0M A; 3TNW A; 3TJC A; 2EVA A; 5EK7 A; 5IH5 A; 5J9L A; 5FGK A; 1JBP E; 2R3L A; 2X1N A; 5CVF A; 5DIA A; 2R3K A; 5B7V A; 4AG8 A; 1IA8 A; 2J90 A; 2YCF A; 1MRU A; 3UOJ A; 2XM9 A; 3O50 A; 4BCF A; 1YWN A; 3COK A; 4XG6 A; 4FKI A; 1XBB A; 4IVA A; 2PZI A; 5CAQ A; 3MVH A; 4O7W A; 4IB1 A; 1H28 A; 4FTQ A; 4QPM A; 4XV2 A; 4XH6 A; 1Y8G A; 3QTR A; 4F9B A; 4MQ2 A; 3QGY A; 3U6I A; 1LPU A; 5ETF A; 1MUO A; 3ACJ A; 3LFN A; 1SM2 A; 4PNK A; 2I1M A; 3PPJ A; 3HUC A; 4L00 A; 4ZZO A; 2VN9 A; 3QXO A; 5EQE A; 3NDM A; 4HW7 A; 3FXW A; 3DBQ A; 5HBE A; 1KSW A; 2YIR A; 4KSQ A; 5FBO A; 4EON A; 1CM8 A; 1OVE A; 1GOL A; 3FXX A; 3KFA A; 2C3L A; 2DQ7 X; 1HCL A; 1P4F A; 3VRZ A; 1JOW B; 4XBU A; 4D4S A; 2VGP A; 1VJY A; 3Q5I A; 4FOB A; 3VUD A; 4FST A; 5EM5 A; 3UP7 A; 5IDP A; 4EH5 A; 5KZI A; 5C4K A; 4FUX A; 4PP9 A; 1ZY4 A; 2Z7L A; 4JL9 A; 2CDZ A; 4E4X A; 2PZ5 A; 3EL7 A; 4IVC A; 3ALO A; 3ZRK A; 3BRB A; 5C9C A; 3PLA E; 3TIY A; 4CLJ A; 3NUU A; 2EU9 A; 3Q60 A; 5FEQ A; 1CSN A; 3MS9 A; 3VW8 A; 4IAN A; 3RM7 A; 3UDB A; 4ACM A; 5A46 A; 4H1M A; 1Q5K A; 3KRL A; 3W18 A; 4PDS A; 2OJF E; 2VO3 A; 3CJF A; 4G34 A; 3DB8 A; 4EBW A; 3PIX A; 5DRD A; 2XK4 A; 5J1W A; 3ZDI A; 2BAJ A; 4O6L A; 4QP6 A; 2Q83 A; 4MTA A; 4RGJ A; 5CEN A; 1MP8 A; 4G6O A; 5DEY A; 5BX0 A; 1OIY A; 5E8Z A; 4UN0 C; 5IMX A; 1JST A; 1CKI A; 5DGZ A; 2HOG A; 1ZWS A; 2WMT A; 3P7B A; 3ZXT A; 4ITI A; 4M0Y A; 3MH1 A; 3AC3 A; 3EZV A; 1SMH A; 3H30 A; 4GU9 A; 3I4B A; 2X4F A; 3H9O A; 2OZA B; 4BL1 A; 2QLU A; 3GCP A; 1SVG A; 5E8V A; 4C3F A; 4TN6 A; 5H9B A; 1V1K A; 1T46 A; 3GI3 A; 3QKK A; 2WEI A; 4V01 A; 3RK9 A; 2OG8 A; 5J7H A; 3SOC A; 1OL1 A; 1ZRZ A; 2X8D A; 3FI3 A; 3AT3 A; 4NM0 A; 4FSM A; 4O2P A; 5CEP A; 2IO6 A; 5HIE A; 3WAR A; 5IKW A; 3NZ0 A; 3R9N A; 3DQX A; 3DPK A; 5DA3 A; 5ANE A; 2YIS A; 2EXM A; 4O6E A; 3NNU A; 3QCX A; 3MPA A; 4FTO A; 5CTP A; 1YXT A; 3L9M A; 2I0E A; 5J5T A; 5F20 A; 2IZU A; 4XJ0 A; 3DOG A; 4JVG A; 4K33 A; 3NUA A; 2RIO A; 4F6S A; 3I79 A; 3QHR A; 5IQI A; 4ALV A; 2VWV A; 2HEL A; 3R71 A; 4AZT A; 2Z7Q A; 4RJ7 A; 4BIB A; 1WMK A; 4XG8 A; 3RAI A; 4LV5 A; 1BI7 A; 2UW6 A; 3LQ8 A; 4JBV A; 3HEC A; 2WQM A; 3OTV A; 3OY1 A; 3D5V A; 3TZ8 A; 5JMS A; 1M2P A; 4JR7 A; 3E93 A; 3OUN B; 5HLP A; 3VN9 A; 3LCS A; 2WKM A; 3KA0 A; 3UG1 A; 3EN4 A; 3E63 A; 3VW6 A; 2J5F A; 3DBC A; 1YI4 A; 3KCF A; 3GNI B; 4TYJ A; 4S31 A; 5FXQ A; 2ZDT A; 5K3Y A; 3FBV A; 3TV7 A; 4NM7 A; 3FYK X; 1O9U A; 4QT1 A; 4Z7H A; 2C6L A; 4U43 A; 3E8D A; 4QMU A; 5FXS A; 3QTU A; 3ATS A; 4CG8 A; 4BCM A; 5JT2 A; 4PP7 A; 3PTG A; 2H34 A; 3H0Z A; 1YWR A; 3HNG A; 2O5K A; 2ZB1 A; 4BKJ A; 5AR2 A; 4C02 A; 1M7N A; 2W7X A; 2ITV A; 3C0H A; 5I9U A; 2G15 A; 1FVR A; 3RHK A; 4YZN A; 1IG1 A; 3PSC A; 4MXY A; 4E6C A; 4JDJ A; 3AOX A; 3G9N A; 4YC8 A; 5CF4 A; 2QI8 A; 2OBJ A; 3QQU A; 3GUB A; 5K9I A; 4FTU A; 4QYG A; 5CSP A; 5LVN A; 3QTW A; 5C01 A; 4BDJ A; 2PHK A; 4J52 A; 3G15 A; 3HUB A; 3RWQ A; 5GIP E; 4RVL A; 3IS5 A; 4L01 A; 1GZN A; 2AC3 A; 3CTH A; 3LCO A; 2FGI A; 3GQI A; 5FXR A; 4R7B A; 3M2W A; 1YVJ A; 1P2A A; 4FSZ A; 4GG7 A; 4F7J A; 1UNL A; 2R3F A; 4PL5 A; 4CGA A; 5DR6 A; 4ZY5 A; 3ZON A; 4D0X A; 4AE9 A; 4QMX A; 1ND4 A; 4JAI A; 3MA6 A; 1H25 A; 3HV4 A; 3PA5 A; 4NT4 A; 3TWJ A; 4UMP A; 4ASD A; 5A4E A; 5JRQ A; 2VTL A; 2VWB A; 1Q62 A; 2J4Z A; 3ROY A; 4MH7 A; 4FIE A; 5DH3 A; 1REJ A; 4OTH A; 4U3Y A; 3NYX A; 3ZUU A; 5KMM A; 4O0R A; 4QPS A; 4JOA A; 2YA9 A; 2VTO A; 5EW9 A; 4I93 A; 4OGR A; 3H3C A; 3U6J A; 5AR8 A; 2BR1 A; 3L8S A; 5AP2 A; 4AGD A; 3GT8 A; 2WHB A; 1NXK A; 2XYN A; 4JR3 A; 4LRJ A; 4EYM A; 3UO6 A; 2ITZ A; 4J95 A; 5KHX A; 5J9Y A; 5DPV A; 3HAM A; 4G9C A; 3QLF A; 2O63 A; 4R1V A; 4EQM A; 2WMB A; 3CR0 A; 3DY7 A; 3VHE A; 2O0U A; 3PE1 A; 3D2I A; 2BZH B; 4PL3 A; 3WF5 A; 5C8N A; 5HD4 A; 5HBJ A; 5CAS A; 2GPH A; 4MBJ A; 5CNO A; 5KZ0 A; 4FV0 A; 4QP9 A; 3OW3 A; 2C0O A; 4YZB A; 4BY9 E; 4C2V A; 3LMH A; 3F7Z A; 3ZSG A; 2UZL A; 2ZB0 A; 1CKP A; 5CY3 A; 4CV9 A; 4BBM A; 2CGX A; 1G3N A; 5HU9 A; 4O7R A; 3UVP A; 3AC1 A; 2JIU A; 2YIQ A; 3AD5 A; 4ZEG A; 4OW8 A; 2WZJ A; 4CFF A; 4YUR A; 2PYW A; 3N4U A; 5IH4 A; 3ELJ A; 4GU6 A; 1XBC A; 4EJ7 A; 3GFW A; 3PSB A; 1QPE A; 4Y73 A; 4UMR A; 3IK3 A; 2PMI A; 4I4F A; 5BYZ A; 4FKJ A; 4LQM A; 4D9U A; 3FSK A; 4V0G A; 3ULZ A; 4FYO A; 3FUP A; 1MRV A; 4KIK A; 1H1W A; 4APP A; 2XCK A; 4ANN A; 2ITU A; 4LM5 A; 4ALU A; 5DN3 A; 1E9H A; 4YHF A; 4CNH A; 2W5A A; 4L67 A; 4F99 A; 3BHH A; 4NJ3 A; 2XIK A; 3NW7 A; 5E7R A; 3WZK A; 4AW5 A; 4FNZ A; 4F1T A; 3NPC A; 2XKE A; 2XIY A; 4TRL A; 4JX3 A; 4FL3 A; 4F6W A; 4C33 A; 2OH0 E; 5LOH A; 3C0G A; 4PWN A; 2WTK C; 2IPX A; 4XP3 A; 3S2P A; 1KE8 A; 3MWU A; 2XVD A; 3HKO A; 3I81 A; 5E92 A; 4RVM A; 5I9Z A; 3LXK A; 3OF0 A; 1J1C A; 5HO6 A; 2BFY A; 2W1E A; 4WRG A; 4R3C A; 1XH6 A; 2O2U A; 3FQH A; 3C5U A; 5HU3 A; 2JKQ A; 2QOO A; 4YJR A; 2WO6 A; 2FSO X; 2YCQ A; 2YAB A; 4LUE A; 4EQU A; 1GIH A; 1O6Y A; 2PUU A; 3QAM E; 4AAA A; 3UOK A; 3BU3 A; 2G2F A; 4D1X A; 3U54 A; 2OW3 A; 2BZK B; 5F9E A; 4QRC A; 4QYH A; 2X9F A; 3D94 A; 4IAI A; 4EEV A; 2XM8 A; 4FV2 A; 4IM3 A; 2CLX A; 1JPA A; 1PXM A; 2NRY A; 3BU5 A; 3ZH8 A; 2XJ1 A; 3V5T A; 3BYV A; 4D1Z A; 1UWH A; 4HYU A; 4ASX A; 2UW0 A; 4C35 A; 2VU3 A; 3QYZ A; 5K4J A; 2OJ9 A; 4P2K A; 1M17 A; 4A4L A; 5E4H A; 3EYH A; 4DH5 A; 3SXF A; 4UXQ A; 4C3P A; 4AF3 A; 4M0Z A; 2VWY A; 3FEG A; 2FST X; 4XG2 A; 2IZT A; 1ERK A; 2OFU A; 3HMN A; 4TPT A; 3VRY A; 2UUE A; 2AUH A; 4AGW A; 4EZ7 A; 4CZU A; 3SRV A; 2XNN A; 4CCB A; 4FEW A; 4BHZ A; 3FL5 A; 3MFU A; 4FF8 A; 5GZ8 A; 1QMZ A; 4FI1 A; 3LM0 A; 3R8Z A; 3WZU A; 4BCJ A; 4CZT A; 2BHE A; 3BQC A; 4BF2 A; 1KWP A; 4MF0 A; 3C50 A; 3OT8 A; 4WRS A; 4ID7 A; 1H0W A; 4ZSA A; 3C7Q A; 5AJQ A; 2H9V A; 4FX3 A; 4FIH A; 4YPS A; 5BNJ A; 5DT3 A; 4O7L A; 3MYG A; 2OZO A; 4M68 A; 3CQU A; 4BCG A; 1JQH A; 4XS2 A; 4BCK A; 3SG9 A; 4TWN A; 3QTI A; 3BWJ A; 3NYN A; 4WKQ A; 3GU6 A; 2PTK A; 3JVR A; 2F7Z E; 2IG7 A; 4Y85 A; 4YC3 A; 4EUU A; 3PZH A; 4IVB A; 5L8L A; 5KMI A; 3UNZ A; 4GW8 A; 4KIN A; 3VJO A; 1OPL A; 3LFS A; 2VUW A; 4YJS A; 4C3R A; 3G2F A; 1OIT A; 1PMN A; 1Z57 A; 4CQG A; 3R1N A; 4K8A A; 1H27 A; 2ITY A; 3E3P A; 3LZB A; 4FK6 A; 1YHS A; 3R7U A; 4E6D A; 4F0I A; 5FWK K; 4X6R A; 4YSM A; 1MQB A; 2ZDU A; 3R7E A; 3UG2 A; 1UU3 A; 3QD4 A; 5DFP A; 4B8L A; 4CFV A; 1PYX A; 4I24 A; 3FMH A; 4D2P A; 4E8A A; 2W99 B; 4ACH A; 5H8G A; 2WTJ A; 4EK5 A; 5M51 A; 4V04 A; 3S00 A; 2PE2 A; 4WNO A; 4EOK A; 3W1F A; 1LEW A; 4NK9 A; 3R22 A; 1OBD A; 3R8L A; 4IWD A; 3LLA A; 2E9N A; 4L7S A; 3LOK A; 3QU0 A; 5IQB A; 3R73 A; 5AV4 A; 3P79 A; 3VUG A; 5CLP A; 3ULI A; 4PDP A; 3TV4 A; 3OY3 A; 5LJJ A; 3Q4U A; 3FZ1 A; 4V05 A; 1XQZ A; 3GGF A; 5L4Q A; 4FT3 A; 5FD2 A; 5CAU A; 2VD5 A; 2HZN A; 3R83 A; 2XK9 A; 2CCH A; 2R64 A; 4OBQ A; 1OGU A; 4BGQ A; 1NVS A; 2VTN A; 3OXZ A; 5H2U A; 1M14 A; 4UBA A; 1OPK A; 4QQJ A; 3P5K A; 5HO7 A; 3FLY A; 4O7Y A; 3BYS A; 5HNI X; 1PY5 A; 2BRN A; 5C03 A; 5AEP A; 3NYV A; 4LMU A; 5KO1 A; 3UVR A; 4D2V A; 4ZSE A; 2QKW B; 3G90 X; 4JDH A; 3DFA A; 3V5Q A; 2BZJ A; 2R3I A; 5AV0 A; 4JBO A; 3LMI A; 1WBT A; 2RG5 A; 2PVL A; 4PPC A; 2VTM A; 5AAG A; 3L8P A; 4PV0 A; 4GRB A; 5ACK A; 4QO9 A; 4QP1 A; 3TEI A; 3CPB A; 3E8E A; 4R77 A; 2BAQ A; 3FKN A; 2QKR A; 3D83 A; 4EK6 A; 2PSQ A; 3KN5 A; 4AXA A; 3C9W A; 3FZT A; 4FT0 A; 3T3U A; 4D2R A; 3KRW A; 4WO5 A; 3F3T A; 2WOU A; 3NSZ A; 4K0Y A; 5I0B A; 4IAY A; 2I0V A; 4F1M A; 4PPA A; 3DXQ A; 1DI9 A; 4RJ8 A; 4CRS A; 4UWY A; 3UYT A; 4RG0 A; 4QTA A; 3WI6 A; 2JED A; 2J9M A; 5IH6 A; 4C58 A; 3VQU A; 3UBD A; 4MHA A; 4F65 A; 3A2C A; 4O7S A; 4RBL A; 2QCS A; 3VS6 A; 3A62 A; 4AUA A; 2HWP A; 4YZD A; 3V5J A; 4HVS A; 4J8N A; 4FII A; 4RSS A; 2Y6M A; 3MPM A; 2GNG A; 2W1G A; 4U81 A; 2ITX A; 3VS7 A; 5I9Y A; 5M57 A; 5EDR A; 5EKN A; 3A7J A; 1YDS E; 3VID A; 5IA4 A; 4TW9 A; 2XCH A; 1O6K A; 4MX9 A; 4W4V A; 5M53 A; 4WNP A; 1P5E A; 3NVM B; 3UNK A; 3VBX A; 4IE9 A; 1BL7 A; 4W8E A; 2C3J A; 3BCE A; 2WEV A; 1PRY A; 2H8H A; 1Q8Y A; 4LV8 A; 3UC3 A; 4M69 B; 1O6L A; 2IVU A; 5HTI A; 3TDW A; 3TKU A; 4GKH A; 4CFM A; 4OTF A; 4FEQ A; 4KSP A; 2A2A A; 1URC A; 2HIW A; 4IM2 A; 1H01 A; 4FTC A; 4LOP A; 5F4N A; 4EH6 A; 4OT5 A; 2CGV A; 3FMK A; 4TWO A; 4DBX A; 1IRK A; 3MH3 A; 3KEX A; 3NNX A; 3WZJ A; 3IGG A; 4DYM A; 2NO3 A; 4BI0 A; 2X7O A; 4MNF A; 3ZC6 A; 3F82 A; 2X9E A; 3G33 A; 1W98 A; 3COH A; 4IZ5 A; 3LQ3 A; 1RQQ A; 4HZS A; 4LYN A; 4I41 A; 3FI2 A; 1UU9 A; 4YTC A; 4NYE A; 1OKV A; 5AR7 A; 3LCT A; 4MNE A; 5HE0 A; 4UXL A; 4IB0 A; 2CKP A; 5AR4 A; 3LCD A; 5AA9 A; 4FVQ A; 3FL4 A; 4I3Z A; 2QR7 A; 3JY9 A; 3FKO A; 3LJ1 A; 2R2P A; 3KXH A; 5T3Q A; 2YZA A; 1WBS A; 3TI1 A; 3UQC A; 5DOS A; 3DKF A; 3IW4 A; 3A8X A; 4G6L A; 3ZBF A; 5CXZ A; 4HCT A; 3LCK A; 3D7T A; 1DM2 A; 4X2F A; 2R0I A; 4FKL A; 4CYJ A; 3GP0 A; 3CY3 A; 2BRM A; 2OIC A; 3GCU A; 3TN8 A; 3WZE A; 5LVP A; 3ORO A; 4N70 A; 2GS7 A; 2GTM A; 3E88 A; 3JVS A; 5HVY A; 4K9Y A; 4EK4 A; 4O38 A; 5LVO A; 3AMY A; 1Q8U A; 3MTF A; 2W1H A; 3W2S A; 3DK7 A; 1SYK A; 2WB8 A; 4AN9 A; 2IJM A; 3EOC A; 3ITZ A; 3PY3 A; 5DWR A; 4TYI A; 1H4L A; 3IQ7 A; 4XOZ A; 5LXM A; 3MW1 A; 3WF6 A; 1SNX A; 3DAE A; 3KKV A; 5FED A; 4XRJ A; 4ZZN A; 3NMU F; 3ATT A; 3ZEW A; 2VO6 A; 4EH9 A; 1XH9 A; 3M42 A; 4YLJ A; 5HNB A; 4O81 A; 1AD5 A; 4F7N A; 3MI9 A; 2JC6 A; 4CYI A; 2HEN A; 3O8U A; 3UYS A; 3HDM A; 3TNH A; 3MY1 A; 4AW0 A; 5M4U A; 2UZN A; 2W1I A; 1E1X A; 5JRS A; 2UZU E; 3HMP A; 5M06 A; 5F1Z A; 4QMS A; 4I1Z A; 4FKS A; 3RPS A; 4OAU C; 3GQL A; 2YM6 A; 3BYM A; 5C4L A; 4XOY A; 3QTQ A; 3RZB A; 1Q97 A; 4DIN A; 4FSU A; 1YOM A; 5BUE A; 4DFB A; 3OOG A; 3DXN A; 5CYI A; 3ION A; 4O91 A; 4S30 A; 3C13 A; 4RQV A; 2ITT A; 2FUM A; 3PP1 A; 2BVA A; 5FQD C; 3LQ5 A; 4DGG A; 5EDP A; 5ETA A; 2BRO A; 4FV3 A; 4AFE A; 3I7B A; 3VWA A; 1OIU A; 3VUT A; 3FLS A; 2XK6 A; 3BV3 A; 5CAL A; 4FSW A; 4YBK A; 2BKK A; 2RG6 A; 5DI1 A; 2F7E E; 2XRU A; 2XF0 A; 4EHV A; 5EM8 A; 3ACK A; 4R3R A; 4BWP A; 4YUQ A; 3RGF A; 5TR6 A; 4E4M A; 3OC1 A; 4OR5 A; 4YHT A; 1QL6 A; 4Y72 A; 2Y6O A; 4EOP A; 3C4C A; 1CKJ A; 4J98 A; 2XH5 A; 2VTJ A; 3VO3 A; 3FWQ A; 4O0S A; 3AT4 A; 3K2L A; 4L68 A; 4OTQ A; 4BZD A; 5CU2 A; 1UA2 A; 1R39 A; 3RJC A; 4DFL A; 5KBR A; 4RIX B; 4ZIM A; 1Z9X A; 3FKL A; 4KBC A; 5JGB A; 4YJU A; 4AP7 A; 4EOR A; 5LAR A; 3I6U A; 1Q8W A; 3ORM A; 3KVX A; 4ZP5 A; 3BHV A; 4X21 A; 3GU4 A; 1XH4 A; 4A07 A; 4BDE A; 4BDH A; 4YTF A; 5AUT A; 5JGD A; 1LUF A; 4FNX A; 2E2B A; 5AM6 A; 