The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
70
|
sequence length |
326
|
structure length |
326
|
Chain Sequence |
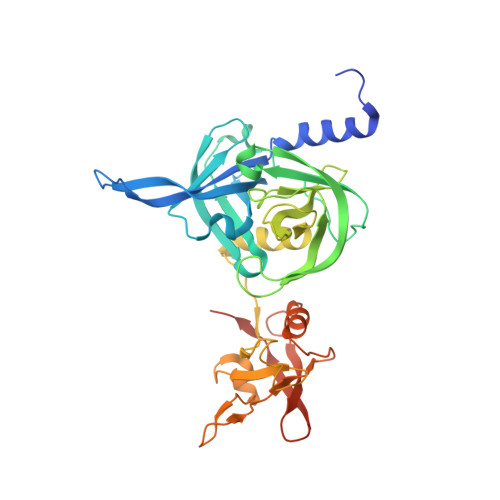
PKKLQTDELATVRLFQENTPSVVYITNLAVRQDAFTLDVLEVPQGSGSGFVWDKQGHIVTNYHVIRGASDLRVTLADQTTFDAKVVGFDQDKDVAVLRIDAPKNKLRPIPVGVSADLLVGQKVFAIGNPFGLDHTLTTGVISGLRREISSAATGRPIQDVIQTDAAINPGNSGGPLLDSSGTLIGINTAIYSPSGASSGVGFSIPVDTVGGIVDQLVRFGKVTRPILGIKFAPDQSVEQLGVSGVLVLDAPPSGPAGKAGLQSTKRDGYGRLVLGDIITSVNGTKVSNGSDLYRILDQCKVGDEVTVEVLRGDHKEKISVTLEPKP
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| molecule tags |
Photosynthesis
|
| molecule keywords |
Protease Do-like 1, chloroplastic
|
| publication title |
Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure.
pubmed doi rcsb |
| source organism |
Arabidopsis thaliana
|
| total genus |
70
|
| structure length |
326
|
| sequence length |
326
|
| chains with identical sequence |
B, C
|
| ec nomenclature |
ec
3.4.21.-: |
| pdb deposition date | 2011-02-09 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF13180 | PDZ_2 | PDZ domain |
| A | PF13365 | Trypsin_2 | Trypsin-like peptidase domain |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Beta | Roll | Pdz3 Domain | Pdz3 Domain | ||
| Mainly Beta | Beta Barrel | Thrombin, subunit H | Trypsin-like serine proteases | ||
| Mainly Beta | Beta Barrel | Thrombin, subunit H | Trypsin-like serine proteases |
#chains in the Genus database with same CATH superfamily 1F5L A; 3BPU A; 1K1L A; 4JOH A; 1ETS H; 2IN2 A; 1HLT H; 1T8N A; 1GBA A; 4F8K A; 2BHG A; 2D8I A; 2Z9I A; 1XUF A; 1YM0 A; 1DLE A; 4J1Y A; 2DE8 A; 1OWE A; 3ZVG A; 3RMO H; 5FCK A; 1ABJ H; 1FI8 A; 4OS6 A; 1N6F A; 2Z17 A; 4WF8 A; 5DP9 A; 5HFD A; 3BG8 A; 3LGY A; 2A4Q A; 1XUK A; 2PNT A; 5E0G A; 5G1D A; 2JH5 D; 3SNA A; 1Y8T A; 1LHC H; 3A8C A; 1P2J A; 2P59 A; 2A31 A; 4NCY A; 3LJJ A; 3GDV A; 3EA8 A; 1TX7 A; 3RU4 E; 4D9R A; 2UWL A; 1AMH A; 1E38 B; 2ZP0 H; 5C5N A; 3LNY A; 1CT2 E; 3VQF A; 2V90 A; 5FBE A; 2ANW A; 2F91 A; 2K1Q A; 3ZZD A; 1C4Y 2; 1ZRK A; 2REY A; 2CF9 H; 2ODY B; 3MH4 A; 3EGG C; 5A2P A; 2MHO A; 5FCR A; 1O3G A; 1UTM A; 2FTM A; 2FOA A; 1W9Q A; 5EU8 A; 1E37 B; 4NIY A; 2BD3 A; 5G1E A; 4Q6H A; 2UUK B; 1QRZ A; 1GI9 B; 1QL9 A; 3ZZ8 A; 3BEU A; 3PYH A; 2HPP H; 1N6X A; 2D2D A; 2DAZ A; 2ZFR H; 3P92 A; 1KWA A; 1ZOK A; 4FU7 A; 5A2M H; 4Z8J A; 3ELA H; 3DIW A; 4Y71 A; 3AAU A; 6GCH G; 4OS5 A; 1QIX B; 1AND A; 1P2Q A; 4TPI Z; 4MPA A; 1Z6E A; 2ZTY A; 4LZ1 H; 1C1V H; 1B7X B; 4ZHA A; 3ID3 A; 1NRR H; 1S81 A; 5BRR E; 1SB1 H; 4MNW A; 2RG3 A; 3QX5 H; 4E06 H; 1O2R A; 4YOI A; 2YNB A; 2J9N A; 2WPM S; 1KXA A; 4O3U A; 3BV9 B; 1UVO A; 1JIR A; 2AFQ B; 1C5N H; 1UJV A; 1U39 A; 5JBB S; 1W2K H; 2C90 B; 3EA9 A; 3O8R A; 3BTH E; 5T3H A; 2AIN A; 1GCT C; 2OIN A; 4CRC A; 1RTF B; 1OBX A; 1GI1 A; 2JET C; 1HBT H; 2QF3 A; 3SHU A; 3LPR A; 1CA8 B; 1ZHP A; 3OY6 U; 1TLD A; 2W4F A; 3O5N A; 2LC6 A; 1H8D H; 3RXC A; 2XC5 A; 1BRB E; 4GVU A; 4K5Z A; 2Q6F A; 1ELE E; 1SGN E; 3HPM A; 1BRC E; 1SGF G; 3VGC B; 1O5C B; 1W9E A; 1AN1 E; 1O3C A; 3P70 B; 1KLT A; 3ATI A; 2LOB A; 3A7X A; 3NKK A; 1EP6 A; 3SU4 A; 1QAU A; 2BD7 A; 2I0L A; 2XNI A; 1CE5 A; 1PFX C; 5HEB A; 2ODQ A; 1BOQ A; 4K1E A; 1LVO A; 1LHF H; 4B75 A; 2KOH A; 1GBD A; 3AXA A; 3GY7 A; 3NCL A; 1V62 A; 5HET A; 4MPV A; 4HGC A; 2F9N A; 3ODF A; 1QRX A; 4NNM A; 1TP3 A; 1T2M A; 1L1J A; 1KDQ A; 2HNT E; 3RL8 A; 2QXJ A; 3K65 B; 3U9A H; 1V2M T; 2E7K A; 1JIM A; 3BTE E; 4ZAE A; 3UFA A; 2FTL E; 1MKW H; 4WYU A; 2ZC9 H; 2B9L A; 1MZA A; 3IG6 B; 3RXF A; 1OOK B; 1MKX K; 1GJD B; 2EAQ A; 2V1W A; 2R3U A; 3A88 A; 4Y0Z E; 2GD4 B; 1WF7 A; 2VIV A; 2Z3E A; 4GCH F; 1V2O T; 1UEQ A; 3EAJ A; 2GGV B; 3A7V A; 5TCC A; 4CRG A; 1XVO A; 3ZVF A; 1YBW A; 4L8N A; 4HZH A; 3B23 B; 2KL1 A; 3A7T A; 2FOB A; 2R2W U; 2ZU5 A; 2VWL A; 4IC6 A; 4XDE A; 1BML A; 1BTZ A; 1UCY E; 4ZKS U; 1HIA A; 2IWO A; 1GBI A; 2B8O H; 1H1B A; 2Z9L A; 4H4F A; 1AD8 H; 1W7G H; 1V2P T; 1L6O A; 3RL7 A; 2XXL A; 2QYI A; 2BDY A; 5HGG A; 3ZYE A; 1G3D A; 2BVX H; 1NRN H; 2A4G A; 2I0I A; 3SV2 H; 4X2Y A; 3RML H; 1GPZ A; 3PV4 A; 1IOE A; 1QAV A; 1QNJ A; 4PQW A; 1NT1 A; 1C1U H; 4I31 A; 1F5K U; 1GHW H; 2OQS A; 1GJ7 B; 1GJC B; 1TPO A; 2HAL A; 2ZU1 A; 3LGI A; 4TWY A; 4WYT A; 1A4W H; 1LVY A; 1DSU A; 3MHW U; 3VGC C; 2FI4 E; 1RTL A; 4OS4 A; 1UJ1 A; 1O33 A; 2OGP A; 2P16 A; 5D13 A; 4X2V A; 3S9A A; 4BNR A; 2VSV A; 4NXQ A; 1FIW A; 3MNB A; 3TJV A; 2Z9J A; 3QGJ A; 1TYN A; 5E1Y A; 1EB1 H; 4IC5 A; 4WMD A; 1B0F A; 4P0C A; 1ZPC A; 1XXD A; 1TNJ A; 1AKS A; 1FC6 A; 1BRA A; 1CA0 B; 1MTW A; 1MD7 A; 1HDT H; 5LPE A; 5GCH F; 2F9V A; 2DLS A; 3MU5 A; 1GBJ A; 3UWJ H; 1ZSJ A; 2BVR H; 3F9F A; 5EXN A; 3I78 A; 1S6F A; 3UY9 A; 1UJD A; 2DJT A; 1WOF A; 1GGD C; 1UEZ A; 1C5Y B; 2BDH A; 1F0U A; 1MFG A; 4LMM A; 1X5N A; 1PQ7 A; 8KME 2; 1UTL M; 3GOV B; 2FX4 A; 2EEI A; 2FES H; 2ZU3 A; 2ZDV H; 4ASH A; 7EST E; 1SHH B; 4O3T A; 3M7T A; 4D9Q A; 5EXM A; 3DJA A; 1GBK A; 2AGG X; 4X1Q U; 3AW0 A; 2A7J A; 3KID U; 3TH2 H; 3TPI Z; 2PGB B; 3GY4 A; 4B71 A; 2ZGB H; 4UU5 A; 1WJL A; 3BTG E; 1ZTJ A; 3ZZ7 A; 1C5M D; 4ZKO U; 1Q2W A; 2R2M B; 4FUJ A; 3ZYD A; 2PA1 A; 4Z0K A; 1QQ4 A; 5F3X A; 3QZR A; 1BA8 B; 7LPR A; 1TP5 A; 2AER H; 3NPS A; 1VAE A; 1MMJ N; 2ZQ2 A; 2OCV B; 3RXV A; 1HIA B; 4XE4 A; 2LC7 A; 1CQQ A; 3ZZA A; 4DCD A; 8GCH F; 3HKJ B; 4WY3 A; 1GCD A; 1GHZ A; 3A8A A; 4P2A B; 3RDZ A; 1D9I A; 3GIS B; 1V2N T; 4X8V H; 1NU9 A; 1CSO E; 1PPH E; 3RXK A; 2H1U A; 3V0X A; 4DY7 B; 3DHK H; 1WCZ A; 2CV3 A; 4BTU B; 4FUD A; 2VNT A; 1T31 A; 2P93 A; 4IMQ A; 3L33 A; 2OQ5 A; 3RC5 A; 2WYG A; 3CBX A; 1HAO H; 1TE0 A; 3JZ1 B; 4ABE A; 1Q3P A; 2H6M A; 5TJX A; 5EOD A; 1SGI B; 1LPK B; 4H42 U; 3QTV H; 3V3M A; 3UNQ A; 1QY6 A; 2ZFF H; 1SGE E; 1THS H; 1BIT A; 3ZVB A; 2A2Q H; 4FXG H; 3SUZ A; 1GI4 A; 3CS7 A; 1Y3V A; 1AVW A; 5AFY H; 3T26 A; 4FUE A; 4NMR A; 3MU8 A; 1DX5 M; 1U6Q A; 4FUG A; 1Y7N A; 2OTV A; 5GWZ A; 2PFE A; 8LPR A; 2F3C E; 3FVF B; 3RXO A; 2D26 C; 1QR3 E; 3MH7 A; 1HXF H; 1G9I E; 4NFF A; 2H2Z A; 1CA0 C; 3UTU H; 4B2B A; 2A7C A; 5PTP A; 3MH6 A; 1QA0 A; 1VIT G; 3OYP A; 3QDZ B; 1U3B A; 4NIX A; 1O3D A; 2Y5F A; 3HVQ C; 1EJM A; 2WPI S; 1O5F H; 3CYY A; 3A8B A; 1BTU A; 3UR9 A; 4JZI A; 1C2E A; 1BUI A; 2G51 A; 1DIT H; 3F1S B; 2GCT B; 2O8M A; 1GI2 A; 4IN1 A; 1XZ9 A; 1G30 B; 1JWT A; 2ZDM A; 1LO6 A; 1P2K A; 3W95 A; 2UUF B; 3BEF B; 5CHA B; 1WTG H; 3RXA A; 1NRS H; 2LA8 A; 4NMV A; 1UK3 A; 2UUJ B; 1ANC A; 1P09 A; 2CHA B; 1C5R A; 1G2M A; 1XX9 A; 1Z8J B; 4WVP E; 1IU0 A; 3RLE A; 3A86 A; 1BT7 A; 4UEH H; 1T8L A; 3PDZ A; 1FY8 E; 2ZO3 H; 4WH8 A; 2ZGX H; 2F9B H; 3PWC A; 1LMW B; 3BTF E; 3FY5 A; 3A81 A; 1RJX B; 4WWY A; 1ARC A; 4CHA C; 1P9U A; 3U98 H; 1PPE E; 4I8L A; 2AGI X; 3T5F H; 2FDA A; 3EST A; 1HCG A; 1MAX A; 2Y81 A; 5LH8 A; 1V2S T; 1V2U T; 3RMN H; 1V3X A; 1W0Z U; 1P2I A; 1UVP A; 3SUE A; 1QLC A; 1X5R A; 2SNV A; 3TIU A; 1OWJ A; 1UCY H; 1AVX A; 1RTK A; 3SUF A; 2ZPL A; 1QBV H; 4GSO A; 1XUJ A; 2J94 A; 1ZR0 A; 2FI3 E; 3KHF A; 1QB9 A; 2TBS A; 1NF3 C; 1W8B H; 3TSW A; 3E17 A; 1UTO A; 4GLY A; 2Z9K A; 1AUT C; 1Z8I B; 5FB8 C; 2J38 A; 2GDD A; 1BDA A; 4XBC A; 4YES B; 4JOG A; 2QC2 A; 4ISO A; 3VB5 A; 4I8G A; 2EI8 A; 3SV7 A; 3VXE H; 3T29 A; 1AZ8 A; 1FN6 A; 2ZPS A; 1OBY A; 1QRW A; 4Q2P A; 3E90 B; 3KF2 A; 2OP9 A; 2CSS A; 5JB9 S; 1C5O H; 2JIN A; 1HB0 B; 3K50 A; 1ZLR A; 4X8T H; 3Q3K A; 1PPG E; 2EC9 H; 1O3K A; 3C1K A; 1WF8 A; 4CRB A; 4ABB A; 3QLP H; 1HAZ B; 1K9O E; 2G52 A; 2ZDK A; 4GPG A; 5HM2 A; 5EQQ A; 2ZCH P; 3KN2 A; 2FM2 A; 4HFP B; 3NFK A; 1L4D A; 1E34 B; 1LTO A; 1A3E H; 1NO9 H; 4I8J A; 4X6P A; 4ABD A; 3GJ9 A; 1VJ6 A; 2ZHF H; 1Y59 T; 1JRT A; 1S5S A; 2GCT C; 1AUJ A; 1AWH B; 2DE9 A; 4IN2 A; 1EZS C; 4JCN A; 1O2Q A; 2SGF E; 4X6O A; 5CHA C; 4K76 A; 2H9T H; 3OY5 U; 1TNG A; 4YT7 H; 1AHT H; 2CHA C; 2QAA A; 1PDR A; 1LHG H; 3SQE E; 2GZ8 A; 1FCF A; 4AYY B; 2VRF A; 2Q3G A; 1T8M A; 2GCH F; 2CS5 A; 3DT0 H; 2G5N A; 4X2W A; 1EAI A; 2H9H A; 1J14 A; 2ZEB A; 1QBO A; 1XKA C; 2IWN A; 3UOP A; 1P04 A; 4FVB A; 1PQ8 A; 2J34 A; 2H3L A; 1BJU A; 3GDU A; 3LGU A; 3U1J B; 2IOT A; 2G4T A; 3R68 A; 2PGQ B; 1QJ1 B; 3SI3 H; 1UO6 A; 3RXT A; 3G01 A; 2JOA A; 1MZD A; 2VJ1 A; 2RCZ A; 1ZGV A; 4X1S U; 3ZZ4 A; 1O36 A; 1B0E A; 1I16 A; 3R3G B; 4Q80 A; 1O2H A; 2BD4 A; 5GDS H; 4JOF A; 3BTM E; 2CSJ A; 5JPM H; 3UQV A; 2SGP E; 1ZJD A; 2FOD A; 2ZG0 H; 4ABF A; 1HJ9 A; 2YT7 A; 1V2V T; 4E7N A; 3K82 A; 2CGA A; 1ZHM A; 1K1N A; 1R6J A; 1GBM A; 2PKU A; 5J4Q A; 1INC A; 2W3I A; 1GMC