4C4E A; 4RIY A; 3DT1 A; 4MXO A; 1TKI A; 2WQO A; 2ZV9 A; 4L52 A; 4IBM A; 2PMN X; 4D55 A; 3BQR A; 3QA0 A; 2PPQ A; 2XNB A; 2R0U A; 2IN6 A; 5I9W A; 3ZM9 A; 3U9C A; 2R3R A; 4C59 A; 4KUJ A; 4MXC A; 3KC3 A; 5EYM A; 3ZS5 A; 4IX4 A; 2B0Q A; 2HK5 A; 4JIA A; 2Y0A A; 4U42 A; 2CKQ A; 1U54 B; 3BX5 A; 5K0X A; 3OD6 X; 2JIV A; 2J0I A; 2XMY A; 1GJO A; 1WBW A; 2G01 A; 3UPX A; 3E87 A; 3JYA A; 1DAY A; 4O0V A; 3O51 A; 4R8Q A; 3S4Q A; 4I6Q A; 2JDV A; 4P4C A; 4RC2 A; 4O7V A; 1YXV A; 3LKM A; 3P1A A; 4QP8 A; 5AUZ A; 2AYP A; 3EQP A; 4YHJ A; 3QZI A; 3A60 A; 3AGL A; 4KBK A; 3HGK A; 5JQ8 A; 2IVT A; 3WYX A; 3QC9 A; 1QPJ A; 2R7I A; 4XV1 A; 4CMU A; 2WMQ A; 5BMS A; 3CTJ A; 4UEU A; 4AW2 A; 5E9E A; 1DAW A; 3ZTX A; 3C5I A; 3FY2 A; 5FTG A; 1NY3 A; 2UVY A; 2ZAZ A; 1RE8 A; 3EN7 A; 4DEB A; 3MN3 A; 3QA8 A; 3KF7 A; 2W9F B; 3TL8 A; 3W2O A; 4ZJI A; 4L45 A; 4IAZ A; 4IX3 A; 5L8J A; 3EZR A; 4BRX A; #chains in the Genus database with same CATH topology 1A06 A; 5BVW A; 1H1Q A; 2EB3 A; 4UMQ A; 2VAG A; 3BE9 A; 3QWJ A; 3SVV A; 2O8Y A; 5AP7 A; 3C4E A; 5DRB A; 5ITA A; 2GQR A; 4ERW A; 1U5Q A; 4XMO A; 5EOB A; 3VS3 A; 5CAV A; 2PUL A; 3U8W A; 3HV6 A; 3FML A; 3QLG A; 1YHV A; 3GCQ A; 1ZAR A; 3CQW A; 1B6C B; 2H6D A; 2J0K A; 4MXX A; 2JDS A; 5D41 A; 5AUV A; 2QC6 A; 5CU6 A; 4EHG A; 1OUY A; 3D5W A; 2YWV A; 4O4B B; 5KMN A; 2FSM X; 3FC1 X; 4BZO A; 3RWP A; 3BKB A; 3KK9 A; 1E1V A; 3JUH A; 5J8I A; 3FDN A; 5HD7 A; 4DCE A; 2XEZ A; 4NUS A; 1UU7 A; 2QOL A; 1Q61 A; 3UNJ A; 1W0X C; 3BEA A; 2BIL B; 2C1A A; 2YIT A; 3EKK A; 4G6N A; 2ZM4 A; 1XH7 A; 5IDN A; 3TDV A; 3WF7 A; 3PIZ A; 3LLT A; 3PZE A; 3QQK A; 2HW7 A; 4XHK B; 4D2T A; 1ZYJ A; 4DCA A; 4J8M A; 3DAJ A; 4IZA A; 1R3C A; 2BDF A; 1GNG A; 5C8K A; 4A4O A; 3WIL A; 4BTM A; 4UYA A; 3FPM A; 4W4Y A; 3TKH A; 4TWP A; 1XWS A; 3CD3 A; 4RIW A; 4UV0 A; 3ZUT A; 2J2I B; 1H1S A; 4JRV A; 3DJ7 A; 3QRI A; 3FE3 A; 2BIY A; 3IW7 A; 3Q4C A; 3ZYA A; 2KUL A; 3F3W A; 5KE0 A; 5A6N A; 5JFV A; 5CEM A; 3IDB A; 4CMT A; 4NM3 A; 1WVW A; 5CI7 A; 5CSW A; 5HBH A; 4MED A; 4BIE A; 3OWP A; 5FP5 A; 3VBQ A; 5LWN A; 4B9D A; 3EQI A; 4BB4 A; 5I4N A; 3EL8 A; 5LVL A; 1J3H A; 1KV1 A; 3BLR A; 3PVU A; 4AGC A; 4DHF A; 2W0J A; 5HVK A; 3V01 A; 2F57 A; 3HVC A; 3RM6 A; 4HVG A; 4LI5 A; 2ACX A; 5EQY A; 2V5Q A; 3QRJ A; 4EZ5 A; 3V8S A; 3UTO A; 1XR1 A; 3ZUV A; 3DLS A; 2X8E A; 5FM2 A; 4C61 A; 2PVY A; 4D5H A; 1GAG A; 4DHZ A; 4NTT A; 1PXO A; 4II5 A; 1PKG A; 2VTI A; 3HV3 A; 4JI9 A; 4I0S A; 1Y8Y A; 5AA8 A; 3SQQ A; 3CC6 A; 3E3B X; 4I92 A; 4QNY A; 4ZLO A; 3K5V A; 2IVV A; 3QRK A; 4FC0 A; 2JKO A; 2A27 A; 3GU7 A; 5EDQ A; 3FMD A; 3HMO A; 3S95 A; 5M09 A; 5JGA A; 4L44 A; 4UZD A; 5D12 A; 3BBW A; 3OOM A; 5BVK A; 4FYN A; 2OI4 X; 3I6W A; 2W4K A; 2C68 A; 1WFC A; 4F9A A; 4DFU A; 1F0Q A; 4IDV A; 3PSD A; 4FSN A; 3PY0 A; 4X3F C; 5IEY A; 3TM0 A; 4CFN A; 3ENH A; 3JPV A; 4RC3 A; 2CCI A; 3BBT B; 3UJG A; 1SVH A; 5J7B A; 2OXX A; 2OF2 A; 3UQF A; 1UKI A; 2VGO A; 4NU1 A; 5HE3 A; 4QYY A; 4GK4 A; 4U41 A; 1PME A; 1G8K A; 3UOH A; 2XNG A; 5EI6 A; 4PMT A; 3GCV A; 3ZZW A; 2VX1 A; 2WIH A; 4OTW A; 1QPC A; 1U4D A; 5E90 A; 5K5N A; 4FBX A; 3OW4 A; 4R3P A; 2YM5 A; 2JAM A; 3U9N A; 2AQX A; 4NZW B; 5EQP A; 3EN9 A; 3QD3 A; 5FRI A; 4X3F A; 4DH7 A; 3R28 A; 3TTJ A; 5IA2 A; 1FGI A; 1RW8 A; 5L2I A; 2OH4 A; 4FIF A; 1FMO E; 4LGG A; 4Q2A A; 4IJP A; 2XJ2 A; 5HTC A; 4G1W A; 4IZY A; 3TAC A; 2V22 A; 4FRF A; 3EN5 A; 4KA3 A; 4AWI A; 1S9I A; 4DEA A; 5I5Z A; 3R1S A; 1FMK A; 4QMV A; 4B0G A; 4HPU E; 3R9R A; 2R3P A; 1XHA A; 4D4Y A; 4U97 A; 1PJK A; 2OJJ A; 4CKI A; 4OTD A; 3OWJ A; 2JDO A; 3DV3 A; 4QQT A; 4ZMF A; 2QOB A; 1R0E A; 4FVP A; 3W16 A; 2G9X A; 2WTI A; 4QMY A; 1BMK A; 5D11 A; 3FZO A; 2UW8 A; 4CCU A; 5LQF A; 4DG0 E; 4IW0 A; 2XP2 A; 5LMK C; 2J6M A; 5E91 A; 2XRW A; 5AP1 A; 2J51 A; 2C69 A; 3AD6 A; 3QUE A; 2ITW A; 2Y4I C; 3H10 A; 3QTS A; 4RWK A; 3EQG A; 4U80 A; 5B2M A; 3CLY A; 2JFM A; 3DK6 A; 4LOO A; 3FI8 A; 3O7L D; 4GK2 A; 3AKJ A; 1UVR A; 2BKZ A; 2C47 A; 2PZP A; 2PVK A; 3VUL A; 2AEG A; 3DCV A; 4W9W A; 2XAR A; 4NJD A; 4H3B A; 3LXL A; 1OMW A; 4QMP A; 4JG6 A; 4ASZ A; 3JY0 A; 5KMO A; 1YOL A; 4J96 A; 5E8Y A; 3R02 A; 5EYD A; 3EB0 A; 4JBQ A; 2PVM A; 3R2Y A; 3FQE A; 5LMK A; 3W0S A; 5EM7 A; 3JR1 A; 2ETM A; 5CVG A; 5DMZ A; 5AAF A; 5DR2 A; 2E9V A; 4XCU A; 4CFW A; 4AAY A; 3QCY A; 2GNL A; 4YJN A; 3P7A A; 4C4J A; 2NP8 A; 4IDT A; 5LVM A; 4I0R A; 3DBE A; 3LFQ A; 4IFG A; 4DGM A; 2XIX A; 3F3U A; 3AKL A; 2Z02 A; 3NFG B; 2YAC A; 2PE1 A; 4XNE A; 1BKX A; 5KNJ A; 1RJB A; 4FSR A; 4ARK A; 1UKH A; 2IF8 A; 2PZY A; 3POO A; 3VC4 A; 2HW6 A; 4M15 A; 1F5Q A; 2ESM A; 3FV8 A; 4ZSL A; 1J91 A; 4XYF A; 5HX8 A; 5AWM A; 3FC2 A; 3FY0 A; 4LG4 A; 4XG7 A; 4YTI A; 3D14 A; 5IME A; 5AV2 A; 3NFF B; 3MGY A; 4F70 A; 4XX9 A; 4HPT E; 4EK8 A; 3AQV A; 4IWO A; 4ZOG A; 4PL4 A; 4YO4 A; 1JKT A; 2VRX A; 3VS5 A; 2ERK A; 3K3J A; 3QZG A; 4F0F A; 5TE0 A; 3F2R A; 4O7T A; 2ITO A; 3EID A; 5BV3 A; 2OFV A; 5DLS A; 4JAJ A; 3NWW A; 5DYJ A; 4Q5E A; 1MQ4 A; 3P4K A; 5L8K A; 4ERK A; 1A9U A; 2XEY A; 4ZAU A; 5AAD A; 3QCQ A; 3F3V A; 3PJ2 A; 4U3Z A; 2QOF A; 3PVW A; 3Q9X A; 3NIZ A; 3QRT A; 4AAC A; 4FTL A; 1ZYS A; 5JN2 A; 4XBR A; 4QD6 A; 4IC8 A; 4QTE A; 3V04 A; 3LFD A; 4CXA A; 2BAK A; 4AU8 A; 3AMB A; 4S32 A; 4C62 A; 4BDD A; 3QZH A; 3ORP A; 4UWC A; 3E92 A; 3ZLY A; 2HZ4 A; 3ODZ X; 3UO4 A; 5HLN A; 4H05 A; 2X7G A; 3DKC A; 4AT4 A; 4DED A; 3L8X A; 3VS4 A; 1Q99 A; 2X0P A; 1IAS A; 2WFY A; 3R7Y A; 4ASE A; 4E5A X; 5G6V A; 3V8T A; 3W0R A; 3E8C A; 3LFF A; 2CNQ A; 5BVN A; 2XNP A; 3VBY A; 4AZS A; 4L7F A; 5TF9 A; 2F7X E; 3S1H A; 2C5X A; 3H9R A; 4DHI B; 4C4H A; 5JFS A; 4FOD A; 2QOC A; 2P2I A; 4BCI A; 3PXF A; 2R4B A; 1KOB A; 2XUU A; 2PE0 A; 4N6Y A; 1PMU A; 2UZT A; 3ZDU A; 5T6K A; 2VTH A; 2YJR A; 4WNM A; 4BTF A; 1H08 A; 4P90 A; 4O2Z A; 1AQ1 A; 3QUP A; 3OFM A; 4M13 A; 4M97 A; 5EI8 A; 5AAE A; 4I22 A; 4CVA A; 2G2H A; 3P23 A; 4DEI A; 3W0N A; 3GVU A; 4J99 A; 3DLZ A; 5HNG A; 5BVE A; 4EOI A; 5M4F A; 4DEH A; 1AGW A; 2R9S A; 4ZK5 A; 3RKB A; 4PDY A; 3EKN A; 3D15 A; 2CGU A; 4J53 A; 5AUY A; 2WNT A; 2YIW A; 3L8V A; 3LM5 A; 4TYH B; 3R7V A; 5IPJ A; 3LVP A; 4ZTQ A; 3FZR A; 4GVJ A; 1CTP E; 5E8W A; 4C0T A; 4U9A A; 2B9I A; 2B55 A; 2YHV A; 4K1B A; 3PVG A; 2C0I A; 4L3P A; 4XLI A; 1HCK A; 5J87 A; 5DNR A; 2GQS A; 5L2T A; 3AC4 A; 4UX9 A; 4M3Q A; 5L2W A; 3REP A; 2C5V A; 4O0W A; 2OSC A; 4J97 A; 4CTB A; 1U46 A; 5J5S A; 4IWP A; 4IAA A; 3LFB A; 4IB5 A; 4OTP A; 3R9H A; 4QQ5 A; 3ZRL A; 4RLL A; 3FME A; 1NW1 A; 3O2M A; 4U8Z A; 4DGN A; 5KUP A; 2B9F A; 3MVJ A; 4GEO A; 2A0C X; 4CTC A; 3CKW A; 1VYZ A; 5M08 A; 2BDJ A; 5FDX A; 2GCD A; 1XO2 B; 4FNY A; 3KF4 A; 1PXK A; 4RFY A; 4BCN A; 3F6X A; 4FL1 A; 1SNU A; 4GYG A; 5AFV A; 3QAL E; 4FTT A; 4RX8 A; 3R9D A; 3UOL A; 1IA9 A; 5L2Q A; 3SD0 A; 1KE9 A; 3QX4 A; 5CS6 A; 4FSQ A; 4WT6 A; 4QP3 A; 3LPB A; 2Q0N A; 3UZT A; 4MNE B; 5EYC A; 3CIK A; 4C7T A; 3PJC A; 4O0U A; 4MK0 A; 4XV9 A; 2Z7S A; 3PXY A; 3PG1 A; 3Q32 A; 1WBN A; 2C3I B; 3SKC A; 3BZ3 A; 1LEZ A; 2C6M A; 5CXH A; 4ZSG A; 2CN8 A; 4A9S A; 3QGW A; 2P3G X; 4G9R A; 2Z8C A; 3T9I A; 3AC8 A; 4F08 A; 2PY3 A; 3MSS A; 4R5S A; 3W2Q A; 2R3M A; 4BI1 A; 4QDE A; 2I6L A; 2QHN A; 1ZOG A; 4H3P A; 4CKR A; 3W0Q A; 3V3V A; 2YCS A; 2VX3 A; 3A8W A; 3NUN A; 4Z16 A; 5TT7 A; 3CGO A; 5H8B A; 4OUC A; 5CEO A; 4U7Z A; 5A14 A; 4BJJ A; 3HMI A; 3FLZ A; 4A9U A; 4RA5 A; 3B2W A; 3R7I A; 3ANR A; 2I0Y A; 4UWB A; 3V6R A; 1WCC A; 3V8W A; 2WMS A; 4ZS4 A; 3UQG A; 3FPQ A; 1JKS A; 4RJ5 A; 3BGZ A; 4Y83 A; 4ALW A; 2GNI A; 4MD7 E; 2WQB A; 1XH5 A; 4WUA A; 5M07 A; 4MYG A; 2EWA A; 2B7A A; 3OWK A; 4UMU A; 3I5Z A; 5CU3 A; 4BDA A; 4FG9 A; 4C36 A; 3FHR A; 3CGF A; 3SOA A; 4C4I A; 3BGQ A; 1J1B A; 2R3O A; 3KUL A; 4APC A; 2FH9 A; 5EFQ A; 2G2I A; 3RPY A; 3FXZ A; 1RDQ E; 2C6E A; 2YFX A; 5CX9 A; 5ACB C; 4AFJ A; 5E1E A; 4USF A; 4R7H A; 3Q5Z A; 2Q0B A; 2VNY A; 5AR5 A; 3TXO A; 1OZ1 A; 3UDZ A; 4FOC A; 1M2R A; 1FVT A; 2E9U A; 4WD5 A; 3HP2 A; 3NEW A; 4EOO A; 4RX7 A; 3UMW A; 2Y8O A; 1VLR A; 2ZM3 A; 3GBZ A; 3SGC A; 2WGJ A; 4FV8 A; 2NRU A; 2V55 A; 4BN1 A; 3U51 A; 2YZL A; 1YXU A; 2ETR A; 1MRY A; 3COI A; 3DKO A; 4W4X A; 4C4G A; 2FA2 A; 4LOQ A; 4KIO A; 3CCN A; 3HRC A; 4U79 A; 3V51 A; 4LGH A; 2YLE A; 1JWH A; 3HLL A; 4A4X A; 3N4T A; 1FPU A; 4QYF A; 3BV2 A; 3V5L A; 3UPZ A; 3FQS A; 3DAK A; 2BHH A; 5CZH A; 2XBA A; 3NAY A; 4LFI A; 3RPV A; 2B54 A; 3LAU A; 5FUT A; 5JK3 A; 3EWH A; 4IAF A; 3F5G A; 1I09 A; 3QYW A; 4O4F A; 4A9R A; 5B0X A; 5KKR B; 2GDO A; 2Z2W A; 3F66 A; 4BGH A; 3FZP A; 4KIQ A; 4ITJ A; 4WBO A; 2CKO A; 4NIF B; 4IX5 A; 2GTN A; 2C5N A; 4RFZ A; 2X6E A; 2UW3 A; 2GSF A; 4EWQ A; 5KMJ A; 4G31 A; 4EH4 A; 2KTY A; 2WU7 A; 2AC5 A; 1IAH A; 1PXP A; 4U40 A; 4ONA A; 2JBO A; 2VWZ A; 4FEU A; 4A9Y A; 3R04 A; 4DLJ A; 2CN5 A; 2HY0 A; 1V0O A; 5EHO A; 2Z60 A; 4S34 A; 4DTA A; 2QOI A; 2WMV A; 4ZTS A; 4YZ9 A; 4NH1 A; 1PXJ A; 3KXN A; 2HXQ A; 4XP2 A; 4FKP A; 4JT3 A; 4IM0 A; 3ZXZ A; 5JH6 A; 3KIO C; 2UZO A; 2W5H A; 3Q2M A; 4NW6 A; 4TY1 A; 3RAL A; 3RIN A; 3OCT A; 3W10 A; 4M84 A; 4IC7 A; 4DH8 A; 3UGC A; 5GIO E; 5JZJ A; 1ZZL A; 4UB7 A; 2ZYB A; 2UW4 A; 4FZ7 A; 3PWD A; 3ZHP C; 4KRD A; 4AN2 A; 2XNM A; 4AS0 A; 3V6S A; 1OUK A; 1GZ8 A; 3HV5 A; 2R3G A; 4PUZ A; 4U0I A; 5FP6 A; 3ORX A; 1FIN A; 2YLF A; 4FNW A; 4Q9Z A; 1OL2 A; 5A6O A; 3ODY X; 3QL8 A; 3QZF A; 3PFQ A; 2YN8 A; 2BFX A; 4MBL A; 3SHE A; 3D5X A; 3O5A A; 1FGK A; 3PLS A; 3TUC A; 2GFC A; 4BDB A; 3NR9 A; 3L9L A; 5DHJ A; 4GL9 A; 3DNE A; 4GV1 A; 4NKA A; 5EKO A; 4L9I A; 5AUU A; 5HG9 A; 1NT2 A; 3UP2 A; 2XK3 A; 4LMN A; 5BMM A; 3SLS A; 1UWJ A; 3G0E A; 3LFA A; 3OXI A; 4Y5H A; 5A4C A; 4FV7 A; 2WXV A; 4LL0 A; 4DDI A; 3KRR A; 3ZFX A; 1YOJ A; 2QOH A; 4OBO A; 2UVX A; 3KRJ A; 4ANO A; 4BBE A; 5IA3 A; 4RC4 A; 2GS2 A; 3OCB A; 4ITH A; 2JDR A; 4MXA A; 4USD A; 3VBT A; 2FVD A; 1WBV A; 2X2M A; 3NYO A; 4EOJ A; 1J7L A; 5FEE A; 3H9F A; 5IA0 A; 2HAK A; 2BCJ A; 3MVM A; 3VS1 A; 5LWM A; 5C1Q B; 4I94 A; 3EJ1 A; 2VNW A; 3RPO A; 1BUH A; 3NAX A; 3AG9 A; 3F61 A; 4O1O A; 4YJO A; 3R8V A; 2R3J A; 1YKR A; 3HL7 A; 5B2K A; 4AOJ A; 5AP0 A; 3BHY A; 3P78 A; 4KIP A; 4BTK A; 3IPH A; 4D58 A; 2XKD A; 3F9N A; 4LRK A; 4OLI A; 4ZY6 A; 4JRN A; 2X39 A; 1J7U A; 4BDG A; 4D9T A; 1BYG A; 2GFS A; 2YDK A; 3F2A A; 2UW7 A; 4JXF A; 4MF1 A; 2DYL A; 2NPQ A; 4QOX A; 4Y8D A; 5M44 A; 4MQ1 A; 3Q9W A; 2OF4 A; 2RGP A; 2C6T A; 3ZRM A; 1Q8Z A; 3CY2 A; 3QKM A; 4O7Z A; 5HI2 A; 4R5Y A; 4FL2 A; 5CEI A; 4N57 A; 2C6I A; 2QLQ A; 5FTQ A; 1PKD A; 2ZMC A; 3WIG A; 4X3J A; 1APM E; 5I3O A; 4NM5 A; 2OWB A; 4ZLY A; 3UO5 A; 2X81 A; 3F3Z A; 4PMP A; 5CQU A; 4P5Q A; 3OG7 A; 4FK3 A; 2YIY A; 3E8N A; 3HRF A; 4H58 A; 4RIW B; 3MFT A; 4NKS A; 3F5U A; 5ANK A; 3IW5 A; 3OWL A; 4HYS A; 2JKM A; 4W9X A; 3OVV A; 1ZZ2 A; 4QQC A; 3QC4 A; 2Y7J A; 3ROC A; 1GZO A; 3F7W A; 3BHU A; 3IW8 A; 1OPJ A; 2W9Z B; 2R3N A; 3NIE A; 3OBG A; 3C4Z A; 4O4D A; 4E73 A; 5A54 A; 2X0G A; 4XEY A; 5A4T A; 4I5M A; 3G9L X; 4MD9 E; 4YJP A; 4YMJ A; 4EI4 A; 1H24 A; 3N4V A; 3PRF A; 4Y46 A; 4U6R A; 3G51 A; 2VTA A; 1K3A A; 3RCJ A; 3FZS A; 1F3M C; 4DG3 E; 2YM3 A; 2YDJ A; 2OGV A; 4GT5 A; 5BVO A; 3W0O A; 5GZ9 A; 2UV2 A; 3NCZ A; 3Q96 A; 4BIC A; 5IU2 A; 4HGT A; 3CKX A; 4F4P A; 3E7O A; 3ZFM A; 4DIT A; 3L1S A; 4WG5 A; 3Q3B A; 2EXC X; 5K5X A; 5IQE A; 3HV7 A; 4O82 A; 3P08 A; 3R21 A; 5L3A A; 3SX9 A; 5HEZ A; 4OTG A; 2C5Y A; 2W1F A; 2WEL A; 3DBF A; 3EFW A; 3MB6 A; 5AR3 A; 4AOI A; 3O7L B; 4DH3 A; 4KB8 A; 2PL0 A; 3JXW A; 3MPT A; 4P7E A; 4BI2 A; 3DDP A; 4MAO A; 5GIN E; 3TNQ B; 3L9N A; 4M67 A; 4DG2 E; 3F88 A; 3QQG A; 1SVE A; 2HXL A; 4AXE A; 3CS9 A; 4K11 A; 4PY1 A; 4FTI A; 2PZR A; 3ZMM A; 3R1Y A; 4I6B A; 1JNK A; 1JSV A; 3VJN A; 1YDR E; 5EI2 A; 4N4S A; 3UVQ A; 3B8R A; 4C57 A; 4RPV A; 3TYK A; 5H3Q A; 2RSV A; 5E1S A; 3DFC B; 4MD8 E; 4S2Z A; 5G1X A; 1Q8T A; 4QMM A; 3SV0 A; 4A06 A; 2PWL A; 2UZB A; 2VX0 A; 3NCG A; 4QP2 A; 5EOL A; 5HCY A; 4X3F B; 1S9J A; 2QOD A; 3KRE A; 4BID A; 4AZV A; 3NLB A; 2DUV A; 5E8T A; 4GS6 A; 4QYE A; 4UAL A; 4YLL A; 3CQE A; 4NW5 A; 5EW8 A; 4X2K A; 4XG9 A; 2PVH A; 3K5U A; 4DTB A; 4FVR A; 4PDO A; 3LXN A; 5HCZ A; 5IIS A; 2ERZ E; 3NNW A; 3RAH A; 4XUF A; 3E5A A; 3ZCL A; 4YBJ A; 4MCV A; 4KBA A; 3NX8 A; 1HOW A; 5BPY A; 5GJD A; 1X8B A; 5KCV A; 3IGO A; 4EH2 A; 4IH8 A; 3RCD A; 1TQP A; 4YR8 A; 4CD0 A; 5J9Z A; 5CU4 A; 1BLX A; 3SXR A; 4QMQ A; 3WF8 A; 3QX2 A; 5AV3 A; 3PDT A; 1W8C A; 4AN3 A; 4RJ6 A; 4LDT A; 3SAY A; 5CF6 A; 4Z9L A; 4ACG A; 1V0P A; 4U5J A; 4JMQ A; 4KS8 A; 3R8P A; 1H26 A; 3TJD A; 3BHT A; 3KXG A; 1KE5 A; 1ZXE A; 5CWZ A; 1JVP P; 3P9J A; 3PPK A; 4FIC A; 4KRC A; 4QTB A; 3GOP A; 4YGA A; 4ZS0 A; 2BRH A; 3OCG A; 4ZTL A; 4YZM A; 2X6D A; 5ETC A; 4PTE A; 4GG5 A; 4YNO A; 4NFN A; 5CEQ A; 1P4O A; 2Y4I B; 4HYH A; 4RX9 A; 4XV3 A; 4BR3 A; 1XJD A; 3OTU A; 2LGC A; 3SA0 A; 3ZBX A; 1Y6B A; 1REK A; 1ZYC A; 4BDF A; 3A61 A; 5CI6 A; 4AX8 A; 1NVQ A; 4BCP A; 5IA1 A; 3QD0 A; 4WOV A; 3KMU A; 5B2L A; 4DA5 A; 2HY8 1; 3EQF A; 1I44 A; 2BTR A; 4Z7G A; 1PXI A; 3EN6 A; 3TNP C; 3ZLW A; 3TUD A; 3O8T A; 2JII A; 1PF8 A; 3QBN A; 3R7G A; 4EH7 A; 5AND A; 3ALN A; 2VTT A; 5CQW A; 3LFE A; 5T68 A; 2R3H A; 4JIK A; 4JJR A; 5AP4 A; 3OS3 A; 4BDC A; 1OM1 A; 3SG8 A; 2G1T A; 4E4N A; 3TZ7 A; 2I0H A; 2R5T A; 4GUE A; 4C34 A; 3W0M A; 4E1Z A; 3GXL A; 2W1D A; 2I7Q A; 2EB2 A; 2WU6 A; 4AA4 A; 2XAL A; 3VQH A; 4J71 A; 5CF5 A; 4BWX A; 3PWY A; 4BDK A; 4L43 A; 4RVK A; 5HGI A; 4W4W A; 3RP9 A; 2F2U A; 3DBD A; 4X2N A; 4L53 A; 3WE4 A; 3E7V A; 1TQI A; 3O23 A; 4RMZ A; 4C4F A; 1JSU A; 3AT2 A; 2RF9 A; 5AM7 A; 5BYL A; 2BRG A; 3IEC A; 3ZLX A; 4BKZ A; 2RFN A; 3MY5 A; 4I0T A; 3FMN A; 3EFL A; 3WIK A; 4EOM A; 5IQA A; 4JLC A; 5IUI A; 4R78 A; 4BCO A; 2GQG A; 2OXD A; 3A7G A; 2IVS A; 5IEV A; 3GC0 A; 3G0F A; 4WG3 A; 4NL0 A; 2B9H A; 1XML A; 3RAK A; 1ZMU A; 3PUP A; 2O65 A; 2IZR A; 5ANI A; 4D2S A; 4I6F A; 2PMO X; 2UZV A; 4I5H A; 1V0B A; 1XKK A; 3ZFY A; 4RX5 A; 3B2T A; 3DGK A; 3G5D A; 2P0C A; 3QKL A; 3KCK A; 4I5P A; 4HVH A; 1IAJ A; 3UC4 A; 3HA6 A; 4BC6 A; 3DS6 A; 4NFM A; 1H1R A; 1W82 A; 3GB2 A; 3NUX A; 4O83 A; 4V0G B; 3GC8 A; 5IQH A; 4YTH A; 3SW7 A; 3CEK A; 2F49 A; 4FG7 A; 4KIK B; 3O71 A; 2ITQ A; 4G3E A; 2JAV A; 4O7N A; 5IQD A; 4EKL A; 2WIP A; 1PW2 A; 4X2G A; 4JG7 A; 4M12 A; 4OTI A; 2WTD A; 5CU0 A; 4M8T A; 3I60 A; 1YXS A; 2BMC A; 3F2N A; 5I3R A; 3E64 A; 1PYE A; 2XJ0 A; 4FKR A; 4YLK A; 4EWH A; 1L8T A; 1OIR A; 2XS0 A; 4EK3 A; 4D4R A; 3D0E A; 4AQC A; 4FKV A; 4RT7 A; 3C4F A; 4IAC A; 4BFM A; 1KE7 A; 4TND A; 3R2B A; 3JS2 A; 4FUY A; 3R8U A; 3R30 A; 4NCT C; 4TYG A; 2XIR A; 4KNB A; 4TKS A; 3VUK A; 3BL7 A; 3KMW A; 3N51 A; 4RFM A; 1Y6A A; 1FBN A; 3CSV A; 2F2C B; 2GNJ A; 3SW4 A; 2B4S B; 5IQF A; 3LFC A; 3LHJ A; 4U45 A; 2CSN A; 4ZJJ A; 4B8M A; 2QG7 A; 3PFY A; 3F5X A; 5CNN A; 5JSM A; 4O84 A; 3LJ2 A; 4G5P A; 5HOA A; 2WTK B; 2XKF A; 1Q41 A; 1FVV A; 4FGB A; 4PF4 A; 2C1B A; 4X6Q C; 3ZO4 A; 3DA6 A; 3F69 A; 3L9P A; 4NCT A; 2WQE A; 2PUI A; 5AX3 A; 4EQC A; 4MKC A; 3CP9 A; 4YU2 A; 3UZR A; 3POZ A; 4PNI A; 3D9V A; 3DXP A; 3Q9Y A; 1KE6 A; 3MV5 A; 2X0Q A; 4QNA A; 3MH2 A; 1BI8 A; 4XG4 A; 3U87 A; 4FT7 A; 5E8S A; 4BWK A; 4CZY B; 3ERK A; 2R7B A; 4PMM A; 4QP4 A; 4ENY A; 4B4L A; 1YW2 A; 3KK8 A; 3AGM A; 3D4Q A; 4GJ2 A; 2CHL A; 4Q5H A; 5D1J A; 4EZ3 A; 5GJG A; 4F7L A; 3RTP A; 3ET7 A; 4BTJ A; 3PRI A; 4RIX A; 2YER A; 3UMX A; 2QQ7 A; 2QVS E; 4PRJ A; 2OJI A; 1UNH A; 3EMG A; 5AIK A; 3RNI A; 2ZM1 A; 4X0M A; 3SRV B; 3TKI A; 1PHK A; 4EYJ A; 2IEW A; 4TTH B; 4FGR A; 3A4P A; 2NNW B; 3Q04 A; 1VR2 A; 4ZSJ A; 2F4J A; 4UMT A; 1J7I A; 1WBP A; 3QWK A; 3O8P A; 3K3I A; 4AT5 A; 4O22 A; 4RLK A; 2YWP A; 5IF1 A; 2BRB A; 1GY3 A; 3R00 A; 5HG8 A; 3CTQ A; 4Y5Q A; 1FQ1 B; 5CSX A; 4EJN A; 3HEG A; 3ID6 C; 4GII A; 3TV6 A; 2CKE A; 3NRM A; 3QQF A; 2WQN A; 4QMZ A; 3MFS A; 2WMR A; 3QQJ A; 4CQE A; 5IQC A; 5FLF A; 2ONL C; 3PXK A; 4IB3 A; 3RZF A; 3THB A; 3UE4 A; 3AKK A; 3BI6 A; 3KU2 A; 3OHT A; 4KAB A; 3LW0 A; 4NWM A; 4ZTN A; 5C26 A; 1U54 A; 2W1Z A; 5HZN A; 2LAV A; 3DZQ A; 4RZW A; 4CG9 A; 4DN5 A; 2W4O A; 3ORL A; 5LMA A; 3WE8 A; 3M1S A; 5A3X A; 3EHA A; 4BVU A; 2B53 A; 5A4L A; 3SDJ A; 3Q52 A; 2VTQ A; 3DZO A; 5FDP A; 5AX9 A; 1KV2 A; 1YI3 A; 4IMY A; 3MA3 A; 4WBB B; 4ZY4 A; 5KMK A; 4XP0 A; 5LCJ A; 1EH4 A; 2RL5 A; 2ONL A; 3PA3 A; 4FTA A; 4S33 A; 3RHX A; 4D0W A; 2GNH A; 5KML A; 4M66 A; 1XH8 A; 4GT4 A; 4X2J A; 3P7C A; 3IW6 A; 4GKI A; 3C51 A; 3BIZ A; 3ORN A; 4QMT A; 3NUS A; 4GVA A; 4K77 A; 5C8M A; 1G8A A; 4D5K A; 4PPB A; 2ZV7 A; 3LXP A; 2QON A; 2P4I A; 3KUL B; 4K6Z A; 4CKJ A; 2E9O A; 2JDT A; 4BZN A; 2JKK A; 5E8U A; 3A4O X; 1YRP A; 3W2P A; 4D28 A; 4I4E A; 4F1O A; 3PXZ A; 3VHK A; 2OKR A; 1UNG A; 4KKG A; 5IA5 A; 2I40 A; 3GC7 A; 4IAD A; 1ST0 A; 5E8X A; 3W2R A; 4PX6 A; 4USE A; 3G6H A; 3QTX A; 5TKD A; 4KAO A; 4EC9 A; 4QMN A; 4JQ7 A; 2EUF B; 4CRL A; 1R78 A; 5HES A; 4ANM A; 1PXL A; 1Q3W A; 2B1P A; 4I5C A; 4G3G A; 1SZM A; 3VUH A; 4ENX A; 5IEX A; 2VTS A; 5JEB A; 3UIM A; 4G3F A; 3W8L A; 4ANS A; 1OIQ A; 4IFC A; 3I0Q A; 4I21 A; 4JG8 A; 2YJS A; 4KS7 A; 4E7W A; 4DLI A; 5F95 A; 4UYN A; 4YC6 A; 4WA9 A; 2VWX A; 4FIJ A; 4L46 A; 4PMS A; 3TIZ A; 3BLH A; 3TZM A; 4GK3 A; 2ZOQ A; 4AZE A; 3EFJ A; 5AUW A; 2A98 A; 2OIQ A; 3RK5 A; 2OO8 X; 1U5R A; 3PP0 A; 3V5P A; 2GHM A; 4R1Y A; 3RK7 A; 5I9X A; 3WZD A; 4ZJV A; 1P14 A; 5EM6 A; 1YHW A; 2JBP A; 1YI6 A; 1OKZ A; 2REI A; 3R9O A; 2J0J A; 4K2R A; 1PXN A; 4AE6 A; 3Q2J A; 4EL9 A; 3DTC A; 3QCS A; 4TZR A; 5TOZ A; 3Q9Z A; 4RVT A; 1WBO A; 3ANQ A; 2GMX A; 2WTW A; 3HP5 A; 4GYI A; 3Q4Z A; 2DWB A; 2FSL X; 3A7F A; 2YDI A; 4F63 A; 3VUI A; 2C30 A; 4K18 A; 5ANO A; 1H00 A; 5HX6 A; 1UU8 A; 4LUD A; 5IUH A; 5KX8 A; 2Z7R A; 3CPC A; 4ACD A; 2WD1 A; 2W05 A; 2YAK A; 2ZV2 A; 4B6L A; 3VBV A; 3MFR A; 4BYI A; 4L3L A; 2XIZ A; 3FMM A; 1LP4 A; 4FKG A; 4GCJ A; 4DE4 A; 2Y9Q A; 3ZU7 A; 2QUR A; 4DC2 A; 5L2S A; 2VV9 A; 4FTR A; 4O27 B; 2XK8 A; 2VTR A; 3OEZ A; 5GJF A; 2C5O A; 4KD1 A; 4DGL C; 5HO8 A; 3RMF A; 2XB7 A; 3AC2 A; 4F64 A; 4AW1 A; 4E20 A; 3EQB A; 3UZP A; 3FLN C; 5JFX A; 1H0V A; 4GH2 A; 4MBI A; 3ZLS A; 2WOT A; 4FTN A; 5LMB A; 5CZI A; 2UZD A; 2CPK E; 2WPA A; 5F94 A; 2C6D A; 5HCX A; 2C3K A; 5AMN A; 2E9P A; 2VZ6 A; 4GSB A; 3S3I A; 3VS0 A; 2PVF A; 2VWW A; 2WMW A; 2OZA A; 3AD4 A; 3C1X A; 4EHZ A; 3OUK A; 2ZVA A; 5CEK A; 5DE2 A; 1OEC A; 5EW3 A; 5L6W L; 1W7H A; 1Y57 A; 1K9A A; 2HWO A; 4FT9 A; 4FTK A; 5FG8 A; 4F6U A; 4ZTR A; 4Z3V A; 3RVG A; 4IJ9 A; 3QQL A; 2V7O A; 5D9H A; 2BUJ A; 5LCK A; 3U55 A; 4FKO A; 3ORT A; 4E6A A; 4GMY A; 4HNF A; 5DBX A; 3VUM A; 4AXD A; 4RA4 A; 2PML X; 3I0O A; 4R6V A; 2WMU A; 3IDP A; 4KWP A; 2JLD A; 3EQC A; 4I20 A; 1JKL A; 2XNO A; 3MVL A; 4HYI A; 1URW A; 3FSF A; 4YSJ A; 4YNE A; 2UW9 A; 5JZN A; 1VZO A; 2WAJ A; 3PIY A; 3OT3 A; 4HVI A; 2FO0 A; 4JBP A; 4P5Z A; 4WNK A; 3R8M A; 1OKW A; 4XG3 A; 3GC9 A; 3NUP A; 3Q4T A; 4AA5 A; 4BBF A; 4DEG A; 5HQ0 A; 4IZ7 A; 3BYO A; 4IX6 A; 3ZM4 A; 2HCK A; 4IWQ A; 2PVR A; 1WAK A; 5AP6 A; 4GJ3 A; 2EXE A; 4FMQ A; 2A19 B; 2B9J A; 4RIO A; 1T45 A; 4WSY A; 3FHI A; 3LMG A; 4Z55 A; 2IW6 A; 4YJQ A; 2GU8 A; 5HG7 A; 1IEP A; 4H39 A; 3MBL A; 4FZF B; 2C6O A; 3BPR A; 1WZY A; 1LR4 A; 2RFS A; 2RFE A; 2BTS A; 3ENM A; 4W8D A; 5HIB A; 4IXP A; 4I6L A; 4I23 A; 3DJ5 A; 3R6X A; 5DEW A; 5AP3 A; 1VEB A; 1H1P A; 1YQJ A; 1H07 A; 4LL5 A; 5KHW A; 3D7T B; 3GFE A; 2SRC A; 4JNW A; 3QTZ A; 2W96 B; 5HVJ A; 4YO6 A; 4BCH A; 3D7Z A; 1JKK A; 2GNF A; 1G5S A; 3RPR A; 1OL6 A; 3V5W A; 3RI1 A; 2J50 A; 5HKM A; 5CAP A; 1W84 A; 2QHM A; 3KXM A; 4WSQ A; 3CD8 A; 4O96 A; 3B8Q A; 4QP7 A; 4FEV A; 2YM7 A; 4BDI A; 3LIJ A; 5ANJ A; 3PXQ A; 2J5E A; 5HE2 A; 3BLQ A; 2YIX A; 3KY2 A; 4Y95 A; 4G16 A; 5CSV A; 5I9V A; 4MXZ A; 1U59 A; 4NTS A; 4FIG A; 3GEN A; 2XNE A; 5IUG A; 3F5P A; 3FYJ X; 4ZTM A; 4RWJ A; 4QML A; 2BIK B; 4FZ6 A; 2OJG A; 2V0D A; 3D2K A; 4HCV A; 5CSH A; 4HGS A; 2UZE A; 3IOK A; 5J79 A; 3NS9 A; 4HNI A; 1M7Q A; 4U44 A; 4O21 A; 4AZF A; 1Q3D A; 2CNU A; 3ORI A; 2QU5 A; 3MES A; 2P2H A; 5GHV A; 5HE1 A; 3U6H A; 2YCR A; 4L6Q A; 4TL0 A; 2V7A A; 3KXX A; 4FE2 A; 2PUP A; 5BUI A; 3GU8 A; 3BWF A; 5D7A A; 1KOA A; 3BYU A; 4ZME A; 3NGA A; 5CT0 A; 3A7I A; 3II5 A; 5D7V A; 4BKY A; 4DFZ E; 4E93 A; 3MTL A; 3Q53 A; 4G2F A; 3KVW A; 4BHN A; 3WBL A; 2JGZ A; 2GS6 A; 3FLW A; 5FBN C; 2QOQ A; 4QMW A; 2ETK A; 3UIB A; 3G6G A; 3CXW A; 3HMM A; 5DR9 A; 3W8Q A; 3BU6 A; 3FAA A; 1QPD A; 2WMA A; 2OIB A; 5DT4 A; 3SXS A; 3KQ7 A; 4C2W A; 4CV8 A; 3TNI A; 4MVF A; 5IG1 A; 3TUB A; 1G8S A; 5DG5 A; 1JLU E; 2H96 A; 2BPM A; 2VWU A; 3O0G A; 4N6Z A; 3FLQ A; 3EQR A; 4WIH A; 3VNT A; 3HDN A; 4JDK A; 4E5B A; 3PG3 A; 4DFN A; 3C4X A; 3W55 A; 3QHW A; 4C38 A; 3HRB A; 4GFG A; 4D1S A; 2VO7 A; 3A7H A; 4FKU A; 3I1A A; 3W2C A; 4B7T A; 4J1R A; 4DTK A; 4EOS A; 4RIY B; 4F7S A; 4MWH A; 3MJ1 A; 3NUY A; 3KGA A; 2BZI B; 1UV5 A; 1CMK E; 4ORK