F; 1P2N A; 4NA8 A; 1TAW A; 3LH3 A; 1OPH B; 1GVZ A; 2FLR H; 1J15 A; 5LGO A; 5E0H A; 3ATL A; 1GHY H; 3RXE A; 1UVS H; 2PKA A; 1CHO F; 3SU5 A; 2GV6 A; 2VIP A; 4OGX A; 3MWI U; 3UIR A; 1ESA A; 4OS2 A; 2KA9 A; 3TK6 A; 2OBQ A; 1GBB A; 4X2X A; 5AF9 H; 2OLG A; 4YTA A; 2OBO A; 3ZVA A; 1O35 A; 3ZRT A; 2BZA A; 2J92 A; 2BMG B; 2M5T A; 3NFL A; 1K21 H; 3SU6 A; 2PUQ H; 3LIW A; 1FZZ A; 1C2J A; 2A32 A; 2FZZ A; 1RY4 A; 3TMH A; 2AWU A; 2J4I A; 3BG4 B; 4GHN A; 2TGA A; 3GY5 A; 5C20 A; 3CC0 A; 1EAT A; 5HXF A; 1O2V A; 2PLX A; 2ZGJ A; 2K1Z A; 3C27 B; 1C5X B; 1MTS A; 1O2W A; 1K28 D; 5C5O A; 1NTP A; 2ALV A; 1ABI H; 3QZQ A; 5EQR A; 2QKU A; 1FY1 A; 1NY2 2; 5JXB A; 1F0R A; 1GJ5 H; 1LCY A; 2STB E; 2M0T A; 2Q9V A; 3I1E A; 4RKO B; 5LPR A; 1P1D A; 1N6E A; 4FGM A; 1O3J A; 2JWE A; 1W14 U; 2VVU A; 1L4Z A; 2BYA X; 4KTC A; 3SQH E; 2KQF A; 4REY A; 4RI0 A; 4KTS A; 2CXV A; 3QO6 A; 1IU2 A; 3ZV8 A; 3RXI A; 1F2S E; 4BAM B; 2EGK A; 2EEG A; 4DUR A; 2OPG A; 4G69 A; 4QT8 C; 5HF4 A; 1CO7 E; 3RXB A; 1SPJ A; 3A89 A; 1EP5 A; 3LON A; 3MNS A; 3WKL A; 1T4V H; 4Y6D A; 5GJ4 B; 2P3W A; 3PMJ A; 4BTI B; 2OC0 A; 1P1E A; 3OTP A; 1DUA A; 5EPN A; 5BPE A; 2ZFP H; 3D65 E; 1BE9 A; 4RKJ B; 3K2U A; 1LPZ B; 4UU6 A; 4IS5 A; 4YOJ A; 2Z94 A; 4LOY H; 3RXJ A; 4B73 A; 1OWD A; 4N6X A; 1IQE A; 1PYT C; 3ODD A; 1C2I A; 1C5Z B; 1YC0 A; 2R4H A; 4FU9 A; 1AU8 A; 3S7K B; 3TK9 A; 3BSQ A; 1KXC A; 2BDC A; 2I6Q A; 3F9E A; 1ARB A; 3SNB A; 3VQG A; 4FUI A; 5HMA A; 1WQS A; 2BY8 X; 3BIV H; 5FX6 A; 2PKA B; 1W11 U; 5C3N A; 1D3P B; 1XMN B; 3PLK A; 3DD2 H; 2Y7X A; 5JBT A; 1VA8 A; 2AMD A; 3DJ1 A; 2YUB A; 1VIT H; 3P8F A; 1BTY A; 4AB9 A; 2ZDA H; 3BTD E; 4AG2 A; 1MBQ A; 1N8O B; 1GG6 B; 1FY5 A; 1T7C A; 2F0A A; 1O2U A; 4CH2 B; 1A46 H; 2HPQ H; 3RXR A; 1GI3 A; 4XFQ A; 5L2Y H; 1RD3 B; 1BHX B; 2YTW A; 5EYZ A; 1CGI E; 2BVS H; 1YCP M; 1GMD G; 2B7D H; 2H3M A; 1UVT H; 1BRU P; 5TCA A; 1F0T A; 4CRA A; 4INL A; 1UMA H; 1P02 A; 2HVX A; 1FPH H; 1YCP K; 1FAX A; 3E16 B; 2EV8 A; 4NZQ A; 1N6D A; 1G3B A; 1M9U A; 1UM7 A; 1DPO A; 3GY3 A; 2KPL A; 1BBR K; 1WIF A; 1A5I A; 1C2F A; 1MBM A; 4MPW A; 1YPH C; 4MNY A; 2A1D B; 5HFC A; 4JK5 A; 2STA E; 2WV4 A; 2K20 A; 3T28 A; 2RDL A; 1ELT A; 1HPG A; 4BAO B; 2VIW A; 3IWM A; 4F49 A; 3PM3 A; 1MQ6 A; 5F03 A; 1EQ9 A; 1SOZ A; 2YT8 A; 4JOJ A; 1L2E A; 4FUF A; 3RLW H; 1P57 B; 3ZVD A; 4Q7X A; 4FVD A; 1QJ6 B; 1UHB A; 1V1T A; 3ZZ9 A; 2ZHE H; 1SLW B; 2HE4 A; 3K9X B; 2V0B A; 4AGK A; 1O2X A; 1C1W H; 4NNL A; 2R0K A; 2YNA A; 1O2I A; 3TK5 A; 2OZF A; 4MQA A; 1V2R T; 1TFX A; 3F9H A; 2D8W A; 2HE2 A; 5E8E H; 2NU3 E; 1HAV A; 4H6S B; 3ID4 A; 5GND A; 3GDS A; 1KLI H; 1O3N A; 3SOR A; 2F9U A; 2GDE H; 1BTP A; 1GDQ A; 1TGB A; 2M10 A; 2QF0 A; 2ZHW H; 4GHQ A; 4XBD A; 2FOF A; 1X5Q A; 1GL1 A; 2RA3 A; 3BTQ E; 3A84 A; 3H7T A; 5A8X A; 3D62 A; 1UF1 A; 3STJ A; 4E34 A; 2OC1 A; 1EB2 A; 2GTB A; 4THN H; 3KQC A; 1DFP A; 1I92 A; 1C9T A; 5GIB A; 3AAV A; 3SV8 A; 1CT0 E; 2AYW A; 1Z87 A; 1EJN A; 1TB6 H; 2GV7 A; 4ZXY H; 3PLP A; 1V5L A; 1XVM A; 1S83 A; 2GZ7 A; 1MKX H; 3VEQ B; 1AWF H; 1P12 E; 1O5G H; 2RA0 A; 1HUT H; 4TPY A; 1HJA B; 4Y0Y E; 1RGQ A; 1S82 A; 2ZGC A; 1MTN B; 1AB9 B; 5JB8 S; 1JOU B; 1ELA A; 1RGW A; 1Q7X A; 2DM8 A; 2UWP A; 2WPL S; 4KFV A; 1DE7 H; 1KYN A; 3RXM A; 3UOU A; 2QBW A; 1KSN A; 2A5K A; 3RU4 T; 1N8O C; 1GG6 C; 4UE7 H; 4AG1 A; 1P2O A; 3GY8 A; 3M7U A; 5HEY A; 3TGJ E; 5T6D A; 4U32 A; 5DGJ A; 5HJ1 A; 3Q3X A; 1OYQ A; 1P05 A; 3QE1 A; 1GI5 A; 1QGF A; 3HK6 B; 4FUH A; 3RC4 A; 2ZEC A; 1SL3 A; 4EST E; 2BDI A; 5IYT A; 1OP8 A; 4YT6 H; 2B0F A; 4AMH A; 2FLB H; 4XOJ A; 5FPT A; 4YDP A; 4YYX A; 4K2Y A; 1FN8 A; 1ELG A; 2ANM H; 3EGK H; 1GQ5 A; 1ZTK A; 3EDX B; 3URE A; 1BHX F; 4ABA A; 1A0H B; 1EZU C; 1TNH A; 3TJN A; 2FOM B; 1YPL H; 3LOX A; 4K8Y A; 5DKM A; 2FPZ A; 1TMB H; 3U1I B; 3Q3Y A; 3TSV A; 5A0C A; 2XTT B; 2BM2 A; 1O3M A; 1IHS H; 3TGK E; 2P8O B; 1RRK A; 2G2L A; 1Q3O A; 3F9G A; 1SG8 B; 2VID A; 1KEF A; 1ELD E; 4Y79 A; 1UK4 A; 1UHB B; 4UFG H; 2O9Q A; 1OWK A; 4B2A A; 2NU2 E; 4Y10 E; 2A4R A; 1YF4 A; 2XYA A; 4AFS A; 1P11 E; 1P03 A; 1LDT T; 3A83 A; 1HAV B; 5L6N H; 1MFL A; 1MUE B; 3KL6 A; 3ZZ3 A; 2NU4 E; 1Z71 A; 2AIQ A; 3BN9 A; 4ISL A; 2PUX B; 1O3P B; 3URC A; 5H3J A; 4DVA U; 1NPM A; 2BLO A; 4OS7 A; 1BJV A; 1UTN A; 1HNE E; 2VGC B; 3CS0 A; 4GCH G; 1D5G A; 4R8T B; 1BIO A; 4ZXX H; 1TX6 A; 1F7Z A; 3SUD A; 1GI0 A; 2ZWL H; 3SHC H; 4U30 A; 1L1N A; 1C1Q A; 3M5O A; 1SGQ E; 1KDQ B; 3KNX A; 2OCS A; 4NA9 H; 1HJA C; 2D1J A; 3ZVE A; 5EQS A; 4JYT A; 3UR6 A; 5HFF A; 3K1R A; 4I8H A; 5E6P B; 5ESB A; 4OEO A; 2FXR A; 1CVW H; 1RXP A; 1FMG A; 2Y6T A; 4YLU A; 1SGR E; 4HI3 A; 4I8K A; 4Q2O A; 2XW9 A; 2G55 A; 2AH4 X; 3NZI A; 4ISN A; 3M37 A; 3OX7 U; 4ABG A; 1SVP A; 1M5Z A; 1K1O A; 5J4S A; 1TPP A; 4EDJ A; 2OC7 A; 3SND A; 1YPH E; 4CR9 A; 1JRS A; 2TGD A; 1DXP A; 2VH0 A; 5C67 A; 3SJO A; 2THF B; 1SQT A; 3A7Y A; 1QBN A; 2Q6D A; 4WJG 2; 3S0N A; 2MX6 A; 1BB0 B; 4X6M A; 3A7Z A; 1SC8 U; 1V5Q A; 1EPT A; 4K1T A; 2OMJ A; 4OGY A; 3TNS A; 4JYV H; 2AWX A; 4MNX A; 4YZU A; 5ETX A; 3SGQ E; 3SNE A; 1C5U A; 2KOJ A; 1AQ7 A; 1NN6 A; 1XKB C; 5DP5 A; 3QTO H; 5JDU B; 1AIX H; 1GHB F; 1ZHR A; 1H9I E; 3DA9 B; 4I7Y H; 4O97 A; 5DG6 A; 4AYV B; 1QAV B; 3TGI E; 2FOH A; 3MOC A; 5CMX H; 3RXU A; 4INK A; 2I6V A; 2ANY A; 2GKV E; 4E05 H; 4LK4 A; 2HGA A; 1NRO H; 1QTF A; 4IBL H; 1DWE H; 1U37 A; 3R69 A; 1KXF A; 3TJO A; 2P8O C; 1ZPB A; 2OQU A; 1GJ6 A; 1PPZ A; 3CH8 A; 3RU4 D; 2BY6 X; 1Q3X A; 3KGP A; 5GCH G; 1NFU A; 1MD8 A; 1UTJ A; 1K1P A; 3MI4 A; 4ZRO A; 3BIU H; 4AZ2 B; 1DIC A; 5CE1 A; 4WXI A; 2QXH A; 5DP4 A; 5GLJ A; 4JMY A; 2BX3 A; 2GX4 A; 1UM1 A; 1O32 A; 1SLV B; 1C2H A; 2ZDN A; 1VCP A; 3ATK A; 3A80 A; 1ID5 H; 3GCT F; 5AHG H; 1AVG H; 1AKS B; 1TNL A; 5FPS A; 4UDW H; 4AFU A; 1TGS Z; 4NZL A; 2ZPM A; 4Q2N A; 1UTQ A; 3MO9 A; 3LGW A; 4GHT A; 3BEI B; 4KKD A; 1BTX A; 2W7R A; 1C2L A; 2ASU B; 5K0H A; 2G5V A; 3O46 A; 4FU8 A; 4KTU A; 5DO4 H; 7KME H; 1KXB A; 2Z6B D; 3F6U H; 3MH5 A; 3FP6 E; 3NK8 A; 5L2Z H; 1DM4 B; 1NM6 A; 5C1U A; 1O3O A; 1ZGI A; 4HOP A; 2I6S A; 1LVB A; 1TBZ H; 2KAW A; 2ZHD A; 3HK3 B; 1IQF A; 1N1L A; 1GHX H; 1K32 A; 2PTN A; 1FJS A; 3LKW A; 4J2Y B; 3SU3 A; 1SGC A; 1C5V A; 1W12 U; 4BAN B; 3Q77 A; 4NXP A; 1TAB E; 1V2Q T; 1YP9 A; 1C1N A; 2C8X B; 4Q6S A; 8GCH G; 1UTK A; 3RXD A; 2F9P A; 3K6Z A; 2H9E H; 4KP0 A; 3SI4 H; 5DP3 A; 2EDV A; 5JBC S; 2EST E; 1BTW A; 3E8L A; 1G36 A; 2WYJ A; 1EAX A; 2WUB A; 3V13 A; 3PV2 A; 4JK6 A; 3O8C A; 4A6L A; 2L4S A; 2EA3 A; 1O5B B; 1UJU A; 1MH0 A; 3L3T A; 2EDZ A; 3GGE A; 3NUM A; 6CHA B; 4KGA A; 1IQH A; 1QL8 A; 3ZZ6 A; 5E21 A; 1F92 A; 5K7R A; 1Y5U T; 2W5E A; 1WQV H; 3HKI B; 3PTB A; 1AE5 A; 1EPT B; 2VH6 A; 5EST E; 4WME A; 2QKV A; 2GZ9 A; 4X1P U; 3D23 A; 1GJ9 B; 1AI8 H; 1QB6 A; 3FFG A; 1C1M A; 1ZMN A; 4WH6 A; 1DLK B; 1BZX E; 1IQI A; 2IJO B; 3PO1 B; 1NFY A; 5C1Y A; 3RP2 A; 3QGL A; 1GBE A; 4K75 A; 2QN5 T; 1HYL A; 2ZI2 H; 3M3V A; 4B2C A; 1ELB A; 3RXG A; 3MTY A; 3SHA H; 1V2J T; 1Y5B T; 2PTC E; 3MU1 A; 4X6N A; 1UHP A; 3V12 A; 4CR5 A; 1A5G H; 1AZZ A; 1Z8G A; 1S6H A; 1W3C A; 2WPH S; 3MO3 A; 1GJB B; 3TVJ B; 2ZNK H; 4JZE H; 1GMH F; 3VXF H; 3I29 A; 1WU1 A; 3VPK A; 3GCN A; 1KTT B; 4BAQ B; 1LHE H; 2G8T A; 2QT5 A; 3PMA B; 1BBR E; 1IQG A; 1GQ4 A; 1HAH H; 2BX4 A; 2HRV A; 1PPC E; 2BYG A; 1X45 A; 4YM9 A; 1C5Q A; 3KQD A; 4NWL A; 2KBS A; 4NMT A; 3SGB E; 1WTH D; 2VWO A; 5JYI A; 1O5D H; 2PW8 H; 1OP2 A; 3TIT A; 1A0J A; 4SGA E; 3SOS A; 2PSY A; 3RXP A; 1O3L A; 1O2K A; 5HED A; 1EKB B; 4Y7A A; 2ZF0 H; 5EGM A; 3A85 A; 2EEH A; 5I25 A; 2EJY A; 1HXE H; 1FY3 A; 1DY8 A; 3LC5 A; 3PMH B; 4MVN A; 1DS2 E; 2G00 A; 1YPE H; 3P17 H; 1TBR H; 3I4W A; 3RMM H; 2WPJ S; 3JXT A; 2WL7 A; 4JZD H; 4UD9 H; 3LNX A; 2HWL B; 2G5M B; 2VWM A; 4FUB A; 1VR1 H; 1TRY A; 1TMT H; 1HAI H; 1TNK A; 1MU6 B; 3PLB A; 1Y3U A; 2DUC A; 4I32 A; 4A5T S; 1THR H; 3MFJ A; 1ORF A; 2ENO A; 3ZZ5 A; 2GT8 A; 4NMO A; 2Q1J A; 1OS8 A; 1WFV A; 4Q2Q A; 2VB0 A; 3STI A; 1O2G H; 6CHA C; 3OTJ E; 3TSZ A; 1DWD H; 5DP7 A; 3FAN A; 3CBY A; 1IQJ A; 1ELC A; 4WMF A; 1EPT C; 1TGC A; 4ISI H; 2GP9 B; 2Y5H A; 4UFE H; 4JYU H; 3KQB A; 1UK2 A; 1L1G A; 2BDB A; 3MU4 A; 1GI8 B; 3KHV A; 1WSS H; 3BTT E; 2LUI A; 3PO1 C; 1D3D B; 2VWN A; 3CP7 A; 4KFW A; 3KQE A; 2R0L A; 1Z1I A; 2VVC A; 5EZ0 A; 1OX1 A; 2WV5 A; 3P95 A; 1T8O A; 4AFQ A; 3SU1 A; 1S0R A; 2ZU2 A; 5HDY A; 1NTE A; 1C1O A; 1SQO A; 1BFE A; 4K6Y A; 1C2G A; 4Y8X A; 2G81 E; 4HTC H; 1SQA A; 2A2X H; 2EEJ A; 2A0Q B; 2O8T A; 3ATM A; 4RN6 A; 2VWR A; 3TU7 H; 5E11 A; 1KTS