A; 3QQH A; 3PE2 A; 5CF8 A; 4FV1 A; 3DND A; 2OLC A; 3WYY A; 3HZT A; 4AA0 A; 3T8O A; 3KMM A; 3ZC5 A; 4ZLZ A; 3CJG A; 3H0Y A; 4M14 A; 5D10 A; 4FV9 A; 4RRV A; 5FM3 A; 5A9U A; 2C4G A; 1ZYD A; 4XRL A; 4F9W A; 5AV1 A; 3BGP A; 3T3V A; 1TVO A; 4NEU A; 4KKH A; 4A7C A; 5BYY A; 2RKU A; 2CJM A; 3KN6 A; 5CT7 A; 1L3R E; 4MWI A; 3HAV A; 4BGG A; 4FA2 A; 2QO2 A; 4KKE A; 4R7I A; 4WHZ A; 3P86 A; 3QXP A; 2W4J A; 3W33 A; 2W5B A; 5EG3 A; 3OMV A; 3PJ8 A; 4RJ4 A; 4J7B A; 2PVJ A; 4M7I A; 2V62 A; 3DQW A; 3LE6 A; 4DGO A; 2FB8 A; 1R0P A; 4O86 A; 3SC1 A; 5M4C A; 4LGD A; 2O3P A; 3ZEP A; 4DHJ A; 3NVK I; 2WMX A; 3R63 A; 5HOR A; 3RAW A; 1KUT A; 2HZI A; 4C8B A; 4EUT A; 4HOK A; 4JQE A; 3QUD A; 4FV6 A; 4CFX A; 4DFX E; 3PPZ A; 3O17 A; 5GNK A; 3R0T A; 4JDI A; 4PTC A; 1GIJ A; 4ANL A; 4F09 A; 2O64 A; 2W1C A; 3VF9 A; 4CFU A; 4YJT A; 5CAN A; 3KB7 A; 2JIT A; 4TYH A; 4DBN A; 2CLQ A; 4RWI A; 3MY0 A; 2VWI A; 4QTC A; 1R1W A; 4FTJ A; 4AT3 A; 4UY9 A; 4WUN A; 4CLI A; 1Y91 A; 4RQK A; 2CGW A; 4HVD A; 5ETI A; 2GHG A; 2OU7 A; 2CMW A; 4O0Y A; 4TNB A; 4UZH A; 3C0I A; 2WTV A; 1VYW A; 3DTW A; 5DT0 A; 3VVH A; 4CMO A; 1TQM A; 3NW6 A; 4CT1 A; 3PA4 A; 1QCF A; 3COM A; 3PJ3 A; 3TG1 A; 3RNY A; 3AMA A; 4BCQ A; 4OAV B; 5IZF A; 1BL6 A; 4E26 A; 4EOQ A; 4FKQ A; 2YM4 A; 2JD5 A; 3TMO A; 4LRM A; 3PUF C; 3GCS A; 3DJ6 A; 3OXT A; 4EH3 A; 3U4W A; 4E5W A; 3S0O A; 2ZMD A; 5EH0 A; 5M56 A; 5AP5 A; 1A48 A; 4AOT A; 1DS5 A; 3R01 A; 4IEB A; 3ZSI A; 3DB6 A; 4EKK A; 2VO0 A; 4FSY A; 4FZD B; 4IVD A; 3BEG A; 3OEF X; 2IZS A; 3R1Q A; 2IW8 A; 4WG4 A; 3AC5 A; 5FTO A; 5HG5 A; 1OBG A; 2QO7 A; 4I6H A; 5EHY A; 4GFM A; 5BUJ A; 4ANB A; 2XBJ A; 3AXW A; 3ETA A; 2A1A B; 2X2L A; 1ATP E; 4FTM A; 4A9T A; 5L6P A; 3CE3 A; 4TT7 A; 2CNV A; 1BX6 A; 5M55 A; 1OI9 A; 2JFL A; 4FZA B; 5J95 A; 3TT0 A; 4D2W A; 2PK9 A; 2QG5 A; 2XKC A; 4HJO A; 5TO8 A; 5JKG A; 4CSV A; 4DT9 A; 3PYY A; 1OL7 A; 4DF3 A; 1W2D A; 1Q4L A; 1XBA A; 4EC8 A; 2DS1 A; 3D5U A; 5BVD A; 3P5J C; 3GU5 A; 5DVR A; 2C0T A; 3HYH A; 4FKW A; 4HGL A; 4AGU A; 4E4L A; 4X7Q A; 1CDK A; 2PUN A; 2XAM A; 2R3Q A; 4EH8 A; 5J1V A; 2F9G A; 3MH0 A; 2IW9 A; 2PU8 A; 3U4U A; 2IWI A; 3OZ6 A; 3ZO2 A; 2XK7 A; 1Q24 A; 2QR8 A; 4HCU A; 3KRX A; 4L3J A; 3ORZ A; 4YP8 A; 5A4Q A; 3DK3 A; 3WF9 A; 3TZ9 A; 1YWV A; 3EFK A; 4ZTH A; 2X4Z A; 2PVN A; 3ORK A; 3IDC A; 4FV4 A; 5HID A; 3TTI A; 5IQG A; 5KBQ A; 1M52 A; 3DDQ A; 3KXZ A; 1W83 A; 3ZSH A; 4E3C A; 3IKA A; 3MIY A; 3H4J A; 5AIR A; 3C4W A; 3HA8 A; 4GT3 A; 2OXY A; 2FYS A; 3I5N A; 3NNV A; 4QMO A; 1OB3 A; 4G3C A; 3Q6W A; 3MJ2 A; 2ITN A; 5HHW A; 3IO7 A; 1ZOE A; 4DH1 A; 2YAA A; 3N9X A; 4IAK A; 1DI8 A; 3A99 A; 3QFV A; 2HYY A; 1OL5 A; 4C37 A; 3EH9 A; 4F9Y A; 4GIH A; 3RBW A; 3T9T A; 3CBL A; 4CEG A; 2YM8 A; 1YDT E; 2OK1 A; 4RJ3 A; 3UOD A; 4JQ8 A; 4PQN A; 4DAW A; 5JQ5 A; 2BDW A; 3FI4 A; 4H1J A; 4F9C A; 4O1P A; 3KL8 A; 1B38 A; 3K54 A; 4FV5 A; 5BVF A; 5KKR C; 3FMJ A; 2Y4P A; 3ZZE A; 3VAP A; 5HIC A; 3ML1 A; 4L42 A; 4H36 A; 4H3Q A; 1JAM A; 1ZAO A; 5ICP A; 1FOT A; 1GII A; 4IIR A; 5ANG A; 4ZXT A; 3QD2 B; 3PY1 A; 4FG8 A; 4JX7 A; 4FT5 A; 2P55 A; 5L6O A; 2XYU A; 5GZA A; 1PMV A; 2J0L A; 4YJV A; 4FEX A; 2J7T A; 4OT6 A; 5K4I A; 1NA7 A; 2OID A; 1H8F A; 4G3D A; 1Z5M A; 3E62 A; 4QPA A; 4TWC A; 4ANQ A; 5M4I A; 4JS8 A; 3PVB A; 2QNJ A; 4FR4 A; 1GZK A; 3Q6U A; 1B39 A; 4OBP A; 5CVH A; 4E6Q A; 5K0K A; 4DT8 A; 4DFY A; 1ZY5 A; 4ZZM A; 3IG7 A; 3LJ0 A; 4NIF A; 1ZMW A; 5HLW A; 1M2Q A; 4QTD A; 2YEX A; 1WVX A; 2XA4 A; 3IOP A; 3VBW A; 3GEQ A; 4AZW A; 1STC E; 2QO9 A; 4EOL A; 2X2K A; 1WVY A; 4EHE A; 1NVR A; 3VS2 A; 5C27 A; 2X8I A; 4AXC A; 3MIA A; 3OP5 A; 2X7F A; 3PJ1 A; 3TCP A; 4G5J A; 2C6K A; 4GFO A; 3EQH A; 3MB7 A; 4CT2 A; 4HGE A; 2QOK A; 3WOW A; 3DKG A; 4HZR A; 2ITP A; 2UVZ A; 3OBJ A; 4FKT A; 4N0S A; 3EQD A; 4PTG A; 3VF8 A; 3ZO3 A; 2QU6 A; 4GUB A; 4IQ6 A; 4YPD A; 4O0X A; 4EBV A; 2W06 A; 5AUX A; 3UIU A; 2WTC A; 1ZOH A; 2P33 A; 4ACC A; 4DEE A; 4Q9S A; 2RFD A; 2XZS A; 3BEL A; 4TXC A; 4RWL A; 1IR3 A; 4TYE A; 4G17 A; 3BL9 A; 4L8M A; 4OTR A; 2B52 A; 5JFW A; 2VTP A; 4XLV A; 1ZMV A; 5EYK A; 3M11 A; 3GOK A; 3R7O A; 2ZJW A; 3QF9 A; 3UIX A; 4D4V A; 1OKY A; 5BQ0 A; 4YZC A; 5H8E A; 4F0G A; 4IHP A; 3O96 A; 3ZOS A; 3EYG A; 2W17 A; 3NW5 A; 2HZ0 A; 3DU8 A; 1ZYL A; 3QRU A; 2J0M B; 2ZV8 A; 5CAO A; 3OCS A; 3HX4 A; 5EHL A; 3ZO1 A; 3I7C A; 4O0T A; 2A4L A; 4B99 A; 4BYJ A; 1YXX A; 3MDY A; 1K2P A; 2GHL A; 2BAL A; 2UW5 A; 4RZV A; 5HTB A; 4M69 A; 1ZLT A; 2QD9 A; 2UZW E; 1W2F A; 3W32 A; 3FJQ E; 3PXR A; 4NST A; 3BE2 A; 3P0M A; 3TNW A; 3TJC A; 2EVA A; 5EK7 A; 5IH5 A; 5J9L A; 5FGK A; 1JBP E; 2R3L A; 2X1N A; 5CVF A; 5DIA A; 2R3K A; 5B7V A; 4AG8 A; 1IA8 A; 2J90 A; 2YCF A; 1MRU A; 3UOJ A; 2XM9 A; 3O50 A; 4BCF A; 1YWN A; 3COK A; 4XG6 A; 4FKI A; 1XBB A; 4IVA A; 2PZI A; 5CAQ A; 3MVH A; 4O7W A; 4IB1 A; 1H28 A; 4FTQ A; 4QPM A; 4XV2 A; 4XH6 A; 1Y8G A; 3QTR A; 4F9B A; 4MQ2 A; 3QGY A; 3U6I A; 1LPU A; 5ETF A; 1MUO A; 3ACJ A; 3LFN A; 1SM2 A; 4PNK A; 2I1M A; 3PPJ A; 3HUC A; 4DDG A; 4L00 A; 4ZZO A; 2VN9 A; 3QXO A; 5EQE A; 3UDS A; 3NDM A; 4HW7 A; 3FXW A; 3DBQ A; 5HBE A; 1KSW A; 2YIR A; 4KSQ A; 5FBO A; 4EON A; 1CM8 A; 1OVE A; 1GOL A; 4QDV A; 3FXX A; 3KFA A; 2C3L A; 2DQ7 X; 1HCL A; 1P4F A; 3VRZ A; 1JOW B; 4XBU A; 4D4S A; 2VGP A; 1VJY A; 3Q5I A; 4FOB A; 3VUD A; 4FST A; 5EM5 A; 3UP7 A; 5IDP A; 4EH5 A; 5KZI A; 5C4K A; 4FUX A; 4PP9 A; 1ZY4 A; 2Z7L A; 4JL9 A; 2CDZ A; 4E4X A; 2PZ5 A; 3EL7 A; 4IVC A; 3ALO A; 3ZRK A; 3BRB A; 5C9C A; 3PLA E; 3TIY A; 4CLJ A; 3NUU A; 2EU9 A; 3Q60 A; 5FEQ A; 1CSN A; 3MS9 A; 3VW8 A; 4IAN A; 3RM7 A; 3UDB A; 4ACM A; 5A46 A; 4H1M A; 1Q5K A; 3KRL A; 3W18 A; 4PDS A; 2OJF E; 2VO3 A; 3CJF A; 4G34 A; 3DB8 A; 4EBW A; 3PIX A; 5DRD A; 2XK4 A; 5J1W A; 3ZDI A; 2BAJ A; 4O6L A; 4QP6 A; 2Q83 A; 4MTA A; 4RGJ A; 5CEN A; 1MP8 A; 4G6O A; 5DEY A; 5BX0 A; 1OIY A; 5E8Z A; 4UN0 C; 5IMX A; 1JST A; 1CKI A; 5DGZ A; 2HOG A; 1ZWS A; 2WMT A; 3P7B A; 3ZXT A; 4ITI A; 4M0Y A; 3MH1 A; 3AC3 A; 3EZV A; 1SMH A; 3H30 A; 4GU9 A; 3I4B A; 2X4F A; 3H9O A; 2OZA B; 4BL1 A; 2QLU A; 3GCP A; 1SVG A; 2XAO A; 5E8V A; 4C3F A; 4TN6 A; 5H9B A; 1V1K A; 1T46 A; 3GI3 A; 3QKK A; 2WEI A; 4V01 A; 3RK9 A; 2OG8 A; 5J7H A; 3SOC A; 1OL1 A; 1ZRZ A; 2X8D A; 3FI3 A; 3AT3 A; 4NM0 A; 4FSM A; 4O2P A; 5CEP A; 2IO6 A; 5HIE A; 3WAR A; 2X0O A; 5IKW A; 3NZ0 A; 3R9N A; 2NYA A; 3DQX A; 3DPK A; 5DA3 A; 4FJV A; 5ANE A; 2YIS A; 2EXM A; 4O6E A; 3NNU A; 3QCX A; 3MPA A; 4FTO A; 5CTP A; 1YXT A; 3L9M A; 2I0E A; 5J5T A; 5F20 A; 2IZU A; 4XJ0 A; 3DOG A; 4JVG A; 4K33 A; 3NUA A; 2RIO A; 4F6S A; 3I79 A; 3QHR A; 5IQI A; 4ALV A; 2VWV A; 2HEL A; 3R71 A; 4AZT A; 2Z7Q A; 4RJ7 A; 4BIB A; 1WMK A; 4XG8 A; 3RAI A; 4LV5 A; 1BI7 A; 2UW6 A; 3LQ8 A; 4JBV A; 3HEC A; 2WQM A; 3OTV A; 3OY1 A; 3D5V A; 3TZ8 A; 5JMS A; 1M2P A; 4JR7 A; 3E93 A; 3OUN B; 5HLP A; 3VN9 A; 3LCS A; 2WKM A; 3KA0 A; 3UG1 A; 3EN4 A; 3E63 A; 3VW6 A; 2J5F A; 3DBC A; 1YI4 A; 3KCF A; 3GNI B; 4TYJ A; 4S31 A; 5FXQ A; 2ZDT A; 5K3Y A; 3FBV A; 3TV7 A; 4NM7 A; 3FYK X; 1O9U A; 4QT1 A; 4Z7H A; 2C6L A; 4U43 A; 3E8D A; 4QMU A; 5FXS A; 3QTU A; 3ATS A; 3BLA A; 4CG8 A; 4BCM A; 5JT2 A; 4PP7 A; 3PTG A; 2H34 A; 3H0Z A; 1YWR A; 3HNG A; 2O5K A; 2ZB1 A; 4BKJ A; 5AR2 A; 4C02 A; 1M7N A; 2W7X A; 2ITV A; 3C0H A; 4BJJ B; 5I9U A; 2G15 A; 1FVR A; 3RHK A; 4YZN A; 1IG1 A; 3PSC A; 4MXY A; 4E6C A; 4JDJ A; 3AOX A; 3G9N A; 4YC8 A; 5CF4 A; 2QI8 A; 2OBJ A; 2XAN A; 3QQU A; 3GUB A; 5K9I A; 4FTU A; 4QYG A; 5CSP A; 5LVN A; 3QTW A; 5C01 A; 4BDJ A; 2PHK A; 4J52 A; 3G15 A; 3HUB A; 3RWQ A; 5GIP E; 4RVL A; 3IS5 A; 4L01 A; 1GZN A; 2AC3 A; 3CTH A; 3LCO A; 2FGI A; 3GQI A; 5FXR A; 4R7B A; 3M2W A; 1YVJ A; 1P2A A; 4FSZ A; 4GG7 A; 4F7J A; 1UNL A; 2R3F A; 4PL5 A; 4CGA A; 5DR6 A; 4ZY5 A; 3ZON A; 4D0X A; 4AE9 A; 4QMX A; 1ND4 A; 4JAI A; 4LV7 A; 3MA6 A; 1H25 A; 3HV4 A; 3PA5 A; 4NT4 A; 3TWJ A; 4UMP A; 4ASD A; 5A4E A; 5JRQ A; 2VTL A; 2VWB A; 1Q62 A; 2J4Z A; 3ROY A; 4MH7 A; 4FIE A; 5DH3 A; 1REJ A; 4OTH A; 4U3Y A; 3NYX A; 3ZUU A; 5KMM A; 4O0R A; 4QPS A; 4JOA A; 2YA9 A; 2VTO A; 5EW9 A; 4I93 A; 4OGR A; 3H3C A; 3U6J A; 1OGY A; 5AR8 A; 2BR1 A; 3L8S A; 5AP2 A; 4AGD A; 3GT8 A; 2WHB A; 1NXK A; 2XYN A; 4JR3 A; 4LRJ A; 4EYM A; 3UO6 A; 2ITZ A; 4J95 A; 5KHX A; 5J9Y A; 5DPV A; 3HAM A; 4G9C A; 3QLF A; 2O63 A; 4R1V A; 4EQM A; 2WMB A; 3CR0 A; 3DY7 A; 3VHE A; 2O0U A; 3PE1 A; 3D2I A; 3W0P A; 4F78 A; 2BZH B; 4PL3 A; 3WF5 A; 4O4E A; 5C8N A; 5HD4 A; 4O4C A; 5HBJ A; 5CAS A; 2GPH A; 4MBJ A; 5CNO A; 5KZ0 A; 4FV0 A; 4QP9 A; 3OW3 A; 2C0O A; 4YZB A; 4BY9 E; 4C2V A; 3LMH A; 3F7Z A; 3ZSG A; 2UZL A; 2ZB0 A; 1CKP A; 5CY3 A; 4CV9 A; 4BBM A; 2CGX A; 1G3N A; 5HU9 A; 4O7R A; 3UVP A; 3AC1 A; 2JIU A; 2YIQ A; 3AD5 A; 4ZEG A; 4OW8 A; 2WZJ A; 4CFF A; 4YUR A; 2PYW A; 3N4U A; 5IH4 A; 3ELJ A; 4GU6 A; 1XBC A; 4EJ7 A; 3GFW A; 3PSB A; 1QPE A; 4Y73 A; 4UMR A; 3IK3 A; 2PMI A; 4I4F A; 5BYZ A; 4FKJ A; 4LQM A; 4D9U A; 3FSK A; 4V0G A; 3ULZ A; 4FYO A; 3FUP A; 1MRV A; 4KIK A; 1H1W A; 4APP A; 2XCK A; 4ANN A; 2ITU A; 4LM5 A; 4ALU A; 5DN3 A; 1E9H A; 4YHF A; 4CNH A; 2W5A A; 4L67 A; 4F99 A; 3BHH A; 4NJ3 A; 2XIK A; 3NW7 A; 5E7R A; 3WZK A; 4AW5 A; 2ZFY A; 4FNZ A; 4F1T A; 3NPC A; 2XKE A; 2XIY A; 4TRL A; 4JX3 A; 4FL3 A; 4F6W A; 4C33 A; 2OH0 E; 5LOH A; 3C0G A; 4PWN A; 2WTK C; 2IPX A; 4XP3 A; 3S2P A; 1KE8 A; 3MWU A; 2XVD A; 3HKO A; 3I81 A; 5E92 A; 4RVM A; 5I9Z A; 3LXK A; 3OF0 A; 1J1C A; 5HO6 A; 3UDT A; 2BFY A; 2W1E A; 4WRG A; 4R3C A; 1XH6 A; 2O2U A; 3FQH A; 3C5U A; 5HU3 A; 2JKQ A; 1TZD A; 2QOO A; 2WO6 A; 4YJR A; 2FSO X; 2YCQ A; 2YAB A; 4LUE A; 4EQU A; 1GIH A; 1O6Y A; 2PUU A; 3QAM E; 4AAA A; 3UOK A; 3BU3 A; 2G2F A; 4D1X A; 3U54 A; 2OW3 A; 2BZK B; 5F9E A; 4QRC A; 4QYH A; 2X9F A; 3D94 A; 4IAI A; 4EEV A; 2XM8 A; 4FV2 A; 4IM3 A; 2CLX A; 1JPA A; 1PXM A; 2NRY A; 3BU5 A; 3ZH8 A; 2XJ1 A; 3V5T A; 3BYV A; 4D1Z A; 1UWH A; 4HYU A; 4ASX A; 2UW0 A; 4C35 A; 2VU3 A; 3QYZ A; 5K4J A; 2OJ9 A; 4P2K A; 1M17 A; 4A4L A; 5E4H A; 3EYH A; 4DH5 A; 3SXF A; 4UXQ A; 4C3P A; 4AF3 A; 4M0Z A; 2VWY A; 3FEG A; 2FST X; 4XG2 A; 2IZT A; 1ERK A; 2OFU A; 3HMN A; 4TPT A; 3VRY A; 2UUE A; 1XMM A; 2AUH A; 4AGW A; 4EZ7 A; 4CZU A; 3SRV A; 2XNN A; 4CCB A; 4FEW A; 4BHZ A; 3FL5 A; 3MFU A; 4FF8 A; 5GZ8 A; 1QMZ A; 4FI1 A; 3LM0 A; 3R8Z A; 3WZU A; 4BCJ A; 4CZT A; 2BHE A; 3BQC A; 4BF2 A; 1KWP A; 4MF0 A; 3C50 A; 3OT8 A; 4WRS A; 4ID7 A; 1H0W A; 4ZSA A; 3C7Q A; 5AJQ A; 2H9V A; 4FX3 A; 4FIH A; 4YPS A; 5BNJ A; 5DT3 A; 4O7L A; 3MYG A; 2OZO A; 4M68 A; 3CQU A; 4BCG A; 1JQH A; 4XS2 A; 4BCK A; 3SG9 A; 4TWN A; 3QTI A; 3BWJ A; 3NYN A; 4WKQ A; 3GU6 A; 2PTK A; 3JVR A; 2F7Z E; 2IG7 A; 4Y85 A; 4YC3 A; 4EUU A; 3PZH A; 4IVB A; 5L8L A; 5KMI A; 3UNZ A; 4GW8 A; 4KIN A; 3VJO A; 1OPL A; 3LFS A; 2VUW A; 4YJS A; 4C3R A; 3G2F A; 1OIT A; 1PMN A; 1Z57 A; 4CQG A; 3R1N A; 4K8A A; 1H27 A; 2ITY A; 3E3P A; 3LZB A; 4FK6 A; 1YHS A; 3R7U A; 4E6D A; 4F0I A; 5FWK K; 4X6R A; 4YSM A; 1MQB A; 2ZDU A; 3R7E A; 3UG2 A; 1UU3 A; 3QD4 A; 5DFP A; 4B8L A; 4CFV A; 1PYX A; 4I24 A; 3FMH A; 4D2P A; 4E8A A; 2W99 B; 4ACH A; 5H8G A; 2WTJ A; 4EK5 A; 5M51 A; 4V04 A; 3S00 A; 2PE2 A; 4WNO A; 4EOK A; 4MUQ A; 3W1F A; 1LEW A; 4NK9 A; 3R22 A; 1OBD A; 3R8L A; 4IWD A; 3LLA A; 2E9N A; 4L7S A; 3LOK A; 3QU0 A; 5IQB A; 3R73 A; 5AV4 A; 3P79 A; 3VUG A; 5CLP A; 3ULI A; 4PDP A; 3TV4 A; 3OY3 A; 5LJJ A; 3Q4U A; 3FZ1 A; 4V05 A; 1XQZ A; 3GGF A; 5L4Q A; 4FT3 A; 5FD2 A; 5CAU A; 2VD5 A; 2HZN A; 3R83 A; 2XK9 A; 2CCH A; 2R64 A; 4OBQ A; 1OGU A; 4BGQ A; 1NVS A; 2VTN A; 3OXZ A; 5H2U A; 1M14 A; 4UBA A; 1OPK A; 4QQJ A; 3P5K A; 5HO7 A; 3FLY A; 4O7Y A; 3BYS A; 5HNI X; 1PY5 A; 2BRN A; 5C03 A; 5AEP A; 3NYV A; 4LMU A; 5KO1 A; 3UVR A; 4D2V A; 4ZSE A; 2QKW B; 3G90 X; 4JDH A; 3DFA A; 3V5Q A; 2BZJ A; 2R3I A; 5AV0 A; 4JBO A; 3LMI A; 1WBT A; 2RG5 A; 2PVL A; 4PPC A; 2VTM A; 5AAG A; 3L8P A; 4PV0 A; 4GRB A; 5ACK A; 4QO9 A; 4QP1 A; 3TEI A; 3CPB A; 3E8E A; 4R77 A; 2BAQ A; 3FKN A; 2QKR A; 3D83 A; 4EK6 A; 2PSQ A; 3KN5 A; 4AXA A; 3C9W A; 3FZT A; 4FT0 A; 3T3U A; 4D2R A; 3KRW A; 4WO5 A; 3F3T A; 2WOU A; 3NSZ A; 4K0Y A; 5I0B A; 4IAY A; 2I0V A; 4F1M A; 4PPA A; 3DXQ A; 1DI9 A; 4RJ8 A; 4CRS A; 4UWY A; 1G8J A; 3UYT A; 4RG0 A; 4QTA A; 3WI6 A; 2JED A; 2J9M A; 5IH6 A; 4C58 A; 3VQU A; 3UBD A; 4MHA A; 4F65 A; 3A2C A; 4O7S A; 4RBL A; 2QCS A; 3VS6 A; 3A62 A; 4AUA A; 2HWP A; 4YZD A; 3V5J A; 4HVS A; 4J8N A; 4FII A; 4RSS A; 2Y6M A; 3MPM A; 2GNG A; 2W1G A; 4U81 A; 2ITX A; 3VS7 A; 5I9Y A; 5M57 A; 5EDR A; 5EKN A; 3A7J A; 1YDS E; 3VID A; 5IA4 A; 4TW9 A; 2XCH A; 1O6K A; 4MX9 A; 4W4V A; 5M53 A; 4WNP A; 1P5E A; 3NVM B; 3UNK A; 3VBX A; 4IE9 A; 1BL7 A; 4W8E A; 2C3J A; 3BCE A; 2WEV A; 1TFF A; 1PRY A; 2H8H A; 1Q8Y A; 4LV8 A; 3UC3 A; 4M69 B; 1O6L A; 2IVU A; 5HTI A; 3TDW A; 3TKU A; 4GKH A; 4CFM A; 4OTF A; 4FEQ A; 4KSP A; 2A2A A; 1URC A; 2HIW A; 4IM2 A; 1H01 A; 4FTC A; 4LOP A; 5F4N A; 4EH6 A; 4OT5 A; 2CGV A; 3FMK A; 4TWO A; 4DBX A; 1IRK A; 3MH3 A; 3KEX A; 3NNX A; 3WZJ A; 3IGG A; 4DYM A; 2NO3 A; 4BI0 A; 2X7O A; 4MNF A; 3ZC6 A; 3F82 A; 2X9E A; 3G33 A; 1W98 A; 3COH A; 4IZ5 A; 3LQ3 A; 1RQQ A; 4HZS A; 4LYN A; 4I41 A; 3FI2 A; 1UU9 A; 4YTC A; 4NYE A; 1OKV A; 5AR7 A; 3LCT A; 4MNE A; 5HE0 A; 4UXL A; 4IB0 A; 2CKP A; 5AR4 A; 3LCD A; 5AA9 A; 4FVQ A; 3FL4 A; 4I3Z A; 2QR7 A; 3JY9 A; 4AQK A; 3FKO A; 1ST4 A; 3LJ1 A; 2R2P A; 3KXH A; 5T3Q A; 4AXF A; 2YZA A; 1WBS A; 3TI1 A; 3UQC A; 5DOS A; 3DKF A; 3IW4 A; 3A8X A; 4G6L A; 3ZBF A; 5CXZ A; 4HCT A; 3LCK A; 4QEB A; 3D7T A; 1DM2 A; 4X2F A; 2R0I A; 4FKL A; 4CYJ A; 3GP0 A; 3CY3 A; 2BRM A; 2OIC A; 3GCU A; 3TN8 A; 3WZE A; 5LVP A; 3ORO A; 4N70 A; 2GS7 A; 2GTM A; 3E88 A; 3JVS A; 5HVY A; 4K9Y A; 4EK4 A; 4O38 A; 5LVO A; 3AMY A; 1Q8U A; 3MTF A; 2W1H A; 3W2S A; 3DK7 A; 1SYK A; 2WB8 A; 4AN9 A; 2IJM A; 3EOC A; 3ITZ A; 3PY3 A; 5DWR A; 4TYI A; 1H4L A; 3IQ7 A; 4XOZ A; 5LXM A; 3MW1 A; 3WF6 A; 1SNX A; 3DAE A; 3KKV A; 5FED A; 4XRJ A; 4ZZN A; 3NMU F; 3ATT A; 3ZEW A; 2VO6 A; 4EH9 A; 1XH9 A; 3M42 A; 4YLJ A; 5HNB A; 4O81 A; 1AD5 A; 4F7N A; 3MI9 A; 2JC6 A; 4CYI A; 2HEN A; 3O8U A; 3UYS A; 3HDM A; 3TNH A; 3MY1 A; 4AW0 A; 5M4U A; 2UZN A; 2W1I A; 1E1X A; 5JRS A; 2UZU E; 3HMP A; 5M06 A; 5F1Z A; 4QMS A; 4I1Z A; 4FKS A; 3RPS A; 4OAU C; 3GQL A; 2YM6 A; 3BYM A; 5C4L A; 4XOY A; 3QTQ A; 3RZB A; 1Q97 A; 4DIN A; 4FSU A; 1YOM A; 5BUE A; 4DFB A; 3OOG A; 3DXN A; 5CYI A; 3ION A; 4O91 A; 4S30 A; 3C13 A; 4RQV A; 2ITT A; 2FUM A; 3PP1 A; 2BVA A; 5FQD C; 3LQ5 A; 4DGG A; 5EDP A; 5ETA A; 2BRO A; 4FV3 A; 4AFE A; 3I7B A; 3VWA A; 1OIU A; 3VUT A; 3FLS A; 2XK6 A; 3BV3 A; 5CAL A; 4FSW A; 4YBK A; 2BKK A; 2RG6 A; 5DI1 A; 2F7E E; 2XRU A; 2XF0 A; 4EHV A; 5EM8 A; 3ACK A; 4R3R A; 4BWP A; 4YUQ A; 3RGF A; 5TR6 A; 1W2C A; 3OC1 A; 4OR5 A; 4YHT A; 1QL6 A; 4Y72 A; 2Y6O A; 4E4M A; 4EOP A; 3C4C A; 1CKJ A; 4J98 A; 2XH5 A; 2VTJ A; 3VO3 A; 3FWQ A; 4O0S A; 3AT4 A; 3K2L A; 4L68 A; 4OTQ A; 4BZD A; 5CU2 A; 1UA2 A; 1R39 A; 3RJC A; 4DFL A; 5KBR A; 4RIX B; 4ZIM A; 1Z9X A; 3FKL A; 4KBC A; 5JGB A; 4YJU A; 4AP7 A; 4EOR A; 5LAR A; 3I6U A; 1Q8W A; 3ORM A; 3KVX A; 4ZP5 A; 3BHV A; 4X21 A; 3GU4 A; 1XH4 A; 4A07 A; 4BDE A; 4BDH A; 4YTF A; 5AUT A; 5JGD A; 1LUF A; 4FNX A; 2E2B A; 5AM6 A; 4C4E A; 4RIY A; 3DT1 A; 4MXO A; 1TKI A; 2WQO A; 3VON A; 2ZV9 A; 4L52 A; 4IBM A; 2PMN X; 4D55 A; 3BQR A; 3QA0 A; 2PPQ A; 2XNB A; 2R0U A; 2IN6 A; 5I9W A; 3ZM9 A; 3U9C A; 2R3R A; 4C59 A; 4KUJ A; 4MXC A; 3KC3 A; 5EYM A; 3ZS5 A; 4IX4 A; 2B0Q A; 2HK5 A; 4JIA A; 2Y0A A; 4U42 A; 2CKQ A; 1U54 B; 3BX5 A; 5K0X A; 3OD6 X; 2JIV A; 2J0I A; 2XMY A; 1GJO A; 1WBW A; 2G01 A; 3UPX A; 3E87 A; 3JYA A; 1DAY A; 4O0V A; 3O51 A; 4R8Q A; 3S4Q A; 4I6Q A; 2JDV A; 4P4C A; 4RC2 A; 4O7V A; 1YXV A; 3LKM A; 3P1A A; 4QP8 A; 5AUZ A; 2AYP A; 3EQP A; 4YHJ A; 3QZI A; 3A60 A; 3AGL A; 4KBK A; 3HGK A; 5JQ8 A; 2IVT A; 3WYX A; 3QC9 A; 1QPJ A; 2R7I A; 4XV1 A; 4CMU A; 2WMQ A; 5BMS A; 3CTJ A; 4UEU A; 4AW2 A; 5E9E A; 1DAW A; 3ZTX A; 3C5I A; 3FY2 A; 5FTG A; 1NY3 A; 2UVY A; 2ZAZ A; 1RE8 A; 3EN7 A; 4DEB A; 3MN3 A; 3QA8 A; 3KF7 A; 2W9F B; 3TL8 A; 3W2O A; 4ZJI A; 4L45 A; 4IAZ A; 4IX3 A; 5L8J A; 3EZR A; 4BRX A; #chains in the Genus database with same CATH homology 1A06 A; 5BVW A; 1H1Q A; 2EB3 A; 4UMQ A; 2VAG A; 3BE9 A; 3QWJ A; 3SVV A; 2O8Y A; 5AP7 A; 3C4E A; 5DRB A; 5ITA A; 2GQR A; 4ERW A; 1U5Q A; 4XMO A; 5EOB A; 3VS3 A; 5CAV A; 2PUL A; 3U8W A; 3HV6 A; 3FML A; 3QLG A; 1YHV A; 3GCQ A; 1ZAR A; 3CQW A; 1B6C B; 2H6D A; 2J0K A; 4MXX A; 2JDS A; 5D41 A; 5AUV A; 2QC6 A; 5CU6 A; 4EHG A; 1OUY A; 3D5W A; 2YWV A; 5KMN A; 2FSM X; 3FC1 X; 4BZO A; 3RWP A; 3BKB A; 3KK9 A; 1E1V A; 3JUH A; 5J8I A; 3FDN A; 5HD7 A; 4DCE A; 2XEZ A; 4NUS A; 1UU7 A; 2QOL A; 1Q61 A; 3UNJ A; 1W0X C; 3BEA A; 2BIL B; 2C1A A; 2YIT A; 3EKK A; 4G6N A; 2ZM4 A; 1XH7 A; 5IDN A; 3TDV A; 3WF7 A; 3PIZ A; 3LLT A; 3PZE A; 3QQK A; 2HW7 A; 4XHK B; 4D2T A; 1ZYJ A; 4DCA A; 4J8M A; 3DAJ A; 4IZA A; 1R3C A; 2BDF A; 1GNG A; 5C8K A; 4A4O A; 3WIL A; 4BTM A; 4UYA A; 3FPM A; 4W4Y A; 3TKH A; 4TWP A; 1XWS A; 3CD3 A; 4RIW A; 4UV0 A; 3ZUT A; 2J2I B; 1H1S A; 4JRV A; 3DJ7 A; 3QRI A; 3FE3 A; 2BIY A; 3IW7 A; 3Q4C A; 3ZYA A; 2KUL A; 3F3W A; 5KE0 A; 5A6N A; 5JFV A; 5CEM A; 3IDB A; 4CMT A; 4NM3 A; 1WVW A; 5CI7 A; 5CSW A; 5HBH A; 4MED A; 4BIE A; 3OWP A; 5FP5 A; 3VBQ A; 5LWN A; 4B9D A; 3EQI A; 4BB4 A; 5I4N A; 3EL8 A; 5LVL A; 1J3H A; 1KV1 A; 3BLR A; 3PVU A; 4AGC A; 4DHF A; 2W0J A; 5HVK A; 3V01 A; 2F57 A; 3HVC A; 3RM6 A; 4HVG A; 4LI5 A; 2ACX A; 5EQY A; 2V5Q A; 3QRJ A; 4EZ5 A; 3V8S A; 3UTO A; 1XR1 A; 3ZUV A; 3DLS A; 2X8E A; 5FM2 A; 4C61 A; 2PVY A; 4D5H A; 1GAG A; 4DHZ A; 4NTT A; 1PXO A; 4II5 A; 1PKG A; 2VTI A; 3HV3 A; 4JI9 A; 4I0S A; 1Y8Y A; 5AA8 A; 3SQQ A; 3CC6 A; 3E3B X; 4I92 A; 4QNY A; 4ZLO A; 3K5V A; 2IVV A; 3QRK A; 4FC0 A; 2JKO A; 2A27 A; 3GU7 A; 5EDQ A; 3FMD A; 3HMO A; 3S95 A; 5M09 A; 5JGA A; 4L44 A; 4UZD A; 5D12 A; 3BBW A; 3OOM A; 5BVK A; 4FYN A; 2OI4 X; 3I6W A; 2W4K A; 2C68 A; 1WFC A; 4F9A A; 4DFU A; 1F0Q A; 4IDV A; 3PSD A; 4FSN A; 3PY0 A; 4X3F C; 5IEY A; 3TM0 A; 4CFN A; 3ENH A; 3JPV A; 4RC3 A; 2CCI A; 3BBT B; 3UJG A; 1SVH A; 5J7B A; 2OXX A; 2OF2 A; 3UQF A; 1UKI A; 2VGO A; 4NU1 A; 5HE3 A; 4QYY A; 4GK4 A; 4U41 A; 1PME A; 1G8K A; 3UOH A; 2XNG A; 5EI6 A; 4PMT A; 3GCV A; 3ZZW A; 2VX1 A; 2WIH A; 4OTW A; 1QPC A; 1U4D A; 5E90 A; 5K5N A; 4FBX A; 3OW4 A; 4R3P A; 2YM5 A; 2JAM A; 3U9N A; 4NZW B; 5EQP A; 3EN9 A; 3QD3 A; 5FRI A; 4X3F A; 4DH7 A; 3R28 A; 3TTJ A; 5IA2 A; 1FGI A; 1RW8 A; 5L2I A; 2OH4 A; 4FIF A; 1FMO E; 4LGG A; 4Q2A A; 4IJP A; 2XJ2 A; 5HTC A; 4G1W A; 4IZY A; 3TAC A; 2V22 A; 3EN5 A; 4KA3 A; 4AWI A; 1S9I A; 4DEA A; 5I5Z A; 3R1S A; 1FMK A; 4QMV A; 4B0G A; 4HPU E; 3R9R A; 2R3P A; 1XHA A; 4D4Y A; 4U97 A; 1PJK A; 2OJJ A; 4CKI A; 4OTD A; 3OWJ A; 2JDO A; 3DV3 A; 4QQT A; 4ZMF A; 2QOB A; 1R0E A; 4FVP A; 3W16 A; 2G9X A; 2WTI A; 4QMY A; 1BMK A; 5D11 A; 3FZO A; 2UW8 A; 4CCU A; 5LQF A; 4DG0 E; 4IW0 A; 2XP2 A; 5LMK C; 2J6M A; 5E91 A; 2XRW A; 5AP1 A; 2J51 A; 2C69 A; 3AD6 A; 3QUE A; 2ITW A; 2Y4I C; 3H10 A; 3QTS A; 4RWK A; 3EQG A; 4U80 A; 5B2M A; 3CLY A; 2JFM A; 3DK6 A; 4LOO A; 3FI8 A; 3O7L D; 4GK2 A; 3AKJ A; 1UVR A; 2BKZ A; 2C47 A; 2PZP A; 2PVK A; 3VUL A; 2AEG A; 3DCV A; 4W9W A; 2XAR A; 4NJD A; 4H3B A; 3LXL A; 1OMW A; 4QMP A; 4JG6 A; 4ASZ A; 3JY0 A; 5KMO A; 1YOL A; 4J96 A; 5E8Y A; 3R02 A; 5EYD A; 3EB0 A; 4JBQ A; 2PVM A; 3R2Y A; 3FQE A; 5LMK A; 3W0S A; 5EM7 A; 3JR1 A; 2ETM A; 5CVG A; 5DMZ A; 5AAF A; 5DR2 A; 2E9V A; 4XCU A; 4CFW A; 4AAY A; 3QCY A; 2GNL A; 4YJN A; 3P7A A; 4C4J A; 2NP8 A; 4IDT A; 5LVM A; 4I0R A; 3DBE A; 3LFQ A; 4IFG A; 4DGM A; 2XIX A; 3F3U A; 3AKL A; 2Z02 A; 3NFG B; 2YAC A; 2PE1 A; 4XNE A; 1BKX A; 5KNJ A; 1RJB A; 4FSR A; 4ARK A; 1UKH A; 2PZY A; 3POO A; 3VC4 A; 2HW6 A; 4M15 A; 1F5Q A; 2ESM A; 3FV8 A; 4ZSL A; 1J91 A; 4XYF A; 5HX8 A; 5AWM A; 3FC2 A; 3FY0 A; 4LG4 A; 4XG7 A; 4YTI A; 3D14 A; 5IME A; 5AV2 A; 3NFF B; 3MGY A; 4F70 A; 4XX9 A; 4HPT E; 4EK8 A; 3AQV A; 4IWO A; 4ZOG A; 4PL4 A; 4YO4 A; 1JKT A; 2VRX A; 3VS5 A; 2ERK A; 3K3J A; 3QZG A; 4F0F A; 5TE0 A; 3F2R A; 4O7T A; 2ITO A; 3EID A; 5BV3 A; 2OFV A; 5DLS A; 4JAJ A; 3NWW A; 5DYJ A; 4Q5E A; 1MQ4 A; 3P4K A; 5L8K A; 4ERK A; 1A9U A; 2XEY A; 4ZAU A; 5AAD A; 3QCQ A; 3F3V A; 3PJ2 A; 4U3Z A; 2QOF A; 3PVW A; 3Q9X A; 3NIZ A; 3QRT A; 4AAC A; 4FTL A; 1ZYS A; 5JN2 A; 4XBR A; 4QD6 A; 4IC8 A; 4QTE A; 3V04 A; 3LFD A; 4CXA A; 2BAK A; 4AU8 A; 3AMB A; 4S32 A; 4C62 A; 4BDD A; 3QZH A; 3ORP A; 4UWC A; 3E92 A; 3ZLY A; 2HZ4 A; 3ODZ X; 3UO4 A; 5HLN A; 4H05 A; 2X7G A; 3DKC A; 4AT4 A; 4DED A; 3L8X A; 3VS4 A; 1Q99 A; 1IAS A; 2WFY A; 3R7Y A; 4ASE A; 4E5A X; 5G6V A; 3V8T A; 3W0R A; 3E8C A; 3LFF A; 2CNQ A; 5BVN A; 2XNP A; 3VBY A; 4AZS A; 4L7F A; 5TF9 A; 2F7X E; 3S1H A; 2C5X A; 3H9R A; 4DHI B; 4C4H A; 5JFS A; 4FOD A; 2QOC A; 2P2I