B; 1ZTL A; 1TBQ H; 3MNC A; 1IQK A; 2Y5G A; 1N6Y A; 2XC0 A; 4NWK A; 3HAT H; 3URD A; 1TPS A; 3M5N A; 2KJP A; 2EI6 A; 4Q7Z A; 1O37 A; 4TWW A; 1O31 A; 2JKH A; 1EAS A; 2GCH G; 3E6P H; 1VJA U; 1GJ4 H; 4BAH B; 1WYK A; 4M7G A; 4X8U H; 1WHA A; 1NES E; 3UWI A; 1J16 A; 2ZPR A; 2KYL A; 1O0D H; 1QA7 A; 2P3U B; 1O3A A; 4CBN A; 1QFK H; 1XXF A; 4K1S A; 1Z8R A; 3SZN A; 1J9C H; 2ALP A; 1DVA H; 5A8Y A; 2NU0 E; 2ZPQ A; 5FX5 A; 1ZJK A; 1O2J A; 2IWQ A; 3U8T H; 1N99 A; 1U38 A; 3VB7 A; 2J2U A; 1TQ3 A; 6GCH F; 1QQU A; 1ETR H; 2ZQ1 A; 3L6P A; 1O2Y A; 4JZ1 A; 4NG9 H; 3A87 A; 3VFE A; 2JIL A; 2VIQ A; 2RRM A; 6LPR A; 1HAY B; 1Z6J H; 3MO6 A; 1N7E A; 1H9H E; 2R9P A; 9LPR A; 2HNT F; 4ZH8 A; 1O3E A; 1GBC A; 3QDO A; 2JET B; 1IQL A; 1LHD H; 2OD3 B; 4NMP A; 1GMC G; 3M3T A; 2YUY A; 1BCU H; 1C5W B; 5F8T A; 1CHO G; 2Y80 A; 7GCH F; 6EST A; 2DC2 A; 2H5C A; 2Y82 A; 3B9F H; 3ZZB A; 5HM9 A; 3TNT A; 2AWW A; 4MLF B; 5C2Z A; 1RGR A; 1ANB A; 3LU9 B; 1DOJ A; 5A8Z A; 3B8J A; 3HPT B; 2BD2 A; 1QJ7 B; 2TPI Z; 2VSP A; 4D76 A; 2AS9 A; 3SGA E; 1GDU A; 3QK1 A; 4AN7 A; 1YLD A; 2UZC A; 2M0U A; 2LPR A; 1FDP A; 1GWA A; 1S0Q A; 1ZMJ A; 4DOQ A; 1TRN A; 1P3E A; 1O2O A; 5G4B A; 1WUN H; 1GJA B; 2EGO A; 3QWC H; 1EZX C; 4X0W U; 4Y11 E; 2ZHQ H; 3M3S A; 1DST A; 2NWN A; 3UPE A; 3PB1 E; 5C1X A; 1CGJ E; 1J8A A; 1GHA F; 4NA7 A; 3WY8 A; 3QJM A; 1BMM H; 1YPG H; 3M35 A; 2FP7 B; 5HFE A; 1HRT H; 3U8O H; 1MCV A; 4NSV A; 1HAP H; 3TH3 H; 4NXR A; 1SBW A; 2YOL A; 3BG4 C; 2KAI A; 1FLE E; 3RM0 H; 1O5A B; 1C5S A; 5EW1 H; 1XUI A; 1J17 T; 1Y3Y A; 1YCP H; 1SGT A; 4NFE A; 3S9C A; 2A7H A; 5HYO A; 2BLV A; 3QGN B; 5E2O A; 1BBR H; 2BLQ A; 2ZIQ H; 2A4O A; 2BY7 X; 3LDX H; 1WHD A; 1QHR B; 3SFJ A; 3RLY H; 1WFG A; 4TY6 A; 2W3K A; 1A61 H; 1GI6 A; 4OS1 A; 2AEI H; 4I33 A; 2FI5 E; 1DY9 A; 1YKT A; 1AFE H; 3W94 A; 2ZGH A; 1C1P A; 1EJA A; 5SGA E; 1CT4 E; 1TWX B; 2FYQ A; 3SU0 A; 3HGN A; 1H9L B; 3EGH C; 3PMB B; 1SOT A; 2Z9G A; 2C3S A; 2SGA A; 3SU2 A; 2ZCK P; 3MYW A; 2XRC A; 3Q76 A; 5HF1 A; 1DAN H; 2PKT A; 5FPY A; 4B74 A; 3R0F A; 7GCH G; 2QV1 A; 3WKM A; 4DII H; 2FS8 A; 1SGP E; 4MPX A; 3SJ9 A; 4BXW A; 2W26 A; 1K1M A; 4B1T A; 1TX8 A; 2VVV A; 3PDV A; 2UWO A; 2EHR A; 3NGH A; 2P3F H; 2CJI A; 5H4I B; 4ABH A; 1O39 A; 1C5T A; 2EEK A; 3PS4 A; 1GBT A; 2FX6 A; 1O2T A; 3QUM P; 3SJI A; 1DWC H; 3ID1 A; 2XCN A; 3P8G A; 1O38 A; 1V2L T; 1GL0 E; 2EXG A; 1X7A C; 5FBI A; 1P2M A; 1GHA G; 1TGT A; 1W10 U; 1SGD E; 1MCT A; 1FV9 A; 4D7G A; 3QJN A; 1Q31 A; 1C2M A; 5GPI B; 2V6N A; 1FXY A; 3EE0 B; 1C5L H; 1EOJ A; 5ABW A; 2DLU A; 5EPY A; 1Z1J A; 3RXQ A; 1SSX A; 1O2Z A; 2AMP A; 3AAS A; 3M5M A; 1NC6 A; 1C1R A; 1GBF A; 1KXE A; 3AW1 A; 3P8O A; 5F8Z A; 2KV8 A; 2OC8 A; 1C2D A; 1YPK H; 4BTT B; 4E3B A; 2ZKS A; 1EST A; 3SUG A; 4ZMA H; 1AY6 H; 2XCF A; 5EG4 A; 1HFD A; 5I7Z A; 1O2L A; 3HPK A; 1BMN H; 4GVC A; 4OEP A; 1K1I A; 3QN7 A; 2TGP Z; 2ULL A; 3NWU A; 1GGD B; 4CH8 B; 2JIK A; 1ELV A; 2BDG A; 3SOE A; 1YLC A; 2AIP A; 4TY7 A; 5DPA A; 1IHJ A; 2KAI B; 3CEN A; 5B6L A; 3BTK E; 3E3T A; 4JOP A; 4U01 A; 3A82 A; 1DXW A; 1P0S H; 2D90 A; 1PJP A; 1X8S A; 4E35 A; 1PQ5 A; 1B8Q A; 2P3T B; 1FC9 A; 1AFQ B; 1CBW B; 2BD9 A; 1OP0 A; 1NRQ H; 5EXL A; 1MKW K; 3DOR A; 3KZE A; 1HAX B; 1ZSK A; 3ZVC A; 1C1T A; 1D3T B; 1OWH A; 4NIV A; 1VCQ A; 2FOE A; 2QA9 E; 1A0L A; 1O2N A; 4NGA H; 2HOB A; 1Z86 A; 1NS3 A; 2BY5 X; 3LJO A; 2FNE A; 2M3M A; 2O8L A; 1LVM A; 2I1N A; 2BY9 X; 2DMZ A; 2GT7 A; 3KZD A; 2ZFT A; 1YBO A; 1KIG H; 4ZHM U; 3DUX H; 1OWI A; 4AOQ A; 1SLU B; 1EZQ A; 2CN0 H; 3MU0 A; 2FIR H; 3CBZ A; 1Z7K A; 1MTV A; 1SGY E; 3RXH A; 1GJ8 B; 1AGJ A; 4DT7 B; 5JBA S; 1HAG E; 2JXO A; 4DJZ B; 1O3B A; 4K69 A; 5F67 A; 1YYY 1; 3H7O A; 2QGR A; 3SW2 B; 4K78 A; 1OYT H; 2V3H H; 3S7H B; 1TAL A; 2BQ7 B; 1C1S A; 1CHG A; 3LH1 A; 2A45 B; 4MNV A; 2QKT A; 1FC7 A; 2Z3C A; 2ZCL P; 3VB6 A; 3PV5 A; 1F0S A; 2KJK A; 2H2C A; 4YOG A; 1O3H A; 2L97 A; 1ELF A; 2IWP A; 4MG3 A; 3IIT A; 1LPG B; 1EX3 A; 1O2M A; 2BXU H; 1YPM H; 2JRE A; 2QIQ A; 4CBO A; 1MTN C; 1AB9 C; 3FP7 E; 1T4U H; 3HGP A; 4CRD A; 1MQ5 A; 2C8Y B; 1ACB E; 2ZA5 A; 3MNX A; 5EW2 H; 2BLW A; 2FOG A; 3S9B A; 1LKA A; 2TGT A; 1W0Y H; 1YGC H; 2GMT B; 4A1V A; 1ZOM A; 1TMU H; 1ZSL A; 1QUR H; 2C93 B; 1QL7 A; 4ZUH A; 5FAH A; 4GVD A; 4ZKN U; 1VCW A; 5E22 A; 1DDJ A; 2BOH B; 1SFQ B; 4IMZ A; 4D8N A; 3NXP A; 1EXF A; 2ZFS A; 2ZFQ H; 5E2P A; 3A7W A; 3FZZ A; 1VIT F; 1PPB H; 1ANE A; 3RM2 H; 4IGD A; 1VGC B; 4DGJ A; 5EOK A; 4BOH A; 2TRM A; 1WV7 H; 1FON A; 1RZX A; 1UFX A; 3R0H A; 1AFQ C; 1CBW C; 4A1T A; 3GSL A; 1Y3X A; 2F5Y A; 4RSP A; 1TGN A; 2BD5 A; 5DP8 A; 3DJ3 A; 2NU1 E; 3A8D A; 3PTN A; 5HFB A; 2PKS B; 2WUC A; 4CHA B; 1D3Q B; 1MU8 B; 1N7F A; 4FUC A; 2KOM A; 1A5H A; 1AE8 H; 3SV6 A; 1NRP H; 1RIW B; 3GY6 A; 1E0F D; 3UNS A; 1W7X H; 1BTH H; 5FHT A; 3ID2 A; 2FEQ H; 4O9V A; 1C2K A; 4XBB A; 4A92 A; 8EST E; 3AVZ A; 1N7T A; 1ZUB A; 1FAK H; 1G32 B; 1HJ8 A; 2TIO A; 2ZDL A; 2I04 A; 2PDZ A; 5FXL A; 2SGD E; 1OXG A; 4MTB A; 1GHV H; 1Y5A T; 1TON A; 2F83 A; 1XUG A; 3ZZC A; 2XBW A; 4AB8 A; 2PZD A; 1SFI A; 2PSX A; 1NFW A; 3LC3 A; 1FQ3 A; 1V2T T; 4DG4 A; 4KEL A; 1LQD B; 5AVD A; 3N7O A; 4Q2K A; 3ZV9 A; 1QB1 A; 3V7T A; 1ZZZ A; 1G2L A; 1VB7 A; 1GBL A; 1MAY A; 1PQA A; 4UFF H; 4VGC B; 4X1R U; 4O06 A; 3VB4 A; 3GYM A; 2DKR A; 1O5E H; 2WPK S; 3PWB A; 4MPU A; 4AOR A; 3H5C B; 1EAW A; 4BXS A; 3GY2 A; 1HGT H; 1SI5 H; 1YPJ H; 4SGB E; 2X7Z A; 3EYD A; 2EI7 A; 2M0Z A; 2QXI A; 3TH4 H; 2VIO A; 1UEP A; 3I18 A; 1T32 A; 4LZ4 B; 2JH0 D; 2VGC C; 2ZU4 A; 3O8D A; 1KY9 A; 1X6D A; 3L4F D; 3LGV A; 1OKX A; 2G4U A; 4R2Z A; 1WAY B; 4D7F A; 1P9S A; 3QIK A; 3PRO A; 2JH6 D; 2FS9 A; 1QCP A; 1L0Z A; 4F4O C; 4U2W A; 5B6O A; 4Y8Y A; 2EGN A; 1P10 A; 2QG1 A; 1V2W T; 1TPA E; 4UFD H; 3EA7 A; 2PR3 A; 1SHY A; 4H6T A; 4JOR A; 1TQ0 B; 1H4W A; 1GBH A; 1H8I H; 1FNI A; 4B76 A; 2C8Z B; 2KRG A; 1ZRB A; 2GMT C; 3E0N B; 3U8R H; 4Y7B A; 1TOM H; 2BTC E; 3Q00 A; 3M5L A; 2P94 A; 1TA6 A; 3KEE A; 1D6W A; 1FUJ A; 1MC7 A; 2W7S A; 3LT3 A; 1P06 A; 2ILN A; 3M7Q A; 2BZ6 H; 5DK1 A; 1SR5 C; 4NSY A; 2FCF A; 2A5I A; 4PRO A; 1VGC C; 1RFN A; 1GHB G; 3U69 H; 2Z7F E; 1S84 A; 4LXB H; 1ETT H; 2HLC A; 1G9O A; 1SGF A; 2FO9 A; 3T2N A; 1PPF E; 2I4S A; 3T27 A; 1A2C H; 1KXD A; 4WXV A; 2GZV A; 5F6M A; 1TA2 A; 1EAU A; 1TOC B; 5DTH A; 1K22 H; 2R3Y A; 2PKS C; 2KPK A; 2KJD A; 4ABJ A; 2QY0 B; 3GCO A; 2BDA A; 1RIW C; 4Y76 A; 2ZTX A; 2O2T A; 2Q6G A; 1D4P B; 3GCH B; 1RS0 A; 2FN5 A; 2VZ5 A; 3GCT G; 1LKB A; 2V35 A; 3P6Z B; 3OU0 A; 3JZ2 B; 3UUZ A; 5LPF A; 1KLJ H; 3SBK A; 5DJ7 A; 5A09 A; 1W13 U; 1IAU A; 1P3C A; 4Z33 A; 1UCY K; 4AFZ A; 1WI4 A; 1HV7 A; 4DW2 U; 3SJ8 A; 1V6D A; 2CMY A; 2BQW B; 3ITI A; 1XM1 A; 1WG6 A; 3SJK A; 5I46 H; 1VJ9 U; 3VB3 A; 4H11 A; 2F9O A; 4NMS A; 1LQE A; 3BF6 H; 4VGC C; 1K1J A; 4IW4 E; 1JXP A; 1THP B; 1V6B A; 3EQ0 H; 4CRF A; 4Y8Z A; 2XBY A; 3SO3 A; 3S69 A; 3D49 H; 2VPH A; 3RUO A; 5A0A E; 4JOK A; 4GAW A; 1GI7 B; 1JBU H; 4NMQ A; 3UNR A; 4X8S H; 4A7I B; 1G3C A; 1AO5 A; 1O3F A; 2QXG A; 1WBG B; 1A3B H; 3I77 A; 2XBV A; 4HOP B; 2L4T A; 3E0P B; 2EDP A; 1OZI A; 1GVK B; 1IHT H; 1O34 A; 1UIT A; 3DFL A; 3FP8 E; 1DWB H; 2OS6 A; 4K60 A; 3SN8 A; 4UON A; 2SNW A; 3RXL A; 3E91 A; 1P8V C; 3SNC A; 1XUH A; 4XHV A; 4JL7 A; 3K6Y A; 4BAK B; 1ESB A; 2XC4 A; 1DUE A; 1TQ7 B; 1CU1 A; 2SGQ E; 3RXS A; 4ISH H; 4INH A; 5T6G A; 2KG2 A; 2FOC A; 1EOL A; 3BTW E; 4OAJ A; 2BB4 A; 1NFX A; 1OSS A; 2BD8 A; 3RC6 A; 1IQN A; 2HGT H; 2P95 A; 4YO9 A; 4CRE A; 3B76 A; 5L30 H; 2BQ6 B; 1G3E A; 3FAO A; 1G37 A; 2SFA A; 1V2K T; 1IQM A; 4YLQ H; 4AX9 H; 2XBX A; 3DFJ A; 4TYD A; 1JMO H; 4O03 A; 5DP6 A; 1W9O A; 1OBZ A; 4NIW A; 9EST A; 3ATW A; 3P8N A; 1C4V 2; 1EUF A; 1PYT D; 3O8B A; 1SLX B; 1C5P A; 1B5G H; 2FMJ A; 3LGT A; 4JOE A; 2H5D A; 2OSG A; 3KCG H; 1SMF E; 1GMH G; 5AFZ H; 3FZD A; 3GCH C; 2A5A A; 3GYL B; 3TLO A; 4ZHL U; 4XSK U; 1WH1 A; 1FPC H; 2IPH A; 4Q7Y A; 4ABI A; 2ZTZ A; 2M0V A; 1ZPZ A; 1ZML A; 2FYR A; 4JZF H; 2PWX A; 2ODP A; 2PHB A; 2J95 A; 1UTP A; 2SGE E; 2H2B A; 3SV9 A; 1S85 A; 2FE5 A; 2OXS A; 1E35 B; 2Z3D A; 3ENS B; 4GUX A; 1A7S A; 1Y3W A; 3E1X B; 3OSY A; 1O2S A; 1UEW A; 2GVF A; 1D6R A; 2Y7Z A; 5T6F A; 5A0B A; 1NZQ H; 1VZQ H; 5IDK A; 3T62 A; 3M36 A; 5F8X A; 2D92 A; 1A1R A; 1FY4 A; 2RCE A; 3F68 H; 2O8W A; 1WI2 A; 1E36 B; 1TIO A; 2UUY A; 2ANK H; 1O2P A; 1NU7 B; 1MTU A; 2C8W B; 3MMG A; 3T25 A; 2BXT H; 1GDN A; 2FWW A; 4E7R G; 1TNI A; 1C4U 2; 3GIC B; 4R0I A; 2HNT C; 1FIZ A; 5GXP A; 1K2I 1; 4DIH H; 2AMQ A; 5E0J A; 4Q3H A; 4X1N