A; 4BCI A; 3PXF A; 2R4B A; 1KOB A; 2XUU A; 2PE0 A; 4N6Y A; 1PMU A; 2UZT A; 3ZDU A; 5T6K A; 2VTH A; 2YJR A; 4WNM A; 4BTF A; 1H08 A; 4P90 A; 4O2Z A; 1AQ1 A; 3QUP A; 3OFM A; 4M13 A; 4M97 A; 5EI8 A; 5AAE A; 4I22 A; 4CVA A; 2G2H A; 3P23 A; 4DEI A; 3W0N A; 3GVU A; 4J99 A; 3DLZ A; 5HNG A; 5BVE A; 4EOI A; 5M4F A; 4DEH A; 1AGW A; 2R9S A; 4ZK5 A; 3RKB A; 4PDY A; 3EKN A; 3D15 A; 2CGU A; 4J53 A; 5AUY A; 2WNT A; 2YIW A; 3L8V A; 3LM5 A; 4TYH B; 3R7V A; 5IPJ A; 3LVP A; 4ZTQ A; 3FZR A; 4GVJ A; 1CTP E; 5E8W A; 4C0T A; 4U9A A; 2B9I A; 2B55 A; 2YHV A; 4K1B A; 3PVG A; 2C0I A; 4L3P A; 4XLI A; 1HCK A; 5J87 A; 5DNR A; 2GQS A; 5L2T A; 3AC4 A; 4UX9 A; 4M3Q A; 5L2W A; 3REP A; 2C5V A; 4O0W A; 2OSC A; 4J97 A; 4CTB A; 1U46 A; 5J5S A; 4IWP A; 4IAA A; 3LFB A; 4IB5 A; 4OTP A; 3R9H A; 4QQ5 A; 3ZRL A; 4RLL A; 3FME A; 1NW1 A; 3O2M A; 4U8Z A; 4DGN A; 5KUP A; 2B9F A; 3MVJ A; 4GEO A; 2A0C X; 4CTC A; 3CKW A; 1VYZ A; 5M08 A; 2BDJ A; 5FDX A; 2GCD A; 1XO2 B; 4FNY A; 3KF4 A; 1PXK A; 4RFY A; 4BCN A; 3F6X A; 4FL1 A; 1SNU A; 4GYG A; 5AFV A; 3QAL E; 4FTT A; 4RX8 A; 3R9D A; 3UOL A; 1IA9 A; 5L2Q A; 3SD0 A; 1KE9 A; 3QX4 A; 5CS6 A; 4FSQ A; 4WT6 A; 4QP3 A; 3LPB A; 2Q0N A; 3UZT A; 4MNE B; 5EYC A; 3CIK A; 4C7T A; 3PJC A; 4O0U A; 4MK0 A; 4XV9 A; 2Z7S A; 3PXY A; 3PG1 A; 3Q32 A; 1WBN A; 2C3I B; 3SKC A; 3BZ3 A; 1LEZ A; 2C6M A; 5CXH A; 4ZSG A; 2CN8 A; 4A9S A; 3QGW A; 2P3G X; 4G9R A; 2Z8C A; 3T9I A; 3AC8 A; 4F08 A; 2PY3 A; 3MSS A; 4R5S A; 3W2Q A; 2R3M A; 4BI1 A; 4QDE A; 2I6L A; 2QHN A; 1ZOG A; 4H3P A; 4CKR A; 3W0Q A; 3V3V A; 2YCS A; 2VX3 A; 3A8W A; 3NUN A; 4Z16 A; 5TT7 A; 3CGO A; 5H8B A; 4OUC A; 5CEO A; 4U7Z A; 5A14 A; 4BJJ A; 3HMI A; 3FLZ A; 4A9U A; 4RA5 A; 3B2W A; 3R7I A; 3ANR A; 2I0Y A; 4UWB A; 3V6R A; 1WCC A; 3V8W A; 2WMS A; 4ZS4 A; 3UQG A; 3FPQ A; 1JKS A; 4RJ5 A; 3BGZ A; 4Y83 A; 4ALW A; 2GNI A; 4MD7 E; 2WQB A; 1XH5 A; 4WUA A; 5M07 A; 4MYG A; 2EWA A; 2B7A A; 3OWK A; 4UMU A; 3I5Z A; 5CU3 A; 4BDA A; 4FG9 A; 4C36 A; 3FHR A; 3CGF A; 3SOA A; 4C4I A; 3BGQ A; 1J1B A; 2R3O A; 3KUL A; 4APC A; 2FH9 A; 5EFQ A; 2G2I A; 3RPY A; 3FXZ A; 1RDQ E; 2C6E A; 2YFX A; 5CX9 A; 5ACB C; 4AFJ A; 5E1E A; 4USF A; 4R7H A; 3Q5Z A; 2Q0B A; 2VNY A; 5AR5 A; 3TXO A; 1OZ1 A; 3UDZ A; 4FOC A; 1M2R A; 1FVT A; 2E9U A; 4WD5 A; 3HP2 A; 3NEW A; 4EOO A; 4RX7 A; 3UMW A; 2Y8O A; 1VLR A; 2ZM3 A; 3GBZ A; 3SGC A; 2WGJ A; 4FV8 A; 2NRU A; 2V55 A; 4BN1 A; 3U51 A; 2YZL A; 1YXU A; 2ETR A; 1MRY A; 3COI A; 3DKO A; 4W4X A; 4C4G A; 2FA2 A; 4LOQ A; 4KIO A; 3CCN A; 3HRC A; 4U79 A; 3V51 A; 4LGH A; 2YLE A; 1JWH A; 3HLL A; 4A4X A; 3N4T A; 1FPU A; 4QYF A; 3BV2 A; 3V5L A; 3UPZ A; 3FQS A; 3DAK A; 2BHH A; 5CZH A; 2XBA A; 3NAY A; 4LFI A; 3RPV A; 2B54 A; 3LAU A; 5FUT A; 5JK3 A; 3EWH A; 4IAF A; 3F5G A; 1I09 A; 3QYW A; 4A9R A; 5KKR B; 5B0X A; 2GDO A; 2Z2W A; 3F66 A; 4BGH A; 3FZP A; 4KIQ A; 4ITJ A; 4WBO A; 2CKO A; 4NIF B; 4IX5 A; 2GTN A; 2C5N A; 4RFZ A; 2X6E A; 2UW3 A; 2GSF A; 4EWQ A; 5KMJ A; 4G31 A; 4EH4 A; 2KTY A; 2WU7 A; 2AC5 A; 1IAH A; 1PXP A; 4U40 A; 4ONA A; 2JBO A; 2VWZ A; 4FEU A; 4A9Y A; 3R04 A; 4DLJ A; 2CN5 A; 2HY0 A; 1V0O A; 5EHO A; 2Z60 A; 4S34 A; 4DTA A; 2QOI A; 2WMV A; 4ZTS A; 4YZ9 A; 4NH1 A; 1PXJ A; 3KXN A; 2HXQ A; 4XP2 A; 4FKP A; 4JT3 A; 4IM0 A; 3ZXZ A; 5JH6 A; 3KIO C; 2UZO A; 2W5H A; 3Q2M A; 4NW6 A; 4TY1 A; 3RAL A; 3RIN A; 3OCT A; 3W10 A; 4M84 A; 4IC7 A; 4DH8 A; 3UGC A; 5GIO E; 5JZJ A; 1ZZL A; 4UB7 A; 2ZYB A; 2UW4 A; 4FZ7 A; 3PWD A; 3ZHP C; 4KRD A; 4AN2 A; 2XNM A; 4AS0 A; 3V6S A; 1OUK A; 1GZ8 A; 3HV5 A; 2R3G A; 4PUZ A; 4U0I A; 5FP6 A; 3ORX A; 1FIN A; 2YLF A; 4FNW A; 4Q9Z A; 1OL2 A; 5A6O A; 3ODY X; 3QL8 A; 3QZF A; 3PFQ A; 2YN8 A; 2BFX A; 4MBL A; 3SHE A; 3D5X A; 3O5A A; 1FGK A; 3PLS A; 3TUC A; 2GFC A; 4BDB A; 3NR9 A; 3L9L A; 5DHJ A; 4GL9 A; 3DNE A; 4GV1 A; 4NKA A; 5EKO A; 4L9I A; 5AUU A; 5HG9 A; 1NT2 A; 3UP2 A; 2XK3 A; 4LMN A; 5BMM A; 3SLS A; 1UWJ A; 3G0E A; 3LFA A; 3OXI A; 4Y5H A; 5A4C A; 4FV7 A; 2WXV A; 4LL0 A; 4DDI A; 3KRR A; 3ZFX A; 1YOJ A; 2QOH A; 4OBO A; 2UVX A; 3KRJ A; 4ANO A; 4BBE A; 5IA3 A; 4RC4 A; 2GS2 A; 3OCB A; 4ITH A; 2JDR A; 4MXA A; 4USD A; 3VBT A; 2FVD A; 1WBV A; 2X2M A; 3NYO A; 4EOJ A; 1J7L A; 5FEE A; 3H9F A; 5IA0 A; 2HAK A; 2BCJ A; 3MVM A; 3VS1 A; 5LWM A; 5C1Q B; 4I94 A; 3EJ1 A; 2VNW A; 3RPO A; 1BUH A; 3NAX A; 3AG9 A; 3F61 A; 4O1O A; 4YJO A; 3R8V A; 2R3J A; 1YKR A; 3HL7 A; 5B2K A; 4AOJ A; 5AP0 A; 3BHY A; 3P78 A; 4KIP A; 4BTK A; 3IPH A; 4D58 A; 2XKD A; 3F9N A; 4LRK A; 4OLI A; 4ZY6 A; 4JRN A; 2X39 A; 1J7U A; 4BDG A; 4D9T A; 1BYG A; 2GFS A; 2YDK A; 3F2A A; 2UW7 A; 4JXF A; 4MF1 A; 2DYL A; 2NPQ A; 4QOX A; 4Y8D A; 5M44 A; 4MQ1 A; 3Q9W A; 2OF4 A; 2RGP A; 2C6T A; 3ZRM A; 1Q8Z A; 3CY2 A; 3QKM A; 4O7Z A; 5HI2 A; 4R5Y A; 4FL2 A; 5CEI A; 4N57 A; 2C6I A; 2QLQ A; 5FTQ A; 1PKD A; 2ZMC A; 3WIG A; 4X3J A; 1APM E; 5I3O A; 4NM5 A; 2OWB A; 4ZLY A; 3UO5 A; 2X81 A; 3F3Z A; 4PMP A; 5CQU A; 4P5Q A; 3OG7 A; 4FK3 A; 2YIY A; 3E8N A; 3HRF A; 4H58 A; 4RIW B; 3MFT A; 4NKS A; 3F5U A; 5ANK A; 3IW5 A; 3OWL A; 4HYS A; 2JKM A; 4W9X A; 3OVV A; 1ZZ2 A; 4QQC A; 3QC4 A; 2Y7J A; 3ROC A; 1GZO A; 3F7W A; 3BHU A; 3IW8 A; 1OPJ A; 2W9Z B; 2R3N A; 3NIE A; 3OBG A; 3C4Z A; 5AV1 A; 4E73 A; 5A54 A; 2X0G A; 4XEY A; 5A4T A; 4I5M A; 3G9L X; 4MD9 E; 4YJP A; 4YMJ A; 4EI4 A; 1H24 A; 3N4V A; 3PRF A; 4Y46 A; 4U6R A; 3G51 A; 2VTA A; 1K3A A; 3RCJ A; 3FZS A; 1F3M C; 4DG3 E; 2YM3 A; 2YDJ A; 2OGV A; 4GT5 A; 5BVO A; 3W0O A; 5GZ9 A; 2UV2 A; 3NCZ A; 3Q96 A; 4BIC A; 5IU2 A; 4HGT A; 3CKX A; 4F4P A; 3E7O A; 3ZFM A; 4DIT A; 3L1S A; 4WG5 A; 3Q3B A; 2EXC X; 5K5X A; 5IQE A; 3HV7 A; 4O82 A; 3P08 A; 3R21 A; 5L3A A; 3SX9 A; 5HEZ A; 4OTG A; 2C5Y A; 2W1F A; 2WEL A; 3DBF A; 3EFW A; 3MB6 A; 5AR3 A; 4AOI A; 3O7L B; 4DH3 A; 4KB8 A; 2PL0 A; 3JXW A; 3MPT A; 4P7E A; 4BI2 A; 3DDP A; 4MAO A; 5GIN E; 3TNQ B; 3L9N A; 4M67 A; 4DG2 E; 3F88 A; 3QQG A; 1SVE A; 2HXL A; 4AXE A; 3CS9 A; 4K11 A; 4PY1 A; 4FTI A; 2PZR A; 3ZMM A; 3R1Y A; 4I6B A; 1JNK A; 1JSV A; 3VJN A; 1YDR E; 5EI2 A; 4N4S A; 3UVQ A; 3B8R A; 4C57 A; 4RPV A; 3TYK A; 5H3Q A; 2RSV A; 5E1S A; 3DFC B; 4MD8 E; 4S2Z A; 5G1X A; 1Q8T A; 4QMM A; 3SV0 A; 4A06 A; 2PWL A; 2UZB A; 2VX0 A; 3NCG A; 4QP2 A; 5EOL A; 5HCY A; 4X3F B; 1S9J A; 2QOD A; 3KRE A; 4BID A; 4AZV A; 3NLB A; 2DUV A; 5E8T A; 4GS6 A; 4QYE A; 4UAL A; 4YLL A; 3CQE A; 4NW5 A; 5EW8 A; 4X2K A; 4XG9 A; 2PVH A; 3K5U A; 4DTB A; 4FVR A; 4PDO A; 3LXN A; 5HCZ A; 5IIS A; 2ERZ E; 3NNW A; 3RAH A; 4XUF A; 3E5A A; 3ZCL A; 4YBJ A; 4MCV A; 4KBA A; 3NX8 A; 1HOW A; 5BPY A; 5GJD A; 1X8B A; 5KCV A; 3IGO A; 4EH2 A; 4IH8 A; 3RCD A; 1TQP A; 4YR8 A; 4CD0 A; 5J9Z A; 5CU4 A; 1BLX A; 3SXR A; 4QMQ A; 3WF8 A; 3QX2 A; 5AV3 A; 3PDT A; 1W8C A; 4AN3 A; 4RJ6 A; 4LDT A; 3SAY A; 5CF6 A; 4Z9L A; 4ACG A; 1V0P A; 4U5J A; 4JMQ A; 4KS8 A; 3R8P A; 1H26 A; 3TJD A; 3BHT A; 3KXG A; 1KE5 A; 1ZXE A; 5CWZ A; 1JVP P; 3P9J A; 3PPK A; 4FIC A; 4KRC A; 4QTB A; 3GOP A; 4YGA A; 4ZS0 A; 2BRH A; 3OCG A; 4ZTL A; 4YZM A; 2X6D A; 5ETC A; 4PTE A; 4GG5 A; 4YNO A; 4NFN A; 5CEQ A; 1P4O A; 2Y4I B; 4HYH A; 4RX9 A; 4XV3 A; 4BR3 A; 1XJD A; 3OTU A; 2LGC A; 3SA0 A; 3ZBX A; 1Y6B A; 1REK A; 1ZYC A; 4BDF A; 3A61 A; 5CI6 A; 4AX8 A; 1NVQ A; 4BCP A; 5IA1 A; 3QD0 A; 4WOV A; 3KMU A; 5B2L A; 4DA5 A; 2HY8 1; 3EQF A; 1I44 A; 2BTR A; 4Z7G A; 1PXI A; 3EN6 A; 3TNP C; 3ZLW A; 3TUD A; 3O8T A; 2JII A; 1PF8 A; 3QBN A; 3R7G A; 4EH7 A; 5AND A; 3ALN A; 2VTT A; 5CQW A; 3LFE A; 5T68 A; 2R3H A; 4JIK A; 4JJR A; 5AP4 A; 3OS3 A; 4BDC A; 1OM1 A; 3SG8 A; 2G1T A; 4E4N A; 3TZ7 A; 2I0H A; 2R5T A; 4GUE A; 4C34 A; 3W0M A; 4E1Z A; 3GXL A; 2W1D A; 2I7Q A; 2EB2 A; 2WU6 A; 4AA4 A; 2XAL A; 3VQH A; 4J71 A; 5CF5 A; 4BWX A; 3PWY A; 4BDK A; 4L43 A; 4RVK A; 5HGI A; 4W4W A; 3RP9 A; 2F2U A; 3DBD A; 4X2N A; 4L53 A; 3WE4 A; 3E7V A; 1TQI A; 3O23 A; 4RMZ A; 4C4F A; 1JSU A; 3AT2 A; 2RF9 A; 5AM7 A; 5BYL A; 2BRG A; 3IEC A; 3ZLX A; 4BKZ A; 2RFN A; 3MY5 A; 4I0T A; 3FMN A; 3EFL A; 3WIK A; 4EOM A; 5IQA A; 4JLC A; 5IUI A; 4R78 A; 4BCO A; 2GQG A; 2OXD A; 3A7G A; 2IVS A; 5IEV A; 3GC0 A; 3G0F A; 4WG3 A; 4NL0 A; 2B9H A; 1XML A; 3RAK A; 1ZMU A; 3PUP A; 2O65 A; 2IZR A; 5ANI A; 4D2S A; 4I6F A; 2PMO X; 2UZV A; 4I5H A; 1V0B A; 1XKK A; 3ZFY A; 4RX5 A; 3B2T A; 3DGK A; 3G5D A; 2P0C A; 3QKL A; 3KCK A; 4I5P A; 4HVH A; 1IAJ A; 3UC4 A; 3HA6 A; 4BC6 A; 3DS6 A; 4NFM A; 1H1R A; 1W82 A; 3GB2 A; 3NUX A; 4O83 A; 4V0G B; 3GC8 A; 5IQH A; 4YTH A; 3SW7 A; 3CEK A; 2F49 A; 4FG7 A; 4KIK B; 3O71 A; 2ITQ A; 4G3E A; 2JAV A; 4O7N A; 5IQD A; 4EKL A; 2WIP A; 1PW2 A; 4X2G A; 4JG7 A; 4M12 A; 4OTI A; 2WTD A; 5CU0 A; 4M8T A; 3I60 A; 1YXS A; 2BMC A; 3F2N A; 5I3R A; 3E64 A; 1PYE A; 2XJ0 A; 4FKR A; 4YLK A; 4EWH A; 1L8T A; 1OIR A; 2XS0 A; 4EK3 A; 4D4R A; 3D0E A; 4AQC A; 4FKV A; 4RT7 A; 3C4F A; 4IAC A; 4BFM A; 1KE7 A; 4TND A; 3R2B A; 3JS2 A; 4FUY A; 3R8U A; 3R30 A; 4NCT C; 4TYG A; 2XIR A; 4KNB A; 4TKS A; 3VUK A; 3BL7 A; 3KMW A; 3N51 A; 4RFM A; 1Y6A A; 1FBN A; 3CSV A; 2F2C B; 2GNJ A; 3SW4 A; 2B4S B; 5IQF A; 3LFC A; 3LHJ A; 4U45 A; 2CSN A; 4ZJJ A; 4B8M A; 2QG7 A; 3PFY A; 3F5X A; 5CNN A; 5JSM A; 4O84 A; 3LJ2 A; 4G5P A; 5HOA A; 2WTK B; 2XKF A; 1Q41 A; 1FVV A; 4FGB A; 4PF4 A; 2C1B A; 4X6Q C; 3ZO4 A; 3DA6 A; 3F69 A; 3L9P A; 4NCT A; 2WQE A; 2PUI A; 5AX3 A; 4EQC A; 4MKC A; 3CP9 A; 4YU2 A; 3UZR A; 3POZ A; 4PNI A; 3D9V A; 3DXP A; 3Q9Y A; 1KE6 A; 3MV5 A; 4QNA A; 4XG4 A; 3MH2 A; 1BI8 A; 3U87 A; 4BWK A; 4FT7 A; 5E8S A; 4CZY B; 3ERK A; 2R7B A; 4PMM A; 4QP4 A; 4ENY A; 4B4L A; 1YW2 A; 3KK8 A; 3AGM A; 3D4Q A; 4GJ2 A; 2CHL A; 4Q5H A; 5D1J A; 4EZ3 A; 5GJG A; 4F7L A; 3RTP A; 3ET7 A; 4BTJ A; 3PRI A; 4RIX A; 2YER A; 3UMX A; 2QQ7 A; 2QVS E; 4PRJ A; 2OJI A; 1UNH A; 3EMG A; 5AIK A; 3RNI A; 2ZM1 A; 4X0M A; 3SRV B; 3TKI A; 1PHK A; 4EYJ A; 4TTH B; 4FGR A; 3A4P A; 2NNW B; 3Q04 A; 1VR2 A; 4ZSJ A; 2F4J A; 4UMT A; 1J7I A; 1WBP A; 3QWK A; 3O8P A; 3K3I A; 4AT5 A; 4O22 A; 4RLK A; 2YWP A; 5IF1 A; 2BRB A; 1GY3 A; 3R00 A; 5HG8 A; 3CTQ A; 4Y5Q A; 1FQ1 B; 5CSX A; 4EJN A; 3HEG A; 3ID6 C; 4GII A; 3TV6 A; 2CKE A; 3NRM A; 3QQF A; 2WQN A; 4QMZ A; 3MFS A; 2WMR A; 3QQJ A; 4CQE A; 5IQC A; 5FLF A; 2ONL C; 3PXK A; 4IB3 A; 3RZF A; 3THB A; 3UE4 A; 3AKK A; 3BI6 A; 3KU2 A; 3OHT A; 4KAB A; 3LW0 A; 4NWM A; 4ZTN A; 5C26 A; 1U54 A; 2W1Z A; 5HZN A; 2LAV A; 3DZQ A; 4RZW A; 4CG9 A; 4DN5 A; 2W4O A; 3ORL A; 5LMA A; 3WE8 A; 3M1S A; 5A3X A; 3EHA A; 4BVU A; 2B53 A; 5A4L A; 3SDJ A; 3Q52 A; 2VTQ A; 3DZO A; 5FDP A; 5AX9 A; 1KV2 A; 1YI3 A; 4IMY A; 3MA3 A; 4WBB B; 4ZY4 A; 5KMK A; 4XP0 A; 5LCJ A; 1EH4 A; 2RL5 A; 2ONL A; 3PA3 A; 4FTA A; 4S33 A; 3RHX A; 4D0W A; 2GNH A; 5KML A; 4M66 A; 1XH8 A; 4GT4 A; 4X2J A; 3P7C A; 3IW6 A; 4GKI A; 3C51 A; 3BIZ A; 3ORN A; 4QMT A; 3NUS A; 4GVA A; 4K77 A; 5C8M A; 1G8A A; 4D5K A; 4PPB A; 2ZV7 A; 3LXP A; 2QON A; 2P4I A; 3KUL B; 4K6Z A; 4CKJ A; 2E9O A; 2JDT A; 4BZN A; 2JKK A; 5E8U A; 3A4O X; 1YRP A; 3W2P A; 4D28 A; 4I4E A; 4F1O A; 3PXZ A; 3VHK A; 2OKR A; 1UNG A; 4KKG A; 5IA5 A; 2I40 A; 3GC7 A; 4IAD A; 1ST0 A; 5E8X A; 3W2R A; 4PX6 A; 4USE A; 3G6H A; 3QTX A; 5TKD A; 4KAO A; 4EC9 A; 4QMN A; 4JQ7 A; 2EUF B; 4CRL A; 1R78 A; 5HES A; 4ANM A; 1PXL A; 1Q3W A; 2B1P A; 4I5C A; 4G3G A; 1SZM A; 3VUH A; 4ENX A; 5IEX A; 2VTS A; 5JEB A; 3UIM A; 4G3F A; 3W8L A; 4ANS A; 1OIQ A; 4IFC A; 3I0Q A; 4I21 A; 4JG8 A; 2YJS A; 4KS7 A; 4E7W A; 4DLI A; 5F95 A; 4UYN A; 4YC6 A; 4WA9 A; 2VWX A; 4FIJ A; 4L46 A; 4PMS A; 3TIZ A; 3BLH A; 3TZM A; 4GK3 A; 2ZOQ A; 4AZE A; 3EFJ A; 5AUW A; 2OIQ A; 3RK5 A; 2OO8 X; 1U5R A; 3PP0 A; 3V5P A; 2GHM A; 4R1Y A; 3RK7 A; 5I9X A; 3WZD A; 4ZJV A; 1P14 A; 5EM6 A; 1YHW A; 2JBP A; 1YI6 A; 1OKZ A; 2REI A; 3R9O A; 2J0J A; 4K2R A; 1PXN A; 4AE6 A; 3Q2J A; 4EL9 A; 3DTC A; 3QCS A; 4TZR A; 5TOZ A; 3Q9Z A; 4RVT A; 1WBO A; 3ANQ A; 2GMX A; 2WTW A; 3HP5 A; 4GYI A; 3Q4Z A; 2DWB A; 2FSL X; 3A7F A; 2YDI A; 4F63 A; 3VUI A; 2C30 A; 4K18 A; 5ANO A; 1H00 A; 5HX6 A; 1UU8 A; 4LUD A; 5IUH A; 5KX8 A; 2Z7R A; 3CPC A; 4ACD A; 2WD1 A; 2W05 A; 2YAK A; 2ZV2 A; 4B6L A; 3VBV A; 3MFR A; 4BYI A; 4L3L A; 2XIZ A; 3FMM A; 1LP4 A; 4FKG A; 4GCJ A; 4DE4 A; 2Y9Q A; 3ZU7 A; 2QUR A; 4DC2 A; 5L2S A; 2VV9 A; 4FTR A; 4O27 B; 2XK8 A; 2VTR A; 3OEZ A; 5GJF A; 2C5O A; 4KD1 A; 4DGL C; 5HO8 A; 3RMF A; 2XB7 A; 3AC2 A; 4F64 A; 4AW1 A; 4E20 A; 3EQB A; 3UZP A; 3FLN C; 5JFX A; 1H0V A; 4GH2 A; 4MBI A; 3ZLS A; 2WOT A; 4FTN A; 5LMB A; 5CZI A; 2UZD A; 2CPK E; 2WPA A; 5F94 A; 2C6D A; 5HCX A; 2C3K A; 5AMN A; 2E9P A; 2VZ6 A; 4GSB A; 3S3I A; 3VS0 A; 2PVF A; 2VWW A; 2WMW A; 2OZA A; 3AD4 A; 3C1X A; 4EHZ A; 3OUK A; 2ZVA A; 5CEK A; 5DE2 A; 1OEC A; 5EW3 A; 5L6W L; 1W7H A; 1Y57 A; 1K9A A; 2HWO A; 4FT9 A; 4FTK A; 5FG8 A; 4F6U A; 4ZTR A; 4Z3V A; 3RVG A; 4IJ9 A; 3QQL A; 2V7O A; 5D9H A; 2BUJ A; 5LCK A; 3U55 A; 4FKO A; 3ORT A; 4E6A A; 4GMY A; 4HNF A; 5DBX A; 3VUM A; 4AXD A; 4RA4 A; 2PML X; 3I0O A; 4R6V A; 2WMU A; 3IDP A; 4KWP A; 2JLD A; 3EQC A; 4I20 A; 1JKL A; 2XNO A; 3MVL A; 4HYI A; 1URW A; 3FSF A; 4YSJ A; 4YNE A; 2UW9 A; 5JZN A; 1VZO A; 2WAJ A; 3PIY A; 3OT3 A; 4HVI A; 2FO0 A; 4JBP A; 4P5Z A; 4WNK A; 3R8M A; 1OKW A; 4XG3 A; 3GC9 A; 3NUP A; 3Q4T A; 4AA5 A; 4BBF A; 4DEG A; 5HQ0 A; 4IZ7 A; 3BYO A; 4IX6 A; 3ZM4 A; 2HCK A; 4IWQ A; 2PVR A; 1WAK A; 5AP6 A; 4GJ3 A; 2EXE A; 4FMQ A; 2A19 B; 2B9J A; 4RIO A; 1T45 A; 4WSY A; 3FHI A; 3LMG A; 4Z55 A; 2IW6 A; 4YJQ A; 2GU8 A; 5HG7 A; 1IEP A; 4H39 A; 3MBL A; 4FZF B; 2C6O A; 3BPR A; 1WZY A; 1LR4 A; 2RFS A; 2RFE A; 2BTS A; 3ENM A; 4W8D A; 5HIB A; 4IXP A; 4I6L A; 4I23 A; 3DJ5 A; 3R6X A; 5DEW A; 5AP3 A; 1VEB A; 1H1P A; 1YQJ A; 1H07 A; 4LL5 A; 5KHW A; 3D7T B; 3GFE A; 2SRC A; 4JNW A; 3QTZ A; 2W96 B; 5HVJ A; 4YO6 A; 4BCH A; 3D7Z A; 1JKK A; 2GNF A; 1G5S A; 3RPR A; 1OL6 A; 3V5W A; 3RI1 A; 2J50 A; 5HKM A; 5CAP A; 1W84 A; 2QHM A; 3KXM A; 4WSQ A; 3CD8 A; 4O96 A; 3B8Q A; 4QP7 A; 4FEV A; 2YM7 A; 4BDI A; 3LIJ A; 5ANJ A; 3PXQ A; 2J5E A; 5HE2 A; 3BLQ A; 2YIX A; 3KY2 A; 4Y95 A; 4G16 A; 5CSV A; 5I9V A; 4MXZ A; 1U59 A; 4NTS A; 4FIG A; 3GEN A; 2XNE A; 5IUG A; 3F5P A; 3FYJ X; 4ZTM A; 4RWJ A; 4QML A; 2BIK B; 4FZ6 A; 2OJG A; 2V0D A; 3D2K A; 4HCV A; 5CSH A; 4HGS A; 2UZE A; 3IOK A; 5J79 A; 3NS9 A; 4HNI A; 1M7Q A; 4U44 A; 4O21 A; 4AZF A; 1Q3D A; 2CNU A; 3ORI A; 2QU5 A; 3MES A; 2P2H A; 5GHV A; 5HE1 A; 3U6H A; 2YCR A; 4L6Q A; 4TL0 A; 2V7A A; 3KXX A; 4FE2 A; 2PUP A; 5BUI A; 3GU8 A; 3BWF A; 5D7A A; 1KOA A; 3BYU A; 4ZME A; 3NGA A; 5CT0 A; 3A7I A; 3II5 A; 5D7V A; 4BKY A; 4DFZ E; 4E93 A; 3MTL A; 3Q53 A; 4G2F A; 3KVW A; 4BHN A; 3WBL A; 2JGZ A; 2GS6 A; 3FLW A; 5FBN C; 2QOQ A; 4QMW A; 2ETK A; 3UIB A; 3G6G A; 3CXW A; 3HMM A; 5DR9 A; 3W8Q A; 3BU6 A; 3FAA A; 1QPD A; 2WMA A; 2OIB A; 5DT4 A; 3SXS A; 3KQ7 A; 4C2W A; 4CV8 A; 3TNI A; 4MVF A; 5IG1 A; 3TUB A; 1G8S A; 5DG5 A; 1JLU E; 2H96 A; 2BPM A; 2VWU A; 3O0G A; 4N6Z A; 3FLQ A; 3EQR A; 4WIH A; 3VNT A; 3HDN A; 4JDK A; 4E5B A; 3PG3 A; 4DFN A; 3C4X A; 3W55 A; 3QHW A; 4C38 A; 3HRB A; 4GFG A; 4D1S A; 2VO7 A; 3A7H A; 4FKU A; 3I1A A; 3W2C A; 4B7T A; 4J1R A; 4DTK A; 4EOS A; 4RIY B; 4F7S A; 4MWH A; 3MJ1 A; 3NUY A; 3KGA A; 2BZI B; 1UV5 A; 1CMK E; 4ORK A; 3QQH A; 3PE2 A; 5CF8 A; 4FV1 A; 3DND A; 2OLC A; 3WYY A; 3HZT A; 4AA0 A; 3T8O A; 3KMM A; 3ZC5 A; 4ZLZ A; 3CJG A; 3H0Y A; 4M14 A; 5D10 A; 4FV9 A; 4RRV A; 5FM3 A; 5A9U A; 2C4G A; 1ZYD A; 4XRL A; 4F9W A; 3T3V A; 3BGP A; 1TVO A; 4NEU A; 4KKH A; 4A7C A; 5BYY A; 2RKU A; 2CJM A; 3KN6 A; 5CT7 A; 1L3R E; 4MWI A; 3HAV A; 4BGG A; 4FA2 A; 2QO2 A; 4KKE A; 4R7I A; 4WHZ A; 3P86 A; 3QXP A; 2W4J A; 3W33 A; 2W5B A; 5EG3 A; 3OMV A; 3PJ8 A; 4RJ4 A; 4J7B A; 2PVJ A; 4M7I A; 2V62 A; 3DQW A; 3LE6 A; 4DGO A; 2FB8 A; 1R0P A; 4O86 A; 3SC1 A; 5M4C A; 4LGD A; 2O3P A; 3ZEP A; 4DHJ A; 3NVK I; 2WMX A; 3R63 A; 5HOR A; 3RAW A; 1KUT A; 2HZI A; 4C8B A; 4EUT A; 4HOK A; 4JQE A; 3QUD A; 4FV6 A; 4CFX A; 4DFX E; 3PPZ A; 3O17 A; 5GNK A; 3R0T A; 4JDI A; 4PTC A; 1GIJ A; 4ANL A; 4F09 A; 2O64 A; 2W1C A; 3VF9 A; 4CFU A; 4YJT A; 5CAN A; 3KB7 A; 2JIT A; 4TYH A; 4DBN A; 2CLQ A; 4RWI A; 3MY0 A; 2VWI A; 4QTC A; 1R1W A; 4FTJ A; 4AT3 A; 4UY9 A; 4WUN A; 4CLI A; 1Y91 A; 4RQK A; 2CGW A; 4HVD A; 5ETI A; 2GHG A; 2OU7 A; 2CMW A; 4O0Y A; 4TNB A; 4UZH A; 3C0I A; 2WTV A; 1VYW A; 3DTW A; 5DT0 A; 3VVH A; 4CMO A; 1TQM A; 3NW6 A; 4CT1 A; 3PA4 A; 1QCF A; 3COM A; 3PJ3 A; 3TG1 A; 3RNY A; 3AMA A; 4BCQ A; 4OAV B; 5IZF A; 1BL6 A; 4E26 A; 4EOQ A; 4FKQ A; 2YM4 A; 2JD5 A; 3TMO A; 4LRM A; 3PUF C; 3GCS A; 3DJ6 A; 3OXT A; 4EH3 A; 3U4W A; 4E5W A; 3S0O A; 2ZMD A; 5EH0 A; 5M56 A; 5AP5 A; 1A48 A; 4AOT A; 1DS5 A; 3R01 A; 4IEB A; 3ZSI A; 3DB6 A; 4EKK A; 2VO0 A; 4FSY A; 4FZD B; 4IVD A; 3BEG A; 3OEF X; 2IZS A; 3R1Q A; 2IW8 A; 4WG4 A; 3AC5 A; 5FTO A; 5HG5 A; 1OBG A; 2QO7 A; 4I6H A; 5EHY A; 4GFM A; 5BUJ A; 4ANB A; 2XBJ A; 3AXW A; 3ETA A; 2A1A B; 2X2L A; 1ATP E; 4FTM A; 4A9T A; 5L6P A; 3CE3 A; 4TT7 A; 2CNV A; 1BX6 A; 5M55 A; 1OI9 A; 2JFL A; 4FZA B; 5J95 A; 3TT0 A; 4D2W A; 2PK9 A; 2QG5 A; 2XKC A; 4HJO A; 5TO8 A; 5JKG A; 4CSV A; 4DT9 A; 3PYY A; 1OL7 A; 4DF3 A; 1Q4L A; 1XBA A; 4EC8 A; 2DS1 A; 3D5U A; 5BVD A; 3P5J C; 3GU5 A; 5DVR A; 2C0T A; 3HYH A; 4FKW A; 4HGL A; 4AGU A; 4E4L A; 4X7Q A; 1CDK A; 2PUN A; 2XAM A; 2R3Q A; 4EH8 A; 5J1V A; 2F9G A; 3MH0 A; 2IW9 A; 2PU8 A; 3U4U A; 2IWI A; 3OZ6 A; 3ZO2 A; 2XK7 A; 1Q24 A; 2QR8 A; 4HCU A; 3KRX A; 4L3J A; 3ORZ A; 4YP8 A; 5A4Q A; 3DK3 A; 3WF9 A; 3TZ9 A; 1YWV A; 3EFK A; 4ZTH A; 2X4Z A; 2PVN A; 3ORK A; 3IDC A; 4FV4 A; 5HID A; 3TTI A; 5IQG A; 5KBQ A; 1M52 A; 3DDQ A; 3KXZ A; 1W83 A; 3ZSH A; 4E3C A; 3IKA A; 3MIY A; 3H4J A; 5AIR A; 3C4W A; 3HA8 A; 4GT3 A; 2OXY A; 2FYS A; 3I5N A; 3NNV A; 4QMO A; 1OB3 A; 4G3C A; 3Q6W A; 3MJ2 A; 2ITN A; 5HHW A; 3IO7 A; 1ZOE A; 4DH1 A; 2YAA A; 3N9X A; 4IAK A; 1DI8 A; 3A99 A; 3QFV A; 2HYY A; 1OL5 A; 4C37 A; 3EH9 A; 4F9Y A; 4GIH A; 3RBW A; 3T9T A; 3CBL A; 4CEG A; 2YM8 A; 1YDT E; 2OK1 A; 4RJ3 A; 3UOD A; 4JQ8 A; 4PQN A; 4DAW A; 5JQ5 A; 2BDW A; 3FI4 A; 4H1J A; 4F9C A; 4O1P A; 3KL8 A; 1B38 A; 3K54 A; 4FV5 A; 5BVF A; 5KKR C; 3FMJ A; 2Y4P A; 3ZZE A; 3VAP A; 5HIC A; 3ML1 A; 4L42 A; 4H36 A; 4H3Q A; 1JAM A; 1ZAO A; 5ICP A; 1FOT A; 1GII A; 4IIR A; 5ANG A; 4ZXT A; 3QD2 B; 3PY1 A; 4FG8 A; 4JX7 A; 4FT5 A; 2P55 A; 5L6O A; 2XYU A; 5GZA A; 1PMV A; 2J0L A; 4YJV A; 4FEX A; 2J7T A; 4OT6 A; 5K4I A; 1NA7 A; 2OID A; 1H8F A; 4G3D A; 1Z5M A; 3E62 A; 4QPA A; 4TWC A; 4ANQ A; 5M4I A; 4JS8 A; 3PVB A; 2QNJ A; 4FR4 A; 1GZK A; 3Q6U A; 1B39 A; 4OBP A; 5CVH A; 4E6Q A; 5K0K A; 4DT8 A; 4DFY A; 1ZY5 A; 4ZZM A; 3IG7 A; 3LJ0 A; 4NIF A; 1ZMW A; 5HLW A; 1M2Q A; 4QTD A; 2YEX A; 1WVX A; 2XA4 A; 3IOP A; 3VBW A; 3GEQ A; 4AZW A; 1STC E; 2QO9 A; 4EOL A; 2X2K A; 1WVY A; 4EHE A; 1NVR A; 3VS2 A; 5C27 A; 2X8I A; 4AXC A; 3MIA A; 3OP5 A; 2X7F A; 3PJ1 A; 3TCP A; 4G5J A; 2C6K A; 4GFO A; 3EQH A; 3MB7 A; 4CT2 A; 4HGE A; 2QOK A; 3WOW A; 3DKG A; 4HZR A; 2ITP A; 2UVZ A; 3OBJ A; 4FKT A; 4N0S A; 3EQD A; 4PTG A; 3VF8 A; 3ZO3 A; 2QU6 A; 4GUB A; 4IQ6 A; 4YPD A; 4O0X A; 4EBV A; 2W06 A; 5AUX A; 3UIU A; 2WTC A; 1ZOH A; 2P33 A; 4ACC A; 4DEE A; 4Q9S A; 2RFD A; 2XZS A; 3BEL A; 4TXC A; 4RWL A; 1IR3 A; 4TYE A; 4G17 A; 3BL9 A; 4L8M A; 4OTR A; 2B52 A; 5JFW A; 2VTP A; 4XLV A; 1ZMV A; 5EYK A; 3M11 A; 3GOK A; 3R7O A; 2ZJW A; 3QF9 A; 3UIX A; 4D4V A; 1OKY A; 5BQ0 A; 4YZC A; 5H8E A; 4F0G A; 4IHP A; 3O96 A; 3ZOS A; 3EYG A; 2W17 A; 3NW5 A; 2HZ0 A; 3DU8 A; 1ZYL A; 3QRU A; 2J0M B; 2ZV8 A; 5CAO A; 3OCS A; 3HX4 A; 5EHL A; 3ZO1 A; 3I7C A; 4O0T A; 2A4L A; 4B99 A; 4BYJ A; 1YXX A; 3MDY A; 1K2P A; 2GHL A; 2BAL A; 2UW5 A; 4RZV A; 5HTB A; 4M69 A; 1ZLT A; 2QD9 A; 2UZW E; 3W32 A; 3FJQ E; 3PXR A; 4NST A; 3BE2 A; 3P0M A; 3TNW A; 3TJC A; 2EVA A; 5EK7 A; 5IH5 A; 5J9L A; 5FGK A; 1JBP E; 2R3L A; 2X1N A; 5CVF A; 5DIA A; 2R3K A; 5B7V A; 4AG8 A; 1IA8 A; 2J90 A; 2YCF A; 1MRU A; 3UOJ A; 2XM9 A; 3O50 A; 4BCF A; 1YWN A; 3COK A; 4XG6 A; 4FKI A; 1XBB A; 4IVA A; 2PZI A; 5CAQ A; 3MVH A; 4O7W A; 4IB1 A; 1H28 A; 4FTQ A; 4QPM A; 4XV2 A; 4XH6 A; 1Y8G A; 3QTR A; 4F9B A; 4MQ2 A; 3QGY A; 3U6I A; 1LPU A; 5ETF A; 1MUO A; 3ACJ A; 3LFN A; 1SM2 A; 4PNK A; 2I1M A; 3PPJ A; 3HUC A; 4DDG A; 4L00 A; 4ZZO A; 2VN9 A; 3QXO A; 5EQE A; 3UDS A; 3NDM A; 4HW7 A; 3FXW A; 3DBQ A; 5HBE A; 1KSW A; 2YIR A; 4KSQ A; 5FBO A; 4EON A; 1CM8 A; 1OVE A; 1GOL A; 4QDV A; 3FXX A; 3KFA A; 2C3L A; 2DQ7 X; 1HCL A; 1P4F A; 3VRZ A; 1JOW B; 4XBU A; 4D4S A; 2VGP A; 1VJY A; 3Q5I A; 4FOB A; 3VUD A; 4FST A; 5EM5 A; 3UP7 A; 5IDP A; 4EH5 A; 5KZI A; 5C4K A; 4FUX A; 4PP9 A; 1ZY4 A; 2Z7L A; 4JL9 A; 2CDZ A; 4E4X A; 2PZ5 A; 3EL7 A; 4IVC A; 3ALO A; 3ZRK A; 3BRB A; 5C9C A; 3PLA E; 3TIY A; 4CLJ A; 3NUU A; 2EU9 A; 3Q60 A; 5FEQ A; 1CSN A; 3MS9 A; 3VW8 A; 4IAN A; 3RM7 A; 3UDB A; 4ACM A; 5A46 A; 4H1M A; 1Q5K A; 3KRL A; 3W18 A; 4PDS A; 2OJF E; 2VO3 A; 3CJF A; 4G34 A; 3DB8 A; 4EBW