U; 1C9P A; 2O8U A; 2ZZU H; 1O3I A; 1DT2 A; 1UVU H; 2CF8 H; 1CGH A; 3UQO A; 2LNC A; 2PV9 B; 4MDS A; 4K8B A; 2QCY A; 2V3O H; 2XWB I; 1TRM A; 4ZKR U; 4A1X A; 4Z6A H; 2VU8 E; 2VIN A; 2AGE X; 2DB5 A; 3M61 U; 1GVL A; 3SHW A; 1GM1 A; 1F5R A; 4AGJ A; 1GCT B; 2B5T B; 4K72 A; 2OUA A; 4K3J A; 1O30 A; 2XWA A; 2BOK A; 1ZYO A; 2C4F H; 1P01 A; 1BMA A; 1GMD F; 2W7U A; 2WIN I; 3TJU A; 1BQY A; #chains in the Genus database with same CATH topology 1F5L A; 3BPU A; 1K1L A; 4JOH A; 1ETS H; 2IN2 A; 1HLT H; 1T8N A; 1GBA A; 4F8K A; 2BHG A; 2D8I A; 2Z9I A; 1XUF A; 1YM0 A; 1DLE A; 4J1Y A; 2DE8 A; 1OWE A; 3N7D A; 3ZVG A; 4AC9 A; 3RMO H; 5FCK A; 1ABJ H; 1FI8 A; 4OS6 A; 1N6F A; 2Z17 A; 4WF8 A; 5DP9 A; 5HFD A; 3BG8 A; 3LGY A; 2A4Q A; 1XUK A; 2PNT A; 5E0G A; 5G1D A; 2JH5 D; 3SNA A; 1Y8T A; 1LHC H; 3A8C A; 1P2J A; 2P59 A; 1K0H A; 2A31 A; 4NCY A; 3LJJ A; 3GDV A; 3EA8 A; 1TX7 A; 3RU4 E; 4D9R A; 2UWL A; 1AMH A; 1E38 B; 2ZP0 H; 5C5N A; 3LNY A; 1CT2 E; 3VQF A; 2V90 A; 5FBE A; 2ANW A; 2F91 A; 2K1Q A; 3ZZD A; 1C4Y 2; 1ZRK A; 2REY A; 2CF9 H; 2ODY B; 3MH4 A; 3EGG C; 5A2P A; 2MHO A; 5FCR A; 1O3G A; 1UTM A; 2FTM A; 3RXM A; 2FOA A; 1W9Q A; 5EU8 A; 1E37 B; 4NIY A; 2BD3 A; 5G1E A; 4Q6H A; 2UUK B; 1QRZ A; 1GI9 B; 1QL9 A; 3ZZ8 A; 3BEU A; 3PYH A; 2HPP H; 1N6X A; 2D2D A; 2DAZ A; 2ZFR H; 3P92 A; 1KWA A; 1ZOK A; 4FU7 A; 5A2M H; 4Z8J A; 3ELA H; 3DIW A; 4Y71 A; 3AAU A; 6GCH G; 4OS5 A; 1QIX B; 1AND A; 1P2Q A; 4TPI Z; 4MPA A; 1Z6E A; 2ZTY A; 4LZ1 H; 1C1V H; 1B7X B; 4ZHA A; 3ID3 A; 1NRR H; 1S81 A; 5BRR E; 1SB1 H; 4MNW A; 2WPD D; 2RG3 A; 3QX5 H; 5FL7 D; 4E06 H; 1O2R A; 4YOI A; 2YNB A; 2J9N A; 2WPM S; 1KXA A; 4O3U A; 3BV9 B; 1UVO A; 1JIR A; 2AFQ B; 1C5N H; 1UJV A; 1U39 A; 5JBB S; 1W2K H; 2C90 B; 3EA9 A; 3O8R A; 3BTH E; 5T3H A; 2AIN A; 1GCT C; 2OIN A; 4CRC A; 1RTF B; 1OBX A; 1GI1 A; 2JET C; 1HBT H; 2QF3 A; 3SHU A; 3LPR A; 1CA8 B; 1ZHP A; 3OY6 U; 1TLD A; 2W4F A; 3O5N A; 1BMF D; 2LC6 A; 1H8D H; 3RXC A; 2XC5 A; 1BRB E; 4GVU A; 4K5Z A; 2Q6F A; 1ELE E; 5DN6 D; 1SGN E; 3HPM A; 1BRC E; 1SGF G; 3VGC B; 1O5C B; 1W9E A; 1AN1 E; 3DSP A; 1O3C A; 3P70 B; 2WZQ A; 1KLT A; 3ATI A; 2LOB A; 3A7X A; 3NKK A; 1EP6 A; 3SU4 A; 1QAU A; 2BD7 A; 2I0L A; 2XNI A; 1CE5 A; 1PFX C; 5HEB A; 2ODQ A; 1BOQ A; 4K1E A; 1LVO A; 1LHF H; 1SQR A; 4B75 A; 2KOH A; 1GBD A; 3AXA A; 3GY7 A; 3NCL A; 1V62 A; 5HET A; 3KYG A; 4HGC A; 4MPV A; 4Q63 A; 2F9N A; 3ODF A; 1QRX A; 4NNM A; 3CNR A; 5IK2 D; 1TP3 A; 1T2M A; 1L1J A; 4TSF D; 1KDQ A; 2HNT E; 3RL8 A; 2QXJ A; 3K65 B; 3U9A H; 1V2M T; 2E7K A; 1JIM A; 3BTE E; 4ZAE A; 3UFA A; 2FTL E; 1MKW H; 4WYU A; 2ZC9 H; 4HG6 A; 2B9L A; 1MZA A; 3IG6 B; 3RXF A; 1OOK B; 1MKX K; 1GJD B; 2EAQ A; 2V1W A; 2R3U A; 3A88 A; 4Y0Z E; 2GD4 B; 1WF7 A; 2VIV A; 2Z3E A; 4GCH F; 2K0Q A; 1V2O T; 1UEQ A; 4ACA A; 3EAJ A; 1H8E D; 2GGV B; 3A7V A; 5TCC A; 4CRG A; 1XVO A; 3ZVF A; 1YBW A; 4L8N A; 4HZH A; 3B23 B; 2KL1 A; 3A7T A; 2FOB A; 2R2W U; 2ZU5 A; 2VWL A; 4IC6 A; 4XDE A; 1BML A; 1BTZ A; 1UCY E; 3W1E A; 3F0O A; 4ZKS U; 1HIA A; 2IWO A; 1GBI A; 2B8O H; 1H1B A; 2Z9L A; 4H4F A; 1AD8 H; 1W7G H; 1V2P T; 1L6O A; 3RL7 A; 2XXL A; 2QYI A; 2BDY A; 5HGG A; 2KX4 A; 3ZYE A; 1G3D A; 2M2A A; 2BVX H; 1NRN H; 2A4G A; 2I0I A; 5KEC A; 3SV2 H; 4X2Y A; 3RML H; 1GPZ A; 3PV4 A; 1IOE A; 1QAV A; 1QNJ A; 4PQW A; 1NT1 A; 1C1U H; 4I31 A; 1F5K U; 2QF4 A; 1GHW H; 2OQS A; 1GJ7 B; 1GJC B; 1TPO A; 2HAL A; 2ZU1 A; 3LGI A; 4TWY A; 4WYT A; 1A4W H; 1LVY A; 4RT0 A; 1DSU A; 3MHW U; 3VGC C; 2FI4 E; 1RTL A; 4OS4 A; 1UJ1 A; 1O33 A; 2OGP A; 2P16 A; 5D13 A; 4X2V A; 3S9A A; 4BNR A; 2VSV A; 4NXQ A; 1FIW A; 3MNB A; 3TJV A; 2Z9J A; 3QGJ A; 2KCO A; 1TYN A; 5E1Y A; 1EB1 H; 4IC5 A; 4WMD A; 1B0F A; 4P0C A; 1ZPC A; 1XXD A; 1TNJ A; 1AKS A; 1FC6 A; 1BRA A; 1CA0 B; 1MTW A; 1MD7 A; 3FN8 A; 1HDT H; 5LPE A; 5GCH F; 2F9V A; 2DLS A; 3MU5 A; 1GBJ A; 3UWJ H; 1ZSJ A; 2BVR H; 3F9F A; 5EXN A; 3I78 A; 1S6F A; 3UY9 A; 1UJD A; 2DJT A; 1WOF A; 1GGD C; 1UEZ A; 1C5Y B; 2BDH A; 1F0U A; 1MFG A; 4LMM A; 1X5N A; 1PQ7 A; 8KME 2; 1UTL M; 3GOV B; 2FX4 A; 2EEI A; 2FES H; 2ZU3 A; 2ZDV H; 4ASH A; 3WWF A; 7EST E; 4ASU D; 1SHH B; 4O3T A; 3M7T A; 4D9Q A; 5EXM A; 1NBM E; 3DJA A; 1GBK A; 2AGG X; 4X1Q U; 3AW0 A; 2A7J A; 3KID U; 3TH2 H; 3TPI Z; 2PGB B; 3GY4 A; 4B71 A; 2ZGB H; 4UU5 A; 1WJL A; 3BTG E; 3KYF A; 1ZTJ A; 3ZZ7 A; 1C5M D; 4ZKO U; 1Q2W A; 2R2M B; 4FUJ A; 3ZYD A; 2PA1 A; 4Z0K A; 1QQ4 A; 4FOU C; 5F3X A; 3QZR A; 1BA8 B; 7LPR A; 1TP5 A; 2AER H; 3NPS A; 1VAE A; 1MMJ N; 2ZQ2 A; 1YWU A; 2OCV B; 3RXV A; 1HIA B; 2L6O A; 4XE4 A; 2LC7 A; 1CQQ A; 3ZZA A; 4DCD A; 8GCH F; 3HKJ B; 4WY3 A; 1GCD A; 1GHZ A; 3A8A A; 4P2A B; 3RDZ A; 1D9I A; 3GIS B; 1V2N T; 4X8V H; 1NU9 A; 1CSO E; 1PPH E; 3RXK A; 2H1U A; 3V0X A; 4DY7 B; 3DHK H; 1WCZ A; 2CV3 A; 4BTU B; 4FUD A; 2VNT A; 1T31 A; 2P93 A; 4IMQ A; 3L33 A; 2OQ5 A; 3RC5 A; 2WYG A; 3CBX A; 1HAO H; 1TE0 A; 3JZ1 B; 4ABE A; 1Q3P A; 2H6M A; 5TJX A; 5EOD A; 1SGI B; 1LPK B; 4H42 U; 3QTV H; 3V3M A; 3UNQ A; 1E79 D; 1QY6 A; 2ZFF H; 1SGE E; 1THS H; 1BIT A; 3ZVB A; 2A2Q H; 4FET A; 4FXG H; 3SUZ A; 1GI4 A; 3CS7 A; 1Y3V A; 1AVW A; 5AFY H; 3T26 A; 4FUE A; 1EFR D; 4NMR A; 3MU8 A; 1DX5 M; 1U6Q A; 4FUG A; 1Y7N A; 2OTV A; 5GWZ A; 2PFE A; 8LPR A; 2F3C E; 3FVF B; 3RXO A; 2D26 C; 1QR3 E; 3MH7 A; 1HXF H; 1G9I E; 4NFF A; 2H2Z A; 1CA0 C; 2JIZ D; 3UTU H; 4B2B A; 2A7C A; 5PTP A; 3MH6 A; 1QA0 A; 1VIT G; 3OYP A; 3QDZ B; 1U3B A; 4NIX A; 1O3D A; 2Y5F A; 3HVQ C; 1EJM A; 2WPI S; 1O5F H; 3CYY A; 3A8B A; 1BTU A; 3UR9 A; 2GEF A; 4JZI A; 1C2E A; 1BUI A; 2G51 A; 1DIT H; 3F1S B; 2GCT B; 2O8M A; 1GI2 A; 4IN1 A; 1XZ9 A; 1G30 B; 1JWT A; 2ZDM A; 1LO6 A; 1P2K A; 3W95 A; 2UUF B; 3BEF B; 5CHA B; 1WTG H; 3RXA A; 1NRS H; 2JSN A; 2LA8 A; 4NMV A; 1UK3 A; 2UUJ B; 1ANC A; 4XD7 D; 1P09 A; 2CHA B; 1C5R A; 1G2M A; 1XX9 A; 1Z8J B; 4WVP E; 1IU0 A; 3RLE A; 3A86 A; 3VG8 A; 1BT7 A; 4UEH H; 1T8L A; 3PDZ A; 1FY8 E; 2ZO3 H; 4WH8 A; 2XND D; 2K8I A; 2ZGX H; 2F9B H; 3PWC A; 1LMW B; 3BTF E; 3FY5 A; 2JJ2 D; 3A81 A; 1RJX B; 4WWY A; 1ARC A; 4CHA C; 1P9U A; 3U98 H; 1PPE E; 3PRB A; 4I8L A; 2AGI X; 3T5F H; 2FDA A; 3EST A; 1HCG A; 1MAX A; 2Y81 A; 5LH8 A; 1V2S T; 1V2U T; 3RMN H; 1V3X A; 1W0Z U; 1P2I A; 1UVP A; 3SUE A; 1QLC A; 1X5R A; 2SNV A; 3TIU A; 1OWJ A; 1UCY H; 1AVX A; 1RTK A; 3SUF A; 2ZPL A; 1QBV H; 4GSO A; 1XUJ A; 2J94 A; 1ZR0 A; 2FI3 E; 3KHF A; 1QB9 A; 2TBS A; 1NF3 C; 1W8B H; 3TSW A; 3E17 A; 1UTO A; 4GLY A; 2Z9K A; 1AUT C; 1Z8I B; 5FB8 C; 2J38 A; 2GDD A; 1BDA A; 4XBC A; 4YES B; 4JOG A; 2QC2 A; 4ISO A; 3VB5 A; 4I8G A; 2EI8 A; 3SV7 A; 3VXE H; 3T29 A; 1AZ8 A; 1FN6 A; 2ZPS A; 1OBY A; 1QRW A; 2EY4 C; 4Q2P A; 3E90 B; 3KF2 A; 2OP9 A; 2W6I D; 2CSS A; 5JB9 S; 1C5O H; 2JIN A; 1HB0 B; 3K50 A; 1ZLR A; 4X8T H; 3Q3K A; 1PPG E; 2EC9 H; 3F2H A; 1O3K A; 3C1K A; 1WF8 A; 4CRB A; 4ABB A; 3QLP H; 1HAZ B; 1K9O E; 3EE7 A; 4OM9 A; 2G52 A; 2ZDK A; 4GPG A; 5HM2 A; 5EQQ A; 2YMS B; 2ZCH P; 3FKS D; 2FM2 A; 3KN2 A; 4HFP B; 3NFK A; 1L4D A; 1E34 B; 1LTO A; 1A3E H; 1NO9 H; 4I8J A; 4X6P A; 2IJD 1; 4ABD A; 3GJ9 A; 1VJ6 A; 2ZHF H; 1Y59 T; 1JRT A; 1S5S A; 2GCT C; 1AUJ A; 1AWH B; 2DE9 A; 4IN2 A; 1EZS C; 4JCN A; 1O2Q A; 3WWB A; 2SGF E; 4X6O A; 5CHA C; 4K76 A; 2H9T H; 3OY5 U; 1TNG A; 4YT7 H; 1AHT H; 2CHA C; 2QAA A; 1PDR A; 3N99 A; 1LHG H; 3SQE E; 2GZ8 A; 1FCF A; 4AYY B; 2VRF A; 2Q3G A; 1T8M A; 3NRL A; 2GCH F; 2CS5 A; 3DT0 H; 2G5N A; 4X2W A; 1EAI A; 2H9H A; 1J14 A; 4Q4L A; 2ZEB A; 1QBO A; 1XKA C; 3DCL A; 2IWN A; 3UOP A; 1P04 A; 4FVB A; 1PQ8 A; 2J34 A; 2H3L A; 1BJU A; 3GDU A; 3LGU A; 4TT3 D; 3U1J B; 2IOT A; 2G4T A; 3R68 A; 2PGQ B; 1QJ1 B; 3SI3 H; 1UO6 A; 3RXT A; 2WHX A; 3G01 A; 3U28 C; 2JOA A; 1MZD A; 2VJ1 A; 2RCZ A; 1ZGV A; 4X1S U; 3ZZ4 A; 1O36 A; 1B0E A; 1I16 A; 3R3G B; 4Q80 A; 1O2H A; 2BD4 A; 5GDS H; 4JOF A; 3BTM E; 2CSJ A; 2KCY A; 5JPM H; 3UQV A; 2SGP E; 1ZJD A; 2FOD A; 2ZG0 H; 4ABF A; 1HJ9 A; 2YT7 A; 1V2V T; 4E7N A; 3K82 A; 2CGA A; 1ZHM A; 1K1N A; 1R6J A; 1GBM A; 2PKU A; 5J4Q A; 1INC A; 2W3I A; 1GMC F; 1P2N A; 4NA8 A; 1TAW A; 3LH3 A; 1OPH B; 1GVZ A; 2FLR H; 1J15 A; 5LGO A; 5E0H A; 3ATL A; 1GHY H; 3RXE A; 1UVS H; 2PKA A; 1CHO F; 3SU5 A; 2GV6 A; 2VIP A; 4OGX A; 3MWI U; 3UIR A; 1ESA A; 3UB0 A; 4OS2 A; 2KA9 A; 3TK6 A; 2OBQ A; 1GBB A; 4X2X A; 5AF9 H; 2OLG A; 4YTA A; 2OBO A; 3ZVA A; 1O35 A; 3ZRT A; 2BZA A; 2J92 A; 2BMG B; 2M5T A; 3NFL A; 4Z1M D; 1K21 H; 5HIZ A; 2PUQ H; 3SU6 A; 3LIW A; 1FZZ A; 1C2J A; 2A32 A; 2FZZ A; 1RY4 A; 3TMH A; 2AWU A; 2J4I A; 2YMS C; 3BG4 B; 4GHN A; 2TGA A; 