A; 3PIX A; 5DRD A; 2XK4 A; 5J1W A; 3ZDI A; 2BAJ A; 4O6L A; 4QP6 A; 2Q83 A; 4MTA A; 4RGJ A; 5CEN A; 1MP8 A; 4G6O A; 5DEY A; 5BX0 A; 1OIY A; 5E8Z A; 4UN0 C; 5IMX A; 1JST A; 1CKI A; 5DGZ A; 2HOG A; 1ZWS A; 2WMT A; 3P7B A; 3ZXT A; 4ITI A; 4M0Y A; 3MH1 A; 3AC3 A; 3EZV A; 1SMH A; 3H30 A; 4GU9 A; 3I4B A; 2X4F A; 3H9O A; 2OZA B; 4BL1 A; 2QLU A; 3GCP A; 1SVG A; 2XAO A; 5E8V A; 4C3F A; 4TN6 A; 5H9B A; 1V1K A; 1T46 A; 3GI3 A; 3QKK A; 2WEI A; 4V01 A; 3RK9 A; 2OG8 A; 5J7H A; 3SOC A; 1OL1 A; 1ZRZ A; 2X8D A; 3FI3 A; 3AT3 A; 4NM0 A; 4FSM A; 4O2P A; 5CEP A; 2IO6 A; 5HIE A; 3WAR A; 5IKW A; 3NZ0 A; 3R9N A; 2NYA A; 3DQX A; 3DPK A; 5DA3 A; 4FJV A; 5ANE A; 2YIS A; 2EXM A; 4O6E A; 3NNU A; 3QCX A; 3MPA A; 4FTO A; 5CTP A; 1YXT A; 3L9M A; 2I0E A; 5J5T A; 5F20 A; 2IZU A; 4XJ0 A; 3DOG A; 4JVG A; 4K33 A; 3NUA A; 2RIO A; 4F6S A; 3I79 A; 3QHR A; 5IQI A; 4ALV A; 2VWV A; 2HEL A; 3R71 A; 4AZT A; 2Z7Q A; 4RJ7 A; 4BIB A; 1WMK A; 4XG8 A; 3RAI A; 4LV5 A; 1BI7 A; 2UW6 A; 3LQ8 A; 4JBV A; 3HEC A; 2WQM A; 3OTV A; 3OY1 A; 3D5V A; 3TZ8 A; 5JMS A; 1M2P A; 4JR7 A; 3E93 A; 3OUN B; 5HLP A; 3VN9 A; 3LCS A; 2WKM A; 3KA0 A; 3UG1 A; 3EN4 A; 3E63 A; 3VW6 A; 2J5F A; 3DBC A; 1YI4 A; 3KCF A; 3GNI B; 4TYJ A; 4S31 A; 5FXQ A; 2ZDT A; 5K3Y A; 3FBV A; 3TV7 A; 4NM7 A; 3FYK X; 1O9U A; 4QT1 A; 4Z7H A; 2C6L A; 4U43 A; 3E8D A; 4QMU A; 5FXS A; 3QTU A; 3ATS A; 3BLA A; 4CG8 A; 4BCM A; 5JT2 A; 4PP7 A; 3PTG A; 2H34 A; 3H0Z A; 1YWR A; 3HNG A; 2O5K A; 2ZB1 A; 4BKJ A; 5AR2 A; 4C02 A; 1M7N A; 2W7X A; 2ITV A; 3C0H A; 4BJJ B; 5I9U A; 2G15 A; 1FVR A; 3RHK A; 4YZN A; 1IG1 A; 3PSC A; 4MXY A; 4E6C A; 4JDJ A; 3AOX A; 3G9N A; 4YC8 A; 5CF4 A; 2QI8 A; 2OBJ A; 2XAN A; 3QQU A; 3GUB A; 5K9I A; 4FTU A; 4QYG A; 5CSP A; 5LVN A; 3QTW A; 5C01 A; 4BDJ A; 2PHK A; 4J52 A; 3G15 A; 3HUB A; 3RWQ A; 5GIP E; 4RVL A; 3IS5 A; 4L01 A; 1GZN A; 2AC3 A; 3CTH A; 3LCO A; 2FGI A; 3GQI A; 5FXR A; 4R7B A; 3M2W A; 1YVJ A; 1P2A A; 4FSZ A; 4GG7 A; 4F7J A; 1UNL A; 2R3F A; 4PL5 A; 4CGA A; 5DR6 A; 4ZY5 A; 3ZON A; 4D0X A; 4AE9 A; 4QMX A; 1ND4 A; 4JAI A; 4LV7 A; 3MA6 A; 1H25 A; 3HV4 A; 3PA5 A; 4NT4 A; 3TWJ A; 4UMP A; 4ASD A; 5A4E A; 5JRQ A; 2VTL A; 2VWB A; 1Q62 A; 2J4Z A; 3ROY A; 4MH7 A; 4FIE A; 5DH3 A; 1REJ A; 4OTH A; 4U3Y A; 3NYX A; 3ZUU A; 5KMM A; 4O0R A; 4QPS A; 4JOA A; 2YA9 A; 2VTO A; 5EW9 A; 4I93 A; 4OGR A; 3H3C A; 3U6J A; 1OGY A; 5AR8 A; 2BR1 A; 3L8S A; 5AP2 A; 4AGD A; 3GT8 A; 2WHB A; 1NXK A; 2XYN A; 4JR3 A; 4LRJ A; 4EYM A; 3UO6 A; 2ITZ A; 4J95 A; 5KHX A; 5J9Y A; 5DPV A; 3HAM A; 4G9C A; 3QLF A; 2O63 A; 4R1V A; 4EQM A; 2WMB A; 3CR0 A; 3DY7 A; 3VHE A; 2O0U A; 3PE1 A; 3D2I A; 3W0P A; 4F78 A; 2BZH B; 4PL3 A; 3WF5 A; 5C8N A; 5HD4 A; 5HBJ A; 5CAS A; 2GPH A; 4MBJ A; 5CNO A; 5KZ0 A; 4FV0 A; 4QP9 A; 3OW3 A; 2C0O A; 4YZB A; 4BY9 E; 4C2V A; 3LMH A; 3F7Z A; 3ZSG A; 2UZL A; 2ZB0 A; 1CKP A; 5CY3 A; 4CV9 A; 4BBM A; 2CGX A; 1G3N A; 5HU9 A; 4O7R A; 3UVP A; 3AC1 A; 2JIU A; 2YIQ A; 3AD5 A; 4ZEG A; 4OW8 A; 2WZJ A; 4CFF A; 4YUR A; 2PYW A; 3N4U A; 5IH4 A; 3ELJ A; 4GU6 A; 1XBC A; 4EJ7 A; 3GFW A; 3PSB A; 1QPE A; 4Y73 A; 4UMR A; 3IK3 A; 2PMI A; 4I4F A; 5BYZ A; 4FKJ A; 4LQM A; 4D9U A; 3FSK A; 4V0G A; 3ULZ A; 4FYO A; 3FUP A; 1MRV A; 4KIK A; 1H1W A; 4APP A; 2XCK A; 4ANN A; 2ITU A; 4LM5 A; 4ALU A; 5DN3 A; 1E9H A; 4YHF A; 4CNH A; 2W5A A; 4L67 A; 4F99 A; 3BHH A; 4NJ3 A; 2XIK A; 3NW7 A; 5E7R A; 3WZK A; 4AW5 A; 2ZFY A; 4FNZ A; 4F1T A; 3NPC A; 2XKE A; 2XIY A; 4TRL A; 4JX3 A; 4FL3 A; 4F6W A; 4C33 A; 2OH0 E; 5LOH A; 3C0G A; 4PWN A; 2WTK C; 2IPX A; 4XP3 A; 3S2P A; 1KE8 A; 3MWU A; 2XVD A; 3HKO A; 3I81 A; 5E92 A; 4RVM A; 5I9Z A; 3LXK A; 3OF0 A; 1J1C A; 5HO6 A; 3UDT A; 2BFY A; 2W1E A; 4WRG A; 4R3C A; 1XH6 A; 2O2U A; 3FQH A; 3C5U A; 5HU3 A; 2JKQ A; 2QOO A; 4YJR A; 2WO6 A; 2FSO X; 2YCQ A; 2YAB A; 4LUE A; 4EQU A; 1GIH A; 1O6Y A; 2PUU A; 3QAM E; 4AAA A; 3UOK A; 3BU3 A; 2G2F A; 4D1X A; 3U54 A; 2OW3 A; 2BZK B; 5F9E A; 4QRC A; 4QYH A; 2X9F A; 3D94 A; 4IAI A; 4EEV A; 2XM8 A; 4FV2 A; 4IM3 A; 2CLX A; 1JPA A; 1PXM A; 2NRY A; 3BU5 A; 3ZH8 A; 2XJ1 A; 3V5T A; 3BYV A; 4D1Z A; 1UWH A; 4HYU A; 4ASX A; 2UW0 A; 4C35 A; 2VU3 A; 3QYZ A; 5K4J A; 2OJ9 A; 4P2K A; 1M17 A; 4A4L A; 5E4H A; 3EYH A; 4DH5 A; 3SXF A; 4UXQ A; 4C3P A; 4AF3 A; 4M0Z A; 2VWY A; 3FEG A; 2FST X; 4XG2 A; 2IZT A; 1ERK A; 2OFU A; 3HMN A; 4TPT A; 3VRY A; 2UUE A; 1XMM A; 2AUH A; 4AGW A; 4EZ7 A; 4CZU A; 3SRV A; 2XNN A; 4CCB A; 4FEW A; 4BHZ A; 3FL5 A; 3MFU A; 4FF8 A; 5GZ8 A; 1QMZ A; 4FI1 A; 3LM0 A; 3R8Z A; 3WZU A; 4BCJ A; 4CZT A; 2BHE A; 3BQC A; 4BF2 A; 1KWP A; 4MF0 A; 3C50 A; 3OT8 A; 4WRS A; 4ID7 A; 1H0W A; 4ZSA A; 3C7Q A; 5AJQ A; 2H9V A; 4FX3 A; 4FIH A; 4YPS A; 5BNJ A; 5DT3 A; 4O7L A; 3MYG A; 2OZO A; 4M68 A; 3CQU A; 4BCG A; 1JQH A; 4XS2 A; 4BCK A; 3SG9 A; 4TWN A; 3QTI A; 3BWJ A; 3NYN A; 4WKQ A; 3GU6 A; 2PTK A; 3JVR A; 2F7Z E; 2IG7 A; 4Y85 A; 4YC3 A; 4EUU A; 3PZH A; 4IVB A; 5L8L A; 5KMI A; 3UNZ A; 4GW8 A; 4KIN A; 3VJO A; 1OPL A; 3LFS A; 2VUW A; 4YJS A; 4C3R A; 3G2F A; 1OIT A; 1PMN A; 1Z57 A; 4CQG A; 3R1N A; 4K8A A; 1H27 A; 2ITY A; 3E3P A; 3LZB A; 4FK6 A; 1YHS A; 3R7U A; 4E6D A; 4F0I A; 5FWK K; 4X6R A; 4YSM A; 1MQB A; 2ZDU A; 3R7E A; 3UG2 A; 1UU3 A; 3QD4 A; 5DFP A; 4B8L A; 4CFV A; 1PYX A; 4I24 A; 3FMH A; 4D2P A; 4E8A A; 2W99 B; 4ACH A; 5H8G A; 2WTJ A; 4EK5 A; 5M51 A; 4V04 A; 3S00 A; 2PE2 A; 4WNO A; 4EOK A; 4MUQ A; 3W1F A; 1LEW A; 4NK9 A; 3R22 A; 1OBD A; 3R8L A; 4IWD A; 3LLA A; 2E9N A; 4L7S A; 3LOK A; 3QU0 A; 5IQB A; 3R73 A; 5AV4 A; 3P79 A; 3VUG A; 5CLP A; 3ULI A; 4PDP A; 3TV4 A; 3OY3 A; 5LJJ A; 3Q4U A; 3FZ1 A; 4V05 A; 1XQZ A; 3GGF A; 5L4Q A; 4FT3 A; 5FD2 A; 5CAU A; 2VD5 A; 2HZN A; 3R83 A; 2XK9 A; 2CCH A; 2R64 A; 4OBQ A; 1OGU A; 4BGQ A; 1NVS A; 2VTN A; 3OXZ A; 5H2U A; 1M14 A; 4UBA A; 1OPK A; 4QQJ A; 3P5K A; 5HO7 A; 3FLY A; 4O7Y A; 3BYS A; 5HNI X; 1PY5 A; 2BRN A; 5C03 A; 5AEP A; 3NYV A; 4LMU A; 5KO1 A; 3UVR A; 4D2V A; 4ZSE A; 2QKW B; 3G90 X; 4JDH A; 3DFA A; 3V5Q A; 2BZJ A; 2R3I A; 5AV0 A; 4JBO A; 3LMI A; 1WBT A; 2RG5 A; 2PVL A; 4PPC A; 2VTM A; 5AAG A; 3L8P A; 4PV0 A; 4GRB A; 5ACK A; 4QO9 A; 4QP1 A; 3TEI A; 3CPB A; 3E8E A; 4R77 A; 2BAQ A; 3FKN A; 2QKR A; 3D83 A; 4EK6 A; 2PSQ A; 3KN5 A; 4AXA A; 3C9W A; 3FZT A; 4FT0 A; 3T3U A; 4D2R A; 3KRW A; 4WO5 A; 3F3T A; 2WOU A; 3NSZ A; 4K0Y A; 5I0B A; 4IAY A; 2I0V A; 4F1M A; 4PPA A; 3DXQ A; 1DI9 A; 4RJ8 A; 4CRS A; 4UWY A; 1G8J A; 3UYT A; 4RG0 A; 4QTA A; 3WI6 A; 2JED A; 2J9M A; 5IH6 A; 4C58 A; 3VQU A; 3UBD A; 4MHA A; 4F65 A; 3A2C A; 4O7S A; 4RBL A; 2QCS A; 3VS6 A; 3A62 A; 4AUA A; 2HWP A; 4YZD A; 3V5J A; 4HVS A; 4J8N A; 4FII A; 4RSS A; 2Y6M A; 3MPM A; 2GNG A; 2W1G A; 4U81 A; 2ITX A; 3VS7 A; 5I9Y A; 5M57 A; 5EDR A; 5EKN A; 3A7J A; 1YDS E; 3VID A; 5IA4 A; 4TW9 A; 2XCH A; 1O6K A; 4MX9 A; 4W4V A; 5M53 A; 4WNP A; 1P5E A; 3NVM B; 3UNK A; 3VBX A; 4IE9 A; 1BL7 A; 4W8E A; 2C3J A; 3BCE A; 2WEV A; 1TFF A; 1PRY A; 2H8H A; 1Q8Y A; 4LV8 A; 3UC3 A; 4M69 B; 1O6L A; 2IVU A; 5HTI A; 3TDW A; 3TKU A; 4GKH A; 4CFM A; 4OTF A; 4FEQ A; 4KSP A; 2A2A A; 1URC A; 2HIW A; 4IM2 A; 1H01 A; 4FTC A; 4LOP A; 5F4N A; 4EH6 A; 4OT5 A; 2CGV A; 3FMK A; 4TWO A; 4DBX A; 1IRK A; 3MH3 A; 3KEX A; 3NNX A; 3WZJ A; 3IGG A; 4DYM A; 2NO3 A; 4BI0 A; 2X7O A; 4MNF A; 3ZC6 A; 3F82 A; 2X9E A; 3G33 A; 1W98 A; 3COH A; 4IZ5 A; 3LQ3 A; 1RQQ A; 4HZS A; 4LYN A; 4I41 A; 3FI2 A; 1UU9 A; 4YTC A; 4NYE A; 1OKV A; 5AR7 A; 3LCT A; 4MNE A; 5HE0 A; 4UXL A; 4IB0 A; 2CKP A; 5AR4 A; 3LCD A; 5AA9 A; 4FVQ A; 3FL4 A; 4I3Z A; 2QR7 A; 3JY9 A; 4AQK A; 3FKO A; 1ST4 A; 3LJ1 A; 2R2P A; 3KXH A; 5T3Q A; 4AXF A; 2YZA A; 1WBS A; 3TI1 A; 3UQC A; 5DOS A; 3DKF A; 3IW4 A; 3A8X A; 4G6L A; 3ZBF A; 5CXZ A; 4HCT A; 3LCK A; 4QEB A; 3D7T A; 1DM2 A; 4X2F A; 2R0I A; 4FKL A; 4CYJ A; 3GP0 A; 3CY3 A; 2BRM A; 2OIC A; 3GCU A; 3TN8 A; 3WZE A; 5LVP A; 3ORO A; 4N70 A; 2GS7 A; 2GTM A; 3E88 A; 3JVS A; 5HVY A; 4K9Y A; 4EK4 A; 4O38 A; 5LVO A; 3AMY A; 1Q8U A; 3MTF A; 2W1H A; 3W2S A; 3DK7 A; 1SYK A; 2WB8 A; 4AN9 A; 2IJM A; 3EOC A; 3ITZ A; 3PY3 A; 5DWR A; 4TYI A; 1H4L A; 3IQ7 A; 4XOZ A; 5LXM A; 3MW1 A; 3WF6 A; 1SNX A; 3DAE A; 3KKV A; 5FED A; 4XRJ A; 4ZZN A; 3NMU F; 3ATT A; 3ZEW A; 2VO6 A; 4EH9 A; 1XH9 A; 3M42 A; 4YLJ A; 5HNB A; 4O81 A; 1AD5 A; 4F7N A; 3MI9 A; 2JC6 A; 4CYI A; 2HEN A; 3O8U A; 3UYS A; 3HDM A; 3TNH A; 3MY1 A; 4AW0 A; 5M4U A; 2UZN A; 2W1I A; 1E1X A; 5JRS A; 2UZU E; 3HMP A; 5M06 A; 5F1Z A; 4QMS A; 4I1Z A; 4FKS A; 3RPS A; 4OAU C; 3GQL A; 2YM6 A; 3BYM A; 5C4L A; 4XOY A; 3QTQ A; 3RZB A; 1Q97 A; 4DIN A; 4FSU A; 1YOM A; 5BUE A; 4DFB A; 3OOG A; 3DXN A; 5CYI A; 3ION A; 4O91 A; 4S30 A; 3C13 A; 4RQV A; 2ITT A; 2FUM A; 3PP1 A; 2BVA A; 5FQD C; 3LQ5 A; 4DGG A; 5EDP A; 5ETA A; 2BRO A; 4FV3 A; 4AFE A; 3I7B A; 3VWA A; 1OIU A; 3VUT A; 3FLS A; 2XK6 A; 3BV3 A; 5CAL A; 4FSW A; 4YBK A; 2BKK A; 2RG6 A; 5DI1 A; 2F7E E; 2XRU A; 2XF0 A; 4EHV A; 5EM8 A; 3ACK A; 4R3R A; 4BWP A; 4YUQ A; 3RGF A; 5TR6 A; 4E4M A; 3OC1 A; 4OR5 A; 4YHT A; 1QL6 A; 4Y72 A; 2Y6O A; 4EOP A; 3C4C A; 1CKJ A; 4J98 A; 2XH5 A; 2VTJ A; 3VO3 A; 3FWQ A; 4O0S A; 3AT4 A; 3K2L A; 4L68 A; 4OTQ A; 4BZD A; 5CU2 A; 1UA2 A; 1R39 A; 3RJC A; 4DFL A; 5KBR A; 4RIX B; 4ZIM A; 1Z9X A; 3FKL A; 4KBC A; 5JGB A; 4YJU A; 4AP7 A; 4EOR A; 5LAR A; 3I6U A; 1Q8W A; 3ORM A; 3KVX A; 4ZP5 A; 3BHV A; 4X21 A; 3GU4 A; 1XH4 A; 4A07 A; 4BDE A; 4BDH A; 4YTF A; 5AUT A; 5JGD A; 1LUF A; 4FNX A; 2E2B A; 5AM6 A; 4C4E A; 4RIY A; 3DT1 A; 4MXO A; 1TKI A; 2WQO A; 3VON A; 2ZV9 A; 4L52 A; 4IBM A; 2PMN X; 4D55 A; 3BQR A; 3QA0 A; 2PPQ A; 2XNB A; 2R0U A; 2IN6 A; 5I9W A; 3ZM9 A; 3U9C A; 2R3R A; 4C59 A; 4KUJ A; 4MXC A; 3KC3 A; 5EYM A; 3ZS5 A; 4IX4 A; 2B0Q A; 2HK5 A; 4JIA A; 2Y0A A; 4U42 A; 2CKQ A; 1U54 B; 3BX5 A; 5K0X A; 3OD6 X; 2JIV A; 2J0I A; 2XMY A; 1GJO A; 1WBW A; 2G01 A; 3UPX A; 3E87 A; 3JYA A; 1DAY A; 4O0V A; 3O51 A; 4R8Q A; 3S4Q A; 4I6Q A; 2JDV A; 4P4C A; 4RC2 A; 4O7V A; 1YXV A; 3LKM A; 3P1A A; 4QP8 A; 5AUZ A; 2AYP A; 3EQP A; 4YHJ A; 3QZI A; 3A60 A; 3AGL A; 4KBK A; 3HGK A; 5JQ8 A; 2IVT A; 3WYX A; 3QC9 A; 1QPJ A; 2R7I A; 4XV1 A; 4CMU A; 2WMQ A; 5BMS A; 3CTJ A; 4UEU A; 4AW2 A; 5E9E A; 1DAW A; 3ZTX A; 3C5I A; 3FY2 A; 5FTG A; 1NY3 A; 2UVY A; 2ZAZ A; 1RE8 A; 3EN7 A; 4DEB A; 3MN3 A; 3QA8 A; 3KF7 A; 2W9F B; 3TL8 A; 3W2O A; 4ZJI A; 4L45 A; 4IAZ A; 4IX3 A; 5L8J A; 3EZR A; 4BRX A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...