3GY5 A; 5C20 A; 3CC0 A; 1EAT A; 5HXF A; 1O2V A; 2PLX A; 2ZGJ A; 2K1Z A; 3C27 B; 1C5X B; 1MTS A; 1O2W A; 2CK3 D; 1K28 D; 2OK5 A; 5C5O A; 1NTP A; 2ALV A; 1ABI H; 3QZQ A; 5EQR A; 2QKU A; 1FY1 A; 1NY2 2; 5JXB A; 1F0R A; 1GJ5 H; 1LCY A; 2STB E; 2M0T A; 2Q9V A; 3I1E A; 4RKO B; 5LPR A; 3OEE D; 1P1D A; 1N6E A; 4FGM A; 1O3J A; 2JWE A; 1W14 U; 2VVU A; 1L4Z A; 2BYA X; 4KTC A; 3SQH E; 2KQF A; 4REY A; 4RI0 A; 2J98 A; 4KTS A; 2CXV A; 3QO6 A; 1IU2 A; 3ZV8 A; 3RXI A; 1F2S E; 4BAM B; 2EGK A; 2EEG A; 4DUR A; 2OPG A; 4G69 A; 4QT8 C; 5HF4 A; 1CO7 E; 3RXB A; 1SPJ A; 3A89 A; 1EP5 A; 3LON A; 3MNS A; 3WKL A; 1T4V H; 4Y6D A; 1X9Z A; 5GJ4 B; 2P3W A; 3PMJ A; 4BTI B; 2OC0 A; 1P1E A; 3OTP A; 1DUA A; 5EPN A; 5BPE A; 2ZFP H; 3D65 E; 1BE9 A; 4RKJ B; 3K2U A; 1LPZ B; 4UU6 A; 4IS5 A; 4YOJ A; 2Z94 A; 4LOY H; 3RXJ A; 4B73 A; 1OWD A; 4N6X A; 1IQE A; 1PYT C; 3ODD A; 1C2I A; 1C5Z B; 1YC0 A; 5CDF E; 2R4H A; 4FU9 A; 1AU8 A; 3N7E A; 3S7K B; 1UW7 A; 3TK9 A; 3BSQ A; 1KXC A; 2HLD D; 3GAB A; 2BDC A; 2I6Q A; 3F9E A; 1ARB A; 3SNB A; 3VQG A; 4FUI A; 5HMA A; 1WQS A; 2BY8 X; 3BIV H; 5FX6 A; 2PKA B; 1W11 U; 5C3N A; 1D3P B; 1XMN B; 3PLK A; 3DD2 H; 2Y7X A; 5JBT A; 1VA8 A; 2AMD A; 3DJ1 A; 2YUB A; 1VIT H; 3P8F A; 1BTY A; 4AB9 A; 2ZDA H; 3BTD E; 4F55 A; 4AG2 A; 1MBQ A; 1N8O B; 1GG6 B; 1FY5 A; 1T7C A; 2F0A A; 1O2U A; 4CH2 B; 1A46 H; 2HPQ H; 3RXR A; 1GI3 A; 4XFQ A; 5L2Y H; 1RD3 B; 1E1Q D; 1BHX B; 2YTW A; 5EYZ A; 1CGI E; 2BVS H; 2V3M A; 1YCP M; 1GMD G; 2B7D H; 2H3M A; 1UVT H; 1BRU P; 5TCA A; 1F0T A; 4CRA A; 4INL A; 1UMA H; 1P02 A; 2HVX A; 1FPH H; 1YCP K; 1FAX A; 3E16 B; 2EV8 A; 4NZQ A; 1N6D A; 1G3B A; 1M9U A; 1UM7 A; 1DPO A; 3GY3 A; 2KPL A; 1BBR K; 1WIF A; 1A5I A; 1C2F A; 1MBM A; 4MPW A; 1YPH C; 4MNY A; 2A1D B; 5HFC A; 4JK5 A; 2STA E; 2WV4 A; 2K20 A; 3T28 A; 2RDL A; 1ELT A; 1HPG A; 2J98 B; 4BAO B; 2VIW A; 3IWM A; 4F49 A; 4B6F A; 3PM3 A; 1MQ6 A; 5F03 A; 1EQ9 A; 1SOZ A; 2YT8 A; 4JOJ A; 1L2E A; 1YY3 A; 4FUF A; 3RLW H; 1P57 B; 3ZVD A; 4Q7X A; 4FVD A; 1QJ6 B; 1UHB A; 1V1T A; 3ZZ9 A; 4F48 B; 2ZHE H; 1SLW B; 2HE4 A; 3K9X B; 2V0B A; 2IPQ X; 4AGK A; 1O2X A; 1C1W H; 4NNL A; 2R0K A; 2YNA A; 1O2I A; 3TK5 A; 2OZF A; 4MQA A; 1V2R T; 1TFX A; 3F9H A; 2D8W A; 2HE2 A; 5E8E H; 2NU3 E; 1HAV A; 4H6S B; 3ID4 A; 5GND A; 3GDS A; 1KLI H; 1O3N A; 3SOR A; 2F9U A; 2GDE H; 1BTP A; 1GDQ A; 1TGB A; 2M10 A; 2QF0 A; 2ZHW H; 4GHQ A; 4XBD A; 2FOF A; 1X5Q A; 1GL1 A; 2RA3 A; 3BTQ E; 3A84 A; 3H7T A; 5A8X A; 3D62 A; 1UF1 A; 3STJ A; 4E34 A; 2OC1 A; 1EB2 A; 2GTB A; 4THN H; 3KQC A; 1DFP A; 1I92 A; 1C9T A; 5GIB A; 3AAV A; 3SV8 A; 1CT0 E; 2AYW A; 1Z87 A; 1EJN A; 1TB6 H; 2GV7 A; 4ZXY H; 3PLP A; 1V5L A; 1XVM A; 1S83 A; 2GZ7 A; 1MKX H; 3VEQ B; 1AWF H; 1P12 E; 1O5G H; 2RA0 A; 1HUT H; 4TPY A; 1HJA B; 4Y0Y E; 1RGQ A; 1S82 A; 2ZGC A; 1MTN B; 1AB9 B; 5JB8 S; 1JOU B; 1ELA A; 1RGW A; 1Q7X A; 2DM8 A; 2UWP A; 2WPL S; 4KFV A; 1DE7 H; 1KYN A; 2XOK D; 3UOU A; 2QBW A; 1KSN A; 2A5K A; 3RU4 T; 1N8O C; 1GG6 C; 4UE7 H; 4AG1 A; 1P2O A; 3GY8 A; 3M7U A; 5HEY A; 3TGJ E; 5T6D A; 4U32 A; 5DGJ A; 5HJ1 A; 3Q3X A; 1OYQ A; 5C0T A; 1P05 A; 3QE1 A; 1GI5 A; 1QGF A; 3HK6 B; 4FUH A; 3RC4 A; 2ZEC A; 1SL3 A; 4EST E; 2BDI A; 5IYT A; 1OP8 A; 4YT6 H; 2B0F A; 4AMH A; 2FLB H; 4XOJ A; 5FPT A; 4YDP A; 4YYX A; 4K2Y A; 1H8H D; 1FN8 A; 1ELG A; 2ANM H; 3EGK H; 1GQ5 A; 1ZTK A; 3EDX B; 1KMH B; 3URE A; 1BHX F; 4ABA A; 1A0H B; 1EZU C; 1TNH A; 3TJN A; 2FOM B; 1YPL H; 3LOX A; 4K8Y A; 5DKM A; 2FPZ A; 1TMB H; 3U1I B; 3Q3Y A; 3TSV A; 5A0C A; 2XTT B; 2BM2 A; 1O3M A; 1IHS H; 3TGK E; 2P8O B; 1RRK A; 2G2L A; 1Q3O A; 3F9G A; 1SG8 B; 2VID A; 1KEF A; 1ELD E; 4Y79 A; 1UK4 A; 1UHB B; 4UFG H; 2O9Q A; 1OWK A; 4B2A A; 2NU2 E; 4Y10 E; 2A4R A; 1WXR A; 1YF4 A; 2XYA A; 2RDE A; 4AFS A; 1P11 E; 1P03 A; 1LDT T; 3A83 A; 3SZE A; 3OFN D; 1HAV B; 5L6N H; 1MFL A; 1MUE B; 3KL6 A; 3ZZ3 A; 2NU4 E; 1Z71 A; 2AIQ A; 3BN9 A; 4ISL A; 2PUX B; 1O3P B; 3URC A; 5H3J A; 4DVA U; 1NPM A; 2BLO A; 4OS7 A; 1BJV A; 1UTN A; 1HNE E; 2VGC B; 3CS0 A; 4GCH G; 1D5G A; 4R8T B; 1BIO A; 4ZXX H; 1TX6 A; 1F7Z A; 3SUD A; 1GI0 A; 2ZWL H; 3SHC H; 4U30 A; 1L1N A; 1C1Q A; 3M5O A; 1SGQ E; 1KDQ B; 3KNX A; 2OCS A; 4NA9 H; 1HJA C; 2D1J A; 3ZVE A; 5EQS A; 4JYT A; 3UR6 A; 5HFF A; 3K1R A; 4I8H A; 5E6P B; 5IL9 A; 5ESB A; 4OEO A; 2FXR A; 1CVW H; 1RXP A; 1FMG A; 2Y6T A; 4YLU A; 1SGR E; 4HI3 A; 4I8K A; 4Q2O A; 2XW9 A; 2G55 A; 2AH4 X; 3NZI A; 4ISN A; 3M37 A; 3OX7 U; 4ABG A; 1SVP A; 3UAI C; 1M5Z A; 1K1O A; 5J4S A; 1TPP A; 4EDJ A; 2OC7 A; 3SND A; 1YPH E; 4CR9 A; 1JRS A; 2TGD A; 1DXP A; 2VH0 A; 5C67 A; 3SJO A; 2THF B; 1SQT A; 3A7Y A; 1QBN A; 2Q6D A; 4WJG 2; 3S0N A; 2MX6 A; 1BB0 B; 4X6M A; 3A7Z A; 1SC8 U; 1V5Q A; 1EPT A; 4K1T A; 2OMJ A; 4OGY A; 3TNS A; 4JYV H; 2AWX A; 4MNX A; 4YZU A; 5ETX A; 3SGQ E; 3SNE A; 1C5U A; 2KOJ A; 1AQ7 A; 1NN6 A; 1XKB C; 5DP5 A; 3QTO H; 5JDU B; 1AIX H; 1GHB F; 1ZHR A; 1H9I E; 3DA9 B; 4I7Y H; 4O97 A; 5DG6 A; 4AYV B; 1QAV B; 3TGI E; 2FOH A; 3MOC A; 5CMX H; 3RXU A; 4INK A; 2I6V A; 2ANY A; 2GKV E; 4E05 H; 4LK4 A; 2HGA A; 1NRO H; 1QTF A; 4IBL H; 1DWE H; 1U37 A; 3R69 A; 1KXF A; 3TJO A; 2P8O C; 1ZPB A; 2OQU A; 1GJ6 A; 1PPZ A; 3CH8 A; 4XRN A; 2BY6 X; 1Q3X A; 3KGP A; 2EQN A; 5GCH G; 3RU4 D; 2RFK C; 1NFU A; 1MD8 A; 1UTJ A; 1K1P A; 3MI4 A; 4ZRO A; 3BIU H; 4AZ2 B; 1DIC A; 5CE1 A; 4WXI A; 2QXH A; 5DP4 A; 5GLJ A; 4JMY A; 3DSO A; 2BX3 A; 2GX4 A; 1UM1 A; 1O32 A; 1SLV B; 1C2H A; 2ZDN A; 1VCP A; 3ATK A; 3A80 A; 1ID5 H; 3GCT F; 5AHG H; 1AVG H; 1AKS B; 1TNL A; 5FPS A; 4UDW H; 4AFU A; 1TGS Z; 4NZL A; 2ZPM A; 4Q2N A; 1UTQ A; 3MO9 A; 3LGW A; 4GHT A; 3BEI B; 4KKD A; 1BTX A; 2W7R A; 1C2L A; 2ASU B; 5K0H A; 2G5V A; 3O46 A; 4FU8 A; 4KTU A; 5DO4 H; 7KME H; 1KXB A; 5EJL A; 2Z6B D; 3F6U H; 3MH5 A; 3FP6 E; 3NK8 A; 5L2Z H; 1DM4 B; 1NM6 A; 5C1U A; 1O3O A; 1ZGI A; 4HOP A; 2I6S A; 1LVB A; 1TBZ H; 2KAW A; 2ZHD A; 3HK3 B; 1IQF A; 1N1L A; 1GHX H; 1K32 A; 2PTN A; 1FJS A; 3LKW A; 4J2Y B; 3SU3 A; 1SGC A; 1C5V A; 1W12 U; 4BAN B; 3Q77 A; 4NXP A; 1TAB E; 1V2Q T; 1YP9 A; 1C1N A; 2C8X B; 4Q6S A; 8GCH G; 1UTK A; 3RXD A; 3AK5 A; 2F9P A; 3K6Z A; 2H9E H; 4KP0 A; 3SI4 H; 5DP3 A; 2EDV A; 5JBC S; 2EST E; 1BTW A; 1W0K D; 1G36 A; 3E8L A; 2WYJ A; 1EAX A; 2WUB A; 3V13 A; 3PV2 A; 4JK6 A; 3O8C A; 4A6L A; 2L4S A; 2EA3 A; 5HIY A; 1O5B B; 1UJU A; 1MH0 A; 3L3T A; 2EDZ A; 3GGE A; 3NUM A; 2KR7 A; 6CHA B; 4KGA A; 5C17 A; 1IQH A; 1QL8 A; 3ZZ6 A; 5E21 A; 1F92 A; 5K7R A; 1Y5U T; 2W5E A; 1WQV H; 3HKI B; 3PTB A; 1AE5 A; 1EPT B; 2VH6 A; 5EST E; 4WME A; 2QKV A; 2GZ9 A; 4X1P U; 3D23 A; 1GJ9 B; 1AI8 H; 2J5U A; 1QB6 A; 3FFG A; 1C1M A; 1ZMN A; 4WH6 A; 1DLK B; 1BZX E; 2WSS D; 1IQI A; 2F4L A; 2IJO B; 1NFY A; 3PO1 B; 5C1Y A; 3RP2 A; 3QGL A; 1GBE A; 4K75 A; 4I86 A; 3ZIA D; 2QN5 T; 1HYL A; 2ZI2 H; 3M3V A; 4B2C A; 1ELB A; 3RXG A; 3MTY A; 3SHA H; 1V2J T; 1Y5B T; 2PTC E; 3MU1 A; 4X6N A; 1UHP A; 3V12 A; 4CR5 A; 5F53 A; 5HKK D; 1A5G H; 1AZZ A; 1Z8G A; 1S6H A; 1W3C A; 2WPH S; 3MO3 A; 1GJB B; 3TVJ B; 2ZNK H; 4JZE H; 1GMH F; 3VXF H; 1VKY A; 3I29 A; 1WU1 A; 3VPK A; 3GCN A; 1KTT B; 4BAQ B; 1LHE H; 5KED A; 2G8T A; 2QT5 A; 2LEL A; 3PMA B; 1BBR E; 1IQG A; 1GQ4 A; 1HAH H; 2BX4 A; 5EIY A; 2HRV A; 1PPC E; 2KZ4 A; 2BYG A; 1X45 A; 4YM9 A; 1C5Q A; 3KQD A; 4NWL A; 2KBS A; 4NMT A; 3SGB E; 1WTH D; 2VWO A; 5JYI A; 1O5D H; 2PW8 H; 1OP2 A; 3TIT A; 1A0J A; 4SGA E; 3SOS A; 2PSY A; 3RXP A; 1O3L A; 1O2K A; 2PP6 A; 5HED A; 1EKB B; 4Y7A A; 2ZF0 H; 5EGM A; 3A85 A; 2EEH A; 5I25 A; 2EJY A; 1HXE H; 1FY3 A; 1DY8 A; 3LC5 A; 3PMH B; 5CHB A; 4MVN A; 1DS2 E; 2G00 A; 1YPE H; 3P17 H; 1TBR H; 3I4W A; 3RMM H; 2WPJ S; 3JXT A; 2WL7 A; 4JZD H; 4UD9 H; 3LNX A; 2HWL B; 2G5M B; 2AHM E; 2VWM A; 4FUB A; 1VR1 H; 1TRY A; 1TMT H; 1HAI H; 1TNK A; 1MU6 B; 3PLB A; 1Y3U A; 2DUC A; 4I32 A; 4A5T S; 1THR H; 3MFJ A; 1ORF A; 2ENO A; 3ZZ5 A; 2GT8 A; 4NMO A; 2Q1J A; 1OS8 A; 1WFV A; 4Q2Q A; 2VB0 A; 3STI A; 1O2G H; 6CHA C; 5T1V A; 3OTJ E; 3TSZ A; 1DWD H; 5DP7 A; 3FAN A; 3CBY A; 1IQJ A; 1ELC A; 4WMF A; 1EPT C; 1TGC A; 4ISI H; 2GP9 B; 2Y5H A; 4UFE H; 4JYU H; 3KQB A; 1UK2 A; 1L1G A; 2BDB A; 3MU4 A; 1GI8 B; 3KHV A; 1WSS H; 3BTT E; 2LUI A; 3PO1 C; 1D3D B; 2GJG A; 2VWN A; 3CP7 A; 4KFW A; 3KQE A; 2R0L A; 1Z1I A; 2VVC A; 5EZ0 A; 1OX1 A; 2WV5 A; 3P95 A; 1T8O A; 4AFQ A; 3SU1 A; 1S0R A; 2ZU2 A; 5HDY A; 1NTE A; 1C1O A; 1SQO A; 1BFE A; 4K6Y A; 1C2G A; 4Y8X A; 2G81 E; 5DSF A; 4HTC H; 1SQA A; 2A2X H; 2EEJ A; 2A0Q B; 2O8T A; 3ATM A; 4RN6 A; 2VWR A; 3TU7 H; 5E11 A; 1KTS B; 1ZTL A; 1TBQ H; 3MNC A; 1IQK A; 2Y5G A; 1N6Y A; 2XC0 A; 4NWK A; 3HAT H; 3URD A; 1TPS A; 3M5N A; 2KJP A; 2EI6 A; 4Q7Z A; 1O37 A; 4TWW A; 1O31 A; 3SYJ A; 2JKH A; 1EAS A; 2GCH G; 3E6P H; 1VJA U; 1GJ4 H; 4BAH B; 1WYK A; 1NBM D; 4M7G A; 4X8U H; 1WHA A; 1NES E; 3UWI A; 1J16 A; 2ZPR A; 2KYL A; 1O0D H; 1QA7 A; 2P3U B; 1O3A A; 4CBN A; 1QFK H; 1XXF A; 4K1S A; 1Z8R A; 3SZN A; 1J9C H; 2ALP A; 1DVA H; 5A8Y A; 2NU0 E; 2ZPQ A; 5FX5 A; 1ZJK A; 1O2J A; 2IWQ A; 3U8T H; 1N99 A; 1U38 A; 3VB7 A; 2J2U A; 1QZ8 A; 1TQ3 A; 6GCH F; 1QQU A; 1ETR H; 2ZQ1 A; 3L6P A; 1O2Y A; 4JZ1 A; 4NG9 H; 3A87 A; 3VFE A; 2JIL A; 2VIQ A; 2RRM A; 6LPR A; 4M9F A; 1HAY B; 1Z6J H; 3MO6 A; 1N7E A; 1H9H E; 2R9P A; 9LPR A; 2HNT F; 4ZH8 A; 5EJ1 A; 1O3E A; 1GBC A; 3QDO A; 2JET B; 1IQL A; 1LHD H; 2OD3 B; 4NMP A; 1GMC G; 3M3T A; 2VBC A; 2YUY A; 1BCU H; 1C5W B; 5F8T A; 1CHO G; 2Y80 A; 2HVY B; 7GCH F; 6EST A; 2DC2 A; 2H5C A; 2KFW A; 2Y82 A; 3B9F H; 3ZZB A; 3TNT A; 2AWW A; 5HM9 A; 2V7Q D; 4MLF B; 5C2Z A; 1RGR A; 1ANB A; 3LU9 B; 1DOJ A; 5A8Z A; 3B8J A; 3HPT B; 2BD2 A; 1QJ7 B; 2TPI Z; 1COW D; 2VSP A; 4D76 A; 2AS9 A; 3SGA E; 1GDU A; 3QK1 A; 4AN7 A; 1YLD A; 2UZC A; 2M0U A; 2LPR A; 1FDP A; 1GWA A; 1S0Q A; 1ZMJ A; 4DOQ A; 1TRN A; 1P3E A; 5ILB A; 1O2O A; 5G4B A; 1WUN H; 1GJA B; 2EGO A; 3QWC H; 1EZX C; 2KM0 A; 4X0W U; 4Y11 E; 2ZHQ H; 3M3S A; 1DST A; 2NWN A; 3UPE A; 3PB1 E; 5C1X A; 1CGJ E; 1J8A A; 1GHA F; 4NA7 A; 3WY8 A; 3QJM A; 1BMM H; 1YPG H; 3M35 A; 2FP7 B; 5HFE A; 1HRT H; 4ZCN A; 3U8O H; 1MCV A; 4NSV A; 1HAP H; 3TH3 H; 4NXR A; 1SBW A; 2YOL A; 3BG4 C; 2KAI A; 3KQ5 A; 1FLE E; 3RM0 H; 1O5A B; 1C5S A; 5EW1 H; 1XUI A; 1J17 T; 1Y3Y A; 1YCP H; 1SGT A; 4NFE A; 1W0J D; 3S9C A; 2A7H A; 5HYO A; 2BLV A; 3QGN B; 5E2O A; 1BBR H; 2BLQ A; 2ZIQ H; 2A4O A; 2BY7 X; 3LDX H; 1WHD A; 1QHR B; 3SFJ A; 3RLY H; 1WFG A; 4TY6 A; 2W3K A; 1A61 H; 1GI6 A; 4OS1 A; 2AEI H; 4I33 A; 2FI5 E; 1DY9 A; 1YKT A; 1AFE H; 3W94 A; 2ZGH A; 1C1P A; 1EJA A; 5SGA E; 1CT4 E; 1TWX B; 2FYQ A; 3OAA D; 3SU0 A; 3HGN A; 2QF5 A; 1H9L B; 3EGH C; 3PMB B; 1SOT A; 2Z9G A; 2C3S A; 2SGA A; 3SU2 A; 2ZCK P; 3MYW A; 3KDK A; 2XRC A; 3Q76 A; 5HF1 A; 1DAN H; 2F43 B; 2PKT A; 5FPY A; 4B74 A; 3R0F A; 7GCH G; 5B42 A; 2QV1 A; 3WKM A; 4DII H; 2FS8 A; 1SGP E; 4MPX A; 3SJ9 A; 4BXW A; 4YXW D; 2W26 A; 1K1M A; 4B1T A; 1TX8 A; 2VVV A; 3PDV A; 2UWO A; 2EHR A; 3NGH A; 2P3F H; 2CJI A; 5H4I B; 4ABH A; 1O39 A; 1C5T A; 2EEK A; 3PS4 A; 3FOA A; 1GBT A; 2FX6 A; 1O2T A; 3QUM P; 3SJI A; 1DWC H; 3ID1 A; 2XCN A; 3P8G A; 1O38 A; 1V2L T; 1GL0 E; 2EXG A; 1X7A C; 5FBI A; 1P2M A; 1GHA G; 1TGT A; 1W10 U; 1SGD E; 1MCT A; 1FV9 A; 3DSG A; 4D7G A; 3QJN A; 1Q31 A; 1C2M A; 5GPI B; 2V6N A; 1FXY A; 3EE0 B; 1C5L H; 1EOJ A; 5ABW A; 2DLU A; 5EPY A; 1Z1J A; 3F0P A; 3RXQ A; 1SSX A; 1O2Z A; 2AMP A; 3AAS A; 1OHH D; 3M5M A; 1NC6 A; 1C1R A; 1GBF A; 1KXE A; 3AW1 A; 5KGO A; 3P8O A; 5F8Z A; 2KV8 A; 2OC8 A; 1C2D A; 1YPK H; 4BTT B; 4E3B A; 2ZKS A; 1EST A; 1MAB B; 3SUG A; 1E1R D; 4ZMA H; 1AY6 H; 2XCF A; 5EG4 A; 1HFD A; 5I7Z A; 1O2L A; 3HPK A; 1BMN H; 4GVC A; 4OEP A; 1K1I A; 3QN7 A; 2TGP Z; 2ULL A; 3NWU A; 1GGD B; 4CH8 B; 2JIK A; 1ELV A; 2BDG A; 3SOE A; 1YLC A; 2AIP A; 4TY7 A; 5DPA A; 1IHJ A; 2KAI B; 3CEN A; 5B6L A; 3BTK E; 3E3T A; 4JOP A; 4U01 A; 3A82 A; 1DXW A; 1P0S H; 2D90 A; 1PJP A; 2JJ1 D; 1X8S A; 4E35 A; 1PQ5 A; 1B8Q A; 2P3T B; 1FC9 A; 1AFQ B; 1CBW B; 2BD9 A; 4ACB A; 1OP0 A; 1NRQ H; 5EXL A; 1MKW K; 3DOR A; 3KZE A; 1HAX B; 1ZSK A; 3ZVC A; 1C1T A; 1D3T B; 1OWH A; 4NIV A; 1VCQ A; 2FOE A; 2QA9 E; 1A0L A; 1O2N A; 4NGA H; 2HOB A; 1Z86 A; 1NS3 A; 2BY5 X; 3LJO A; 2FNE A; 2M3M A; 2O8L A; 1LVM A; 2I1N A; 2BY9 X; 2DMZ A; 2GT7 A; 3KZD A; 2ZFT A; 1YBO A; 1KIG H; 4ZHM U; 3DUX H; 1OWI A; 4AOQ A; 1SLU B; 1EZQ A; 2CN0 H; 4JJ0 A; 3MU0 A; 2FIR H; 3CBZ A; 1Z7K A; 1MTV A; 1SGY E; 1FX0 B; 3RXH A; 1GJ8 B; 1AGJ A; 4DT7 B; 5JBA S; 1HAG E; 2JXO A; 4DJZ B; 1O3B A; 3OEH D; 4K69 A; 5F67 A; 1YYY 1; 3H7O A; 2QGR A; 3SW2 B; 4K78 A; 1OYT H; 2V3H H; 3S7H B; 1TAL A; 2BQ7 B; 1C1S A; 1CHG A; 3LH1 A; 2A45 B; 1S6L A; 4MNV A; 2QKT A; 1FC7 A; 2Z3C A; 2ZCL P; 3F2G A; 3VB6 A; 3PV5 A; 1F0S A; 2KJK A; 2H2C A; 4YOG A; 1O3H A; 2L97 A; 1ELF A; 2IWP A; 4MG3 A; 3IIT A; 1LPG B; 1EX3 A; 2L74 A; 1O2M A; 2BXU H; 1YPM H; 2JRE A; 2QIQ A; 4CBO A; 1MTN C; 1AB9 C; 3FP7 E; 1T4U H; 3HGP A; 4CRD A; 1MQ5 A; 2C8Y B; 1ACB E; 2ZA5 A; 3MNX A; 5EW2 H; 2BLW A; 2FOG A; 3S9B A; 1LKA A; 2TGT A; 1W0Y H; 1YGC H; 2GMT B; 3GS9 A; 1ZOM A; 1TMU H; 4A1V A; 1ZSL A; 1QUR H; 2C93 B; 1QL7 A; 4ZUH A; 5FAH A; 4GVD A; 4ZKN U; 1VCW A; 5E22 A; 1DDJ A; 2BOH B; 1SFQ B; 4IMZ A; 4D8N A; 3NXP A; 1EXF A; 2ZFS A; 2ZFQ H; 5E2P A; 3A7W A; 3FZZ A; 1VIT F; 1PPB H; 5C0U A; 1ANE A; 3RM2 H; 4IGD A; 1VGC B; 4DGJ A; 5EOK A; 4BOH A; 2TRM A; 1WV7 H; 1FON A; 1RZX A; 1UFX A; 3R0H A; 1AFQ C; 1CBW C; 4A1T A; 3GSL A; 1Y3X A; 2F5Y A; 4RSP A; 1TGN A; 2BD5 A; 5DP8 A; 3DJ3 A; 2NU1 E; 3A8D A; 4P00 A; 3PTN A; 5HFB A; 2PKS B; 2WUC A; 4CHA B; 1D3Q B; 1MU8 B; 1N7F A; 4FUC A; 2KOM A; 1A5H A; 1AE8 H; 3SV6 A; 1NRP H; 1RIW B; 3GY6 A; 1E0F D; 3NCV A; 3UNS A; 1W7X H; 1BTH H; 5EJZ A; 5FHT A; 3ID2 A; 2FEQ H; 4O9V A; 1C2K A; 4XBB A; 4A92 A; 8EST E; 3AVZ A; 1N7T A; 2JDI D; 1ZUB A; 1FAK H; 1G32 B; 1HJ8 A; 2TIO A; 2ZDL A; 2I04 A; 2PDZ A; 5FXL A; 2SGD E; 1OXG A; 4MTB A; 1GHV H; 1Y5A T; 1TON A; 2F83 A; 1XUG A; 3ZZC A; 2XBW A; 4AB8 A; 2PZD A; 1SFI A; 2PSX A; 1NFW A; 3LC3 A; 1FQ3 A; 1V2T T; 4DG4 A; 4KEL A; 1LQD B; 5AVD A; 3N7O A; 4Q2K A; 3ZV9 A; 1QB1 A; 3V7T A; 1ZZZ A; 1G2L A; 1VB7 A; 1GBL A; 1MAY A; 1PQA A; 4UFF H; 4VGC B; 4X1R U; 4O06 A; 3VB4 A; 3GYM A; 2DKR A; 1O5E H; 2WPK S; 3PWB A; 4MPU A; 4AOR A; 3H5C B; 1EAW A; 4BXS A; 3GY2 A; 1HGT H; 1SI5 H; 1YPJ H; 4SGB E; 2X7Z A; 3EYD A; 2EI7 A; 2M0Z A; 2QXI A; 3TH4 H; 2VIO A; 1UEP A; 3I18 A; 1T32 A; 4LZ4 B; 2JH0 D; 2VGC C; 2ZU4 A; 3O8D A; 1KY9 A; 1X6D A; 3L4F D; 3LGV A; 3PRD A; 1OKX A; 2G4U A; 4R2Z A; 1WAY B; 4D7F A; 1P9S A; 3QIK A; 3PRO A; 2JH6 D; 2FS9 A; 1QCP A; 1L0Z A; 4F4O C; 4U2W A; 5B6O A; 4Y8Y A; 2EGN A; 1P10 A; 2QG1 A; 1V2W T; 1TPA E; 4UFD H; 3EA7 A; 2PR3 A; 1SHY A; 4H6T A; 4JOR A; 1TQ0 B; 1H4W A; 1GBH A; 1H8I H; 1FNI A; 4B76 A; 2C8Z B; 2KRG A; 1ZRB A; 2GMT C; 3E0N B; 3H09 A; 4Y7B A; 1TOM H; 3U8R H; 2BTC E; 3Q00 A; 3M5L A; 2P94 A; 1TA6 A; 3KEE A; 1D6W A; 1FUJ A; 1MC7 A; 2W7S A; 3LT3 A; 1P06 A; 2ILN A; 3M7Q A; 2BZ6 H; 5DK1 A; 1SR5 C; 4NSY A; 2FCF A; 2A5I A; 4PRO A; 1VGC C; 2LP6 A; 1RFN A; 1GHB G; 3U69 H; 2Z7F E; 1S84 A; 4LXB H; 1ETT H; 2HLC A; 1G9O A; 1SGF A; 2FO9 A; 3T2N A; 1PPF E; 2I4S A; 3T27 A; 1A2C H; 4RT1 A; 1KXD A; 4WXV A; 2GZV A; 5F6M A; 1EAU A; 1TA2 A; 1TOC B; 5DTH A; 1K22 H; 2R3Y A; 2PKS C; 2KPK A; 3KDG A; 2KJD A; 4ABJ A; 1SKY E; 2QY0 B; 3GCO A; 4DT4 A; 2BDA A; 1RIW C; 2KCP A; 4Y76 A; 2ZTX A; 2O2T A; 2Q6G A; 1D4P B; 3GCH B; 1RS0 A; 2FN5 A; 2VZ5 A; 3GCT G; 1LKB A; 2V35 A; 3P6Z B; 3OU0 A; 3JZ2 B; 3UUZ A; 5LPF A; 1KLJ H; 3SBK A; 5DJ7 A; 5A09 A; 1W13 U; 1IAU A; 1P3C A; 4JJ3 A; 4Z33 A; 1UCY K; 4AFZ A; 1WI4 A; 1HV7 A; 4DW2 U; 3SJ8 A; 1V6D A; 2CMY A; 2BQW B; 3ITI A; 1XM1 A; 1WG6 A; 3SJK A; 5I46 H; 1VJ9 U; 3VB3 A; 4H11 A; 2F9O A; 4NMS A; 1LQE A; 3BF6 H; 4VGC C; 1K1J A; 4IW4 E; 1JXP A; 1THP B; 1V6B A; 3EQ0 H; 4CRF A; 4Y8Z A; 2YMS D; 2XBY A; 3SO3 A; 3S69 A; 3D49 H; 2VPH A; 5C94 A; 1YLN A; 3OE7 D; 3RUO A; 5A0A E; 3WW7 A; 4JOK A; 4GAW A; 3F3B A; 1GI7 B; 1JBU H; 4NMQ A; 3UNR A; 4X8S H; 4A7I B; 1G3C A; 1AO5 A; 1O3F A; 2QXG A; 1WBG B; 1A3B H; 3I77 A; 2XBV A; 4HOP B; 2L4T A; 3E0P B; 2EDP A; 1OZI A; 1GVK B; 1IHT H; 1O34 A; 1UIT A; 3DFL A; 3FP8 E; 4B6E A; 1DWB H; 2OS6 A; 4K60 A; 3SN8 A; 4UON A; 2SNW A; 3RXL A; 3E91 A; 1P8V C; 3SNC A; 1XUH A; 4XHV A; 4JL7 A; 3K6Y A; 4BAK B; 1ESB A; 2XC4 A; 4FLN A; 1DUE A; 1TQ7 B; 1CU1 A; 2SGQ E; 3RXS A; 4ISH H; 4INH A; 5T6G A; 2KG2 A; 2FOC A; 3MQK C; 1EOL A; 2KCA A; 3BTW E; 4OAJ A; 2BB4 A; 1NFX A; 1OSS A; 2BD8 A; 3RC6 A; 1IQN A; 2HGT H; 2P95 A; 4YO9 A; 4CRE A; 3B76 A; 5L30 H; 2BQ6 B; 1G3E A; 3FAO A; 1G37 A; 2SFA A; 4P02 A; 1V2K T; 1IQM A; 4YLQ H; 4AX9 H; 2XBX A; 3DFJ A; 4TYD A; 1JMO H; 2W6J D; 4O03 A; 1W9O A; 5DP6 A; 1OBZ A; 4NIW A; 9EST A; 3ATW A; 3P8N A; 1C4V 2; 1EUF A; 1PYT D; 3O8B A; 1SLX B; 1C5P A; 1B5G H; 2FMJ A; 3LGT A; 4JOE A; 2H5D A; 2OSG A; 3KCG H; 1SMF E; 1GMH G; 5AFZ H; 3FZD A; 3GCH C; 2A5A A; 3GYL B; 3TLO A; 4ZHL U; 4XSK U; 1WH1 A; 1FPC H; 2IPH A; 4Q7Y A; 4ABI A; 2ZTZ A; 3F2F A; 2WV9 A; 1WDI A; 2M0V A; 1ZPZ A; 1ZML A; 2FYR A; 4JZF H; 2PWX A; 2ODP A; 2PHB A; 2J95 A; 1UTP A; 2SGE E; 2H2B A; 3SV9 A; 1S85 A; 2FE5 A; 2OXS A; 1E35 B; 2Z3D A; 3ENS B; 4GUX A; 1A7S A; 1Y3W A; 3E1X B; 3OSY A; 1O2S A; 1UEW A; 2GVF A; 1D6R A; 2Y7Z A; 5T6F A; 5A0B A; 1NZQ H; 1VZQ H; 5IDK A; 3T62 A; 3M36 A; 5F8X A; 2D92 A; 1A1R A; 1FY4 A; 2RCE A; 3F68 H; 2O8W A; 1WI2 A; 1E36 B; 1TIO A; 2UUY A; 2ANK H; 1O2P A; 1NU7 B; 1MTU A; 2C8W B; 3FO8 D; 3MMG A; 3T25 A; 2BXT H; 1GDN A; 2FWW A; 4E7R G; 1TNI A; 1C4U 2; 3GIC B; 4R0I A; 2HNT C; 1FIZ A; 5GXP A; 2QE7 D; 1K2I 1; 4DIH H; 2AMQ A; 5E0J A; 4Q3H A; 4X1N U; 1C9P A; 2O8U A; 2ZZU H; 1O3I A; 1DT2 A; 1UVU H; 2CF8 H; 1CGH A; 3UQO A; 2LNC A; 2PV9 B; 4MDS A; 4K8B A; 2QCY A; 2V3O H; 2XWB I; 1TRM A; 4ZKR U; 4A1X A; 4Z6A H; 2VU8 E; 2VIN A; 2J97 A; 2AGE X; 2DB5 A; 3M61 U; 1GVL A; 3SHW A; 1GM1 A; 1F5R A; 4AGJ A; 1GCT B; 2B5T B; 4K72 A; 2OUA A; 4K3J A; 1O30 A; 2XWA A; 2BOK A; 1ZYO A; 2C4F H; 1P01 A; 1BMA A; 1GMD F; 2W7U A; 2WIN I; 3TJU A; 1BQY A; #chains in the Genus database with same CATH homology 1F5L A; 3BPU A; 1K1L A; 4JOH A; 1ETS H; 2IN2 A; 1HLT H; 1T8N A; 1GBA A; 4F8K A; 2BHG A; 2D8I A; 2Z9I A; 1XUF A; 1YM0 A; 1DLE A; 4J1Y A; 2DE8 A; 1OWE A; 3ZVG A; 3RMO H; 5FCK A; 1ABJ H; 1FI8 A; 4OS6 A; 1N6F A; 2Z17 A; 4WF8 A; 5DP9 A; 5HFD A; 3BG8 A; 3LGY A; 2A4Q A; 1XUK A; 2PNT A; 5E0G A; 5G1D A; 2JH5 D; 3SNA A; 1Y8T A; 1LHC H; 3A8C A; 1P2J A; 2P59 A; 2A31 A; 4NCY A; 3LJJ A; 3GDV A; 3EA8 A; 1TX7 A; 3RU4 E; 4D9R A; 2UWL A; 1AMH A; 1E38 B; 2ZP0 H; 5C5N A; 3LNY A; 1CT2 E; 3VQF A; 2V90 A; 5FBE A; 2ANW A; 2F91 A; 2K1Q A; 3ZZD A; 1C4Y 2; 1ZRK A; 2REY A; 2CF9 H; 2ODY B; 3MH4 A; 3EGG C; 5A2P A; 2MHO A; 5FCR A; 1O3G A; 1UTM A; 2FTM A; 2FOA A; 1W9Q A; 5EU8 A; 1E37 B; 4NIY A; 2BD3 A; 5G1E A; 4Q6H A; 2UUK B; 1QRZ A; 1GI9 B; 1QL9 A; 3ZZ8 A; 3BEU A; 3PYH A; 2HPP H; 1N6X A; 2D2D A; 2DAZ A; 2ZFR H; 3P92 A; 1KWA A; 1ZOK A; 4FU7 A; 5A2M H; 4Z8J A; 3ELA H; 3DIW A; 4Y71 A; 3AAU A; 6GCH G; 4OS5 A; 1QIX B; 1AND A; 1P2Q A; 4TPI Z; 4MPA A; 1Z6E A; 2ZTY A; 4LZ1 H; 1C1V H; 1B7X B; 4ZHA A; 3ID3 A; 1NRR H; 1S81 A; 5BRR E; 1SB1 H; 4MNW A; 2RG3 A; 3QX5 H; 4E06 H; 1O2R A; 4YOI A; 2YNB A; 2J9N A; 2WPM S; 1KXA A; 4O3U A; 3BV9 B; 1UVO A; 1JIR A; 2AFQ B; 1C5N H; 1UJV A; 1U39 A; 5JBB S; 1W2K H; 2C90 B; 3EA9 A; 3O8R A; 3BTH E; 5T3H A; 2AIN A; 1GCT C; 2OIN A; 4CRC A; 1RTF B; 1OBX A; 1GI1 A; 2JET C; 1HBT H; 2QF3 A; 3SHU A; 3LPR A; 1CA8 B; 1ZHP A; 3OY6 U; 1TLD A; 2W4F A; 3O5N A; 2LC6 A; 1H8D H; 3RXC A; 2XC5 A; 1BRB E; 4GVU A; 4K5Z A; 2Q6F A; 1ELE E; 1SGN E; 3HPM A; 1BRC E; 1SGF G; 3VGC B; 1O5C B; 1W9E A; 1AN1 E; 1O3C A; 3P70 B; 1KLT A; 3ATI A; 2LOB A; 3A7X A; 3NKK A; 1EP6 A; 3SU4 A; 1QAU A; 2BD7 A; 2I0L A; 2XNI A; 1CE5 A; 1PFX C; 5HEB A; 2ODQ A; 1BOQ A; 4K1E A; 1LVO A; 1LHF H; 4B75 A; 2KOH A; 1GBD A; 3AXA A; 3GY7 A; 3NCL A; 1V62 A; 5HET A; 4MPV A; 4HGC A; 2F9N A; 3ODF A; 1QRX A; 4NNM A; 1TP3 A; 1T2M A; 1L1J A; 1KDQ A; 2HNT E; 3RL8 A; 2QXJ A; 3K65 B; 3U9A H; 1V2M T; 2E7K A; 1JIM A; 3BTE E; 4ZAE A; 3UFA A; 2FTL E; 1MKW H; 4WYU A; 2ZC9 H; 2B9L A; 1MZA A; 3IG6 B; 3RXF A; 1OOK B; 1MKX K; 1GJD B; 2EAQ A; 2V1W A; 2R3U A; 3A88 A; 4Y0Z E; 2GD4 B; 1WF7 A; 2VIV A; 2Z3E A; 4GCH F; 1V2O T; 1UEQ A; 3EAJ A; 2GGV B; 3A7V A; 5TCC A; 4CRG A; 1XVO A; 3ZVF A; 1YBW A; 4L8N A; 4HZH A; 3B23 B; 2KL1 A; 3A7T A; 2FOB A; 2R2W U; 2ZU5 A; 2VWL A; 4IC6 A; 4XDE A; 1BML A; 1BTZ A; 1UCY E; 4ZKS U; 1HIA A; 2IWO A; 1GBI A; 2B8O H; 1H1B A; 2Z9L A; 4H4F A; 1AD8 H; 1W7G H; 1V2P T; 1L6O A; 3RL7 A; 2XXL A; 2QYI A; 2BDY A; 5HGG A; 3ZYE A; 1G3D A; 2BVX H; 1NRN H; 2A4G A; 2I0I A; 3SV2 H; 4X2Y A; 3RML H; 1GPZ A; 3PV4 A; 1IOE A; 1QAV A; 1QNJ A; 4PQW A; 1NT1 A; 1C1U H; 4I31 A; 1F5K U; 1GHW H; 2OQS A; 1GJ7 B; 1GJC B; 1TPO A; 2HAL A; 2ZU1 A; 3LGI A; 4TWY A; 4WYT A; 1A4W H; 1LVY A; 1DSU A; 3MHW U; 3VGC C; 2FI4 E; 1RTL A; 4OS4 A; 1UJ1 A; 1O33 A; 2OGP A; 2P16 A; 5D13 A; 4X2V A; 3S9A A; 4BNR A; 2VSV A; 4NXQ A; 1FIW A; 3MNB A; 3TJV A; 2Z9J A; 3QGJ A; 1TYN A; 5E1Y A; 1EB1 H; 4IC5 A; 4WMD A; 1B0F A; 4P0C A; 1ZPC A; 1XXD A; 1TNJ A; 1AKS A; 1FC6 A; 1BRA A; 1CA0 B; 1MTW A; 1MD7 A; 1HDT H; 5LPE A; 5GCH F; 2F9V A; 2DLS A; 3MU5 A; 1GBJ A; 3UWJ H; 1ZSJ A; 2BVR H; 3F9F A; 5EXN A; 3I78 A; 1S6F A; 3UY9 A; 1UJD A; 2DJT A; 1WOF A; 1GGD C; 1UEZ A; 1C5Y B; 2BDH A; 1F0U A; 1MFG A; 4LMM A; 1X5N A; 1PQ7 A; 8KME 2; 1UTL M; 3GOV B; 2FX4 A; 2EEI A; 2FES H; 2ZU3 A; 2ZDV H; 4ASH A; 7EST E; 1SHH B; 4O3T A; 3M7T A; 4D9Q A; 5EXM A; 3DJA A; 1GBK A; 2AGG X; 4X1Q U; 3AW0 A; 2A7J A; 3KID U; 3TH2 H; 3TPI Z; 2PGB B; 3GY4 A; 4B71 A; 2ZGB H; 4UU5 A; 1WJL A; 3BTG E; 1ZTJ A; 3ZZ7 A; 1C5M D; 4ZKO U; 1Q2W A; 2R2M B; 4FUJ A; 3ZYD A; 2PA1 A; 4Z0K A; 1QQ4 A; 5F3X A; 3QZR A; 1BA8 B; 7LPR A; 1TP5 A; 2AER H; 3NPS A; 1VAE A; 1MMJ N; 2ZQ2 A; 2OCV B; 3RXV A; 1HIA B; 4XE4 A; 2LC7 A; 1CQQ A; 3ZZA A; 4DCD A; 8GCH F; 3HKJ B; 4WY3 A; 1GCD A; 1GHZ A; 3A8A A; 4P2A B; 3RDZ A; 1D9I A; 3GIS B; 1V2N T; 4X8V H; 1NU9 A; 1CSO E; 1PPH E; 3RXK A; 2H1U A; 3V0X A; 4DY7 B; 3DHK H; 1WCZ A; 2CV3 A; 4BTU B; 4FUD A; 2VNT A; 1T31 A; 2P93 A; 4IMQ A; 3L33 A; 2OQ5 A; 3RC5 A; 2WYG A; 3CBX A; 1HAO H; 1TE0 A; 3JZ1 B; 4ABE A; 1Q3P A; 2H6M A; 5TJX A; 5EOD A; 1SGI B; 1LPK B; 4H42 U; 3QTV H; 3V3M A; 3UNQ A; 1QY6 A; 2ZFF H; 1SGE E; 1THS H; 1BIT A; 3ZVB A; 2A2Q H; 4FXG H; 3SUZ A; 1GI4 A; 3CS7 A; 1Y3V A; 1AVW A; 5AFY H; 3T26 A; 4FUE A; 4NMR A; 3MU8 A; 1DX5 M; 1U6Q A; 4FUG A; 1Y7N A; 2OTV A; 5GWZ A; 2PFE A; 8LPR A; 2F3C E; 3FVF B; 3RXO A; 2D26 C; 1QR3 E; 3MH7 A; 1HXF H; 1G9I E; 4NFF A; 2H2Z A; 1CA0 C; 3UTU H; 4B2B A; 2A7C A; 5PTP A; 3MH6 A; 1QA0 A; 1VIT G; 3OYP A; 3QDZ B; 1U3B A; 4NIX A; 1O3D A; 2Y5F A; 3HVQ C; 1EJM A; 2WPI S; 1O5F H; 3CYY A; 3A8B A; 1BTU A; 3UR9 A; 2GEF A; 4JZI A; 1C2E A; 1BUI A; 2G51 A; 1DIT H; 3F1S B; 2GCT B; 2O8M A; 1GI2 A; 4IN1 A; 1XZ9 A; 1G30 B; 1JWT A; 2ZDM A; 1LO6 A; 1P2K A; 3W95 A; 2UUF B; 3BEF B; 5CHA B; 1WTG H; 3RXA A; 1NRS H; 2JSN A; 2LA8 A; 4NMV A; 1UK3 A; 2UUJ B; 1ANC A; 1P09 A; 2CHA B; 1C5R A; 1G2M A; 1XX9 A; 1Z8J B; 4WVP E; 1IU0 A; 3RLE A; 3A86 A; 1BT7 A; 4UEH H; 1T8L A; 3PDZ A; 1FY8 E; 2ZO3 H; 4WH8 A; 2ZGX H; 2F9B H; 3PWC A; 1LMW B; 3BTF E; 3FY5 A; 3A81 A; 1RJX B; 4WWY A; 1ARC A; 4CHA C; 1P9U A; 3U98 H; 1PPE E; 4I8L A; 2AGI X; 3T5F H; 2FDA A; 3EST A; 1HCG A; 1MAX A; 2Y81 A; 5LH8 A; 1V2S T; 1V2U T; 3RMN H; 1V3X A; 1W0Z U; 1P2I A; 1UVP A; 3SUE A; 1QLC A; 1X5R A; 2SNV A; 3TIU A; 1OWJ A; 1UCY H; 1AVX A; 1RTK A; 3SUF A; 2ZPL A; 1QBV H; 4GSO A; 1XUJ A; 2J94 A; 1ZR0 A; 2FI3 E; 3KHF A; 1QB9 A; 2TBS A; 1NF3 C; 1W8B H; 3TSW A; 3E17 A; 1UTO A; 4GLY A; 2Z9K A; 1AUT C; 1Z8I B; 5FB8 C; 2J38 A; 2GDD A; 1BDA A; 4XBC A; 4YES B; 4JOG A; 2QC2 A; 4ISO A; 3VB5 A; 4I8G A; 2EI8 A; 3SV7 A; 3VXE H; 3T29 A; 1AZ8 A; 1FN6 A; 2ZPS A; 1OBY A; 1QRW A; 4Q2P A; 3E90 B; 3KF2 A; 2OP9 A; 2CSS A; 5JB9 S; 1C5O H; 2JIN A; 1HB0 B; 3K50 A; 1ZLR A; 4X8T H; 3Q3K A; 1PPG E; 2EC9 H; 1O3K A; 3C1K A; 1WF8 A; 4CRB A; 4ABB A; 3QLP H; 1HAZ B; 1K9O E; 2G52 A; 2ZDK A; 4GPG A; 5HM2 A; 5EQQ A; 2ZCH P; 3KN2 A; 2FM2 A; 4HFP B; 3NFK A; 1L4D A; 1E34 B; 1LTO A; 1A3E H; 1NO9 H; 4I8J A; 4X6P A; 4ABD A; 3GJ9 A; 1VJ6 A; 2ZHF H; 1Y59 T; 1JRT A; 1S5S A; 2GCT C; 1AUJ A; 1AWH B; 2DE9 A; 4IN2 A; 1EZS C; 4JCN A; 1O2Q A; 2SGF E; 4X6O A; 5CHA C; 4K76 A; 2H9T H; 3OY5 U; 1TNG A; 4YT7 H; 1AHT H; 2CHA C; 2QAA A; 1PDR A; 1LHG H; 3SQE E; 2GZ8 A; 1FCF A; 4AYY B; 2VRF A; 2Q3G A; 1T8M A; 2GCH F; 2CS5 A; 3DT0 H; 2G5N A; 4X2W A; 1EAI A; 2H9H A; 1J14 A; 2ZEB A; 1QBO A; 1XKA C; 2IWN A; 3UOP A; 1P04 A; 4FVB A; 1PQ8 A; 2J34 A; 2H3L A; 1BJU A; 3GDU A; 3LGU A; 3U1J B; 2IOT A; 2G4T A; 3R68 A; 2PGQ B; 1QJ1 B; 3SI3 H; 1UO6 A; 3RXT A; 3G01 A; 2JOA A; 1MZD A; 2VJ1 A; 2RCZ A; 1ZGV A; 4X1S U; 3ZZ4 A; 1O36 A; 1B0E A; 1I16 A; 3R3G B; 4Q80 A; 1O2H A; 2BD4 A; 5GDS H; 4JOF A; 3BTM E; 2CSJ A; 5JPM H; 3UQV A; 2SGP E; 1ZJD A; 2FOD A; 2ZG0 H; 4ABF A; 1HJ9 A; 2YT7 A; 1V2V T; 4E7N A; 3K82 A; 2CGA A; 1ZHM A; 1K1N A; 1R6J A; 1GBM A; 2PKU A; 5J4Q A; 1INC A; 2W3I A; 1GMC F; 1P2N A; 4NA8 A; 1TAW A; 3LH3 A; 1OPH B; 1GVZ A; 2FLR H; 1J15 A; 5LGO A; 5E0H A; 3ATL A; 1GHY H; 3RXE A; 1UVS H; 2PKA A; 1CHO F; 3SU5 A; 2GV6 A; 2VIP A; 4OGX A; 3MWI U; 3UIR A; 1ESA A; 4OS2 A; 2KA9 A; 3TK6 A; 2OBQ A; 1GBB A; 4X2X A; 5AF9 H; 2OLG A; 4YTA A; 2OBO A; 3ZVA A; 1O35 A; 3ZRT A; 2BZA A; 2J92 A; 2BMG B; 2M5T A; 3NFL A; 1K21 H; 3SU6 A; 2PUQ H; 3LIW A; 1FZZ A; 1C2J A; 2A32 A; 2FZZ A; 1RY4 A; 3TMH A; 2AWU A; 2J4I A; 3BG4 B; 4GHN A; 2TGA A; 3GY5 A; 5C20 A; 3CC0 A; 1EAT A; 5HXF A; 1O2V A; 2PLX A; 2ZGJ A; 2K1Z A; 3C27 B; 1C5X B; 1MTS A; 1O2W A; 1K28 D; 5C5O A; 1NTP A; 2ALV A; 1ABI H; 3QZQ A; 5EQR A; 2QKU A; 1FY1 A; 1NY2 2; 5JXB A; 1F0R A; 1GJ5 H; 1LCY A; 2STB E; 2M0T A; 2Q9V A; 3I1E A; 4RKO B; 5LPR A; 1P1D A; 1N6E A; 4FGM A; 1O3J A; 2JWE A; 1W14 U; 2VVU A; 1L4Z A; 2BYA X; 4KTC A; 3SQH E; 2KQF A; 4REY A; 4RI0 A; 4KTS A; 2CXV A; 3QO6 A; 1IU2 A; 3ZV8 A; 3RXI A; 1F2S E; 4BAM B; 2EGK A; 2EEG A; 4DUR A; 2OPG A; 4G69 A; 4QT8 C; 5HF4 A; 1CO7 E; 3RXB A; 1SPJ A; 3A89 A; 1EP5 A; 3LON A; 3MNS A; 3WKL A; 1T4V H; 4Y6D A; 1X9Z A; 5GJ4 B; 2P3W A; 3PMJ A; 4BTI B; 2OC0 A; 1P1E A; 3OTP A; 1DUA A; 5EPN A; 5BPE A; 2ZFP H; 3D65 E; 1BE9 A; 4RKJ B; 3K2U A; 1LPZ B; 4UU6 A; 4IS5 A; 4YOJ A; 2Z94 A; 4LOY H; 3RXJ A; 4B73 A; 1OWD A; 4N6X A; 1IQE A; 1PYT C; 3ODD A; 1C2I A; 1C5Z B; 1YC0 A; 2R4H A; 4FU9 A; 1AU8 A; 3S7K B; 3TK9 A; 3BSQ A; 1KXC A; 3GAB A; 2BDC A; 2I6Q A; 3F9E A; 1ARB A; 3SNB A; 3VQG A; 4FUI A; 5HMA A; 1WQS A; 2BY8 X; 3BIV H; 5FX6 A; 2PKA B; 1W11 U; 5C3N A; 1D3P B; 1XMN B; 3PLK A; 3DD2 H; 2Y7X A; 5JBT A; 1VA8 A; 2AMD A; 3DJ1 A; 2YUB A; 1VIT H; 3P8F A; 1BTY A; 4AB9 A; 2ZDA H; 3BTD E; 4AG2 A; 1MBQ A; 1N8O B; 1GG6 B; 1FY5 A; 1T7C A; 2F0A A; 1O2U A; 4CH2 B; 1A46 H; 2HPQ H; 3RXR A; 1GI3 A; 4XFQ A; 5L2Y H; 1RD3 B; 1BHX B; 2YTW A; 5EYZ A; 1CGI E; 2BVS H; 1YCP M; 1GMD G; 2B7D H; 2H3M A; 1UVT H; 1BRU P; 5TCA A; 1F0T A; 4CRA A; 4INL A; 1UMA H; 1P02 A; 2HVX A; 1FPH H; 1YCP K; 1FAX A; 3E16 B; 2EV8 A; 4NZQ A; 1N6D A; 1G3B A; 1M9U A; 1UM7 A; 1DPO A; 3GY3 A; 2KPL A; 1BBR K; 1WIF A; 1A5I A; 1C2F A; 1MBM A; 4MPW A; 1YPH C; 4MNY A; 2A1D B; 5HFC A; 4JK5 A; 2STA E; 2WV4 A; 2K20 A; 3T28 A; 2RDL A; 1ELT A; 1HPG A; 4BAO B; 2VIW A; 3IWM A; 4F49 A; 3PM3 A; 1MQ6 A; 5F03 A; 1EQ9 A; 1SOZ A; 2YT8 A; 4JOJ A; 1L2E A; 4FUF A; 3RLW H; 1P57 B; 3ZVD A; 4Q7X A; 4FVD A; 1QJ6 B; 1UHB A; 1V1T A; 3ZZ9 A; 2ZHE H; 1SLW B; 2HE4 A; 3K9X B; 2V0B A; 4AGK A; 1O2X A; 1C1W H; 4NNL A; 2R0K A; 2YNA A; 1O2I A; 3TK5 A; 2OZF A; 4MQA A; 1V2R T; 1TFX A; 3F9H A; 2D8W A; 2HE2 A; 5E8E H; 2NU3 E; 1HAV A; 4H6S B; 3ID4 A; 5GND A; 3GDS A; 1KLI H; 1O3N A; 3SOR A; 2F9U A; 2GDE H; 1BTP A; 1GDQ A; 1TGB A; 2M10 A; 2QF0 A; 2ZHW H; 4GHQ A; 4XBD A; 2FOF A; 1X5Q A; 1GL1 A; 2RA3 A; 3BTQ E; 3A84 A; 3H7T A; 5A8X A; 3D62 A; 1UF1 A; 3STJ A; 4E34 A; 2OC1 A; 1EB2 A; 2GTB A; 4THN H; 3KQC A; 1DFP A; 1I92 A; 1C9T A; 5GIB A; 3AAV A; 3SV8 A; 1CT0 E; 2AYW A; 1Z87 A; 1EJN A; 1TB6 H; 2GV7 A; 4ZXY H; 3PLP A; 1V5L A; 1XVM A; 1S83 A; 2GZ7 A; 1MKX H; 3VEQ B; 1AWF H; 1P12 E; 1O5G H; 2RA0 A; 1HUT H; 4TPY A; 1HJA B; 4Y0Y E; 1RGQ A; 1S82 A; 2ZGC A; 1MTN B; 1AB9 B; 5JB8 S; 1JOU B; 1ELA A; 1RGW A; 1Q7X A; 2DM8 A; 2UWP A; 2WPL S; 4KFV A; 1DE7 H; 1KYN A; 3RXM A; 3UOU A; 2QBW A; 1KSN A; 2A5K A; 3RU4 T; 1N8O C; 1GG6 C; 4UE7 H; 4AG1 A; 1P2O A; 3GY8 A; 3M7U A; 5HEY A; 3TGJ E; 5T6D A; 4U32 A; 5DGJ A; 5HJ1 A; 3Q3X A; 1OYQ A; 1P05 A; 3QE1 A; 1GI5 A; 1QGF A; 3HK6 B; 4FUH A; 3RC4 A; 2ZEC A; 1SL3 A; 4EST E; 2BDI A; 5IYT A; 1OP8 A; 4YT6 H; 2B0F A; 4AMH A; 2FLB H; 4XOJ A; 5FPT A; 4YDP A; 4YYX A; 4K2Y A; 1FN8 A; 1ELG A; 2ANM H; 3EGK H; 1GQ5 A; 1ZTK A; 3EDX B; 3URE A; 1BHX F; 4ABA A; 1A0H B; 1EZU C; 1TNH A; 3TJN A; 2FOM B; 1YPL H; 3LOX A; 4K8Y A; 5DKM A; 2FPZ A; 1TMB H; 3U1I B; 3Q3Y A; 3TSV A; 5A0C A; 2XTT B; 2BM2 A; 1O3M A; 1IHS H; 3TGK E; 2P8O B; 1RRK A; 2G2L A; 1Q3O A; 3F9G A; 1SG8 B; 2VID A; 1KEF A; 1ELD E; 4Y79 A; 1UK4 A; 1UHB B; 4UFG H; 2O9Q A; 1OWK A; 4B2A A; 2NU2 E; 4Y10 E; 2A4R A; 1YF4 A; 2XYA A; 4AFS A; 1P11 E; 1P03 A; 1LDT T; 3A83 A; 1HAV B; 5L6N H; 1MFL A; 1MUE B; 3KL6 A; 3ZZ3 A; 2NU4 E; 1Z71 A; 2AIQ A; 3BN9 A; 4ISL A; 2PUX B; 1O3P B; 3URC A; 5H3J A; 4DVA U; 1NPM A; 2BLO A; 4OS7 A; 1BJV A; 1UTN A; 1HNE E; 2VGC B; 3CS0 A; 4GCH G; 1D5G A; 4R8T B; 1BIO A; 4ZXX H; 1TX6 A; 1F7Z A; 3SUD A; 1GI0 A; 2ZWL H; 3SHC H; 4U30 A; 1L1N A; 1C1Q A; 3M5O A; 1SGQ E; 1KDQ B; 3KNX A; 2OCS A; 4NA9 H; 1HJA C; 2D1J A; 3ZVE A; 5EQS A; 4JYT A; 3UR6 A; 5HFF A; 3K1R A; 4I8H A; 5E6P B; 5IL9 A; 5ESB A; 4OEO A; 2FXR A; 1CVW H; 1RXP A; 1FMG A; 2Y6T A; 4YLU A; 1SGR E; 4HI3 A; 4I8K A; 4Q2O A; 2XW9 A; 2G55 A; 2AH4 X; 3NZI A; 4ISN A; 3M37 A; 3OX7 U; 4ABG A; 1SVP A; 1M5Z A; 1K1O A; 5J4S A; 1TPP A; 4EDJ A; 2OC7 A; 3SND A; 1YPH E; 4CR9 A; 1JRS A; 2TGD A; 1DXP A; 2VH0 A; 5C67 A; 3SJO A; 2THF B; 1SQT A; 3A7Y A; 1QBN A; 2Q6D A; 4WJG 2; 3S0N A; 2MX6 A; 1BB0 B; 4X6M A; 3A7Z A; 1SC8 U; 1V5Q A; 1EPT A; 4K1T A; 2OMJ A; 4OGY A; 3TNS A; 4JYV H; 2AWX A; 4MNX A; 4YZU A; 5ETX A; 3SGQ E; 3SNE A; 1C5U A; 2KOJ A; 1AQ7 A; 1NN6 A; 1XKB C; 5DP5 A; 3QTO H; 5JDU B; 1AIX H; 1GHB F; 1ZHR A; 1H9I E; 3DA9 B; 4I7Y H; 4O97 A; 5DG6 A; 4AYV B; 1QAV B; 3TGI E; 2FOH A; 3MOC A; 5CMX H; 3RXU A; 4INK A; 2I6V A; 2ANY A; 2GKV E; 4E05 H; 4LK4 A; 2HGA A; 1NRO H; 1QTF A; 4IBL H; 1DWE H; 1U37 A; 3R69 A; 1KXF A; 3TJO A; 2P8O C; 1ZPB A; 2OQU A; 1GJ6 A; 1PPZ A; 3CH8 A; 3RU4 D; 2BY6 X; 1Q3X A; 3KGP A; 5GCH G; 1NFU A; 1MD8 A; 1UTJ A; 1K1P A; 3MI4 A; 4ZRO A; 3BIU H; 4AZ2 B; 1DIC A; 5CE1 A; 4WXI A; 2QXH A; 5DP4 A; 5GLJ A; 4JMY A; 2BX3 A; 2GX4 A; 1UM1 A; 1O32 A; 1SLV B; 1C2H A; 2ZDN A; 1VCP A; 3ATK A; 3A80 A; 1ID5 H; 3GCT F; 5AHG H; 1AVG H; 1AKS B; 1TNL A; 5FPS A; 4UDW H; 4AFU A; 1TGS Z; 4NZL A; 2ZPM A; 4Q2N A; 1UTQ A; 3MO9 A; 3LGW A; 4GHT A; 3BEI B; 4KKD A; 1BTX A; 2W7R A; 1C2L A; 2ASU B; 5K0H A; 2G5V A; 3O46 A; 4FU8 A; 4KTU A; 5DO4 H; 7KME H; 1KXB A; 2Z6B D; 3F6U H; 3MH5 A; 3FP6 E; 3NK8 A; 5L2Z H; 1DM4 B; 1NM6 A; 5C1U A; 1O3O A; 1ZGI A; 4HOP A; 2I6S A; 1LVB A; 1TBZ H; 2KAW A; 2ZHD A; 3HK3 B; 1IQF A; 1N1L A; 1GHX H; 1K32 A; 2PTN A; 1FJS A; 3LKW A; 4J2Y B; 3SU3 A; 1SGC A; 1C5V A; 1W12 U; 4BAN B; 3Q77 A; 4NXP A; 1TAB E; 1V2Q T; 1YP9 A; 1C1N A; 2C8X B; 4Q6S A; 8GCH G; 1UTK A; 3RXD A; 2F9P A; 3K6Z A; 2H9E H; 4KP0 A; 3SI4 H; 5DP3 A; 2EDV A; 5JBC S; 2EST E; 1BTW A; 3E8L A; 1G36 A; 2WYJ A; 1EAX A; 2WUB A; 3V13 A; 3PV2 A; 4JK6 A; 3O8C A; 4A6L A; 2L4S A; 2EA3 A; 1O5B B; 1UJU A; 1MH0 A; 3L3T A; 2EDZ A; 3GGE A; 3NUM A; 6CHA B; 4KGA A; 1IQH A; 1QL8 A; 3ZZ6 A; 5E21 A; 1F92 A; 5K7R A; 1Y5U T; 2W5E A; 1WQV H; 3HKI B; 3PTB A; 1AE5 A; 1EPT B; 2VH6 A; 5EST E; 4WME A; 2QKV A; 2GZ9 A; 4X1P U; 3D23 A; 1GJ9 B; 1AI8 H; 1QB6 A; 3FFG A; 1C1M A; 1ZMN A; 4WH6 A; 1DLK B; 1BZX E; 1IQI A; 2IJO B; 3PO1 B; 1NFY A; 5C1Y A; 3RP2 A; 3QGL A; 1GBE A; 4K75 A; 2QN5 T; 1HYL A; 2ZI2 H; 3M3V A; 4B2C A; 1ELB A; 3RXG A; 3MTY A; 3SHA H; 1V2J T; 1Y5B T; 2PTC E; 3MU1 A; 4X6N A; 1UHP A; 3V12 A; 4CR5 A; 1A5G H; 1AZZ A; 1Z8G A; 1S6H A; 1W3C A; 2WPH S; 3MO3 A; 1GJB B; 3TVJ B; 2ZNK H; 4JZE H; 1GMH F; 3VXF H; 3I29 A; 1WU1 A; 3VPK A; 3GCN A; 1KTT B; 4BAQ B; 1LHE H; 2G8T A; 2QT5 A; 3PMA B; 1BBR E; 1IQG A; 1GQ4 A; 1HAH H; 2BX4 A; 2HRV A; 1PPC E; 2BYG A; 1X45 A; 4YM9 A; 1C5Q A; 3KQD A; 4NWL A; 2KBS A; 4NMT A; 3SGB E; 1WTH D; 2VWO A; 5JYI A; 1O5D H; 2PW8 H; 1OP2 A; 3TIT A; 1A0J A; 4SGA E; 3SOS A; 2PSY A; 3RXP A; 1O3L A; 1O2K A; 5HED A; 1EKB B; 4Y7A A; 2ZF0 H; 5EGM A; 3A85 A; 2EEH A; 5I25 A; 2EJY A; 1HXE H; 1FY3 A; 1DY8 A; 3LC5 A; 3PMH B; 4MVN A; 1DS2 E; 2G00 A; 1YPE H; 3P17 H; 1TBR H; 3I4W A; 3RMM H; 2WPJ S; 3JXT A; 2WL7 A; 4JZD H; 4UD9 H; 3LNX A; 2HWL B; 2G5M B; 2VWM A; 4FUB A; 1VR1 H; 1TRY A; 1TMT H; 1HAI H; 1TNK A; 1MU6 B; 3PLB A; 1Y3U A; 2DUC A; 4I32 A; 4A5T S; 1THR H; 3MFJ A; 1ORF A; 2ENO A; 3ZZ5 A; 2GT8 A; 4NMO A; 2Q1J A; 1OS8 A; 1WFV A; 4Q2Q A; 2VB0 A; 3STI A; 1O2G H; 6CHA C; 3OTJ E; 3TSZ A; 1DWD H; 5DP7 A; 3FAN A; 3CBY A; 1IQJ A; 1ELC A; 4WMF A; 1EPT C; 1TGC A; 4ISI H; 2GP9 B; 2Y5H A; 4UFE H; 4JYU H; 3KQB A; 1UK2 A; 1L1G A; 2BDB A; 3MU4 A; 1GI8 B; 3KHV A; 1WSS H; 3BTT E; 2LUI A; 3PO1 C; 1D3D B; 2VWN A; 3CP7 A; 4KFW A; 3KQE A; 2R0L A; 1Z1I A; 2VVC A; 5EZ0 A; 1OX1 A; 2WV5 A; 3P95 A; 1T8O A; 4AFQ A; 3SU1 A; 1S0R A; 2ZU2 A; 5HDY A; 1NTE A; 1C1O A; 1SQO A; 1BFE A; 4K6Y A; 1C2G A; 4Y8X A; 2G81 E; 4HTC H; 1SQA A; 2A2X H; 2EEJ A; 2A0Q B; 2O8T A; 3ATM A; 4RN6 A; 2VWR A; 3TU7 H; 5E11 A; 1KTS B; 1ZTL A; 1TBQ H; 3MNC A; 1IQK A; 2Y5G A; 1N6Y A; 2XC0 A; 4NWK A; 3HAT H; 3URD A; 1TPS A; 3M5N A; 2KJP A; 2EI6 A; 4Q7Z A; 1O37 A; 4TWW A; 1O31 A; 2JKH A; 1EAS A; 2GCH G; 3E6P H; 1VJA U; 1GJ4 H; 4BAH B; 1WYK A; 4M7G A; 4X8U H; 1WHA A; 1NES E; 3UWI A; 1J16 A; 2ZPR A; 2KYL A; 1O0D H; 1QA7 A; 2P3U B; 1O3A A; 4CBN A; 1QFK H; 1XXF A; 4K1S A; 1Z8R A; 3SZN A; 1J9C H; 2ALP A; 1DVA H; 5A8Y A; 2NU0 E; 2ZPQ A; 5FX5 A; 1ZJK A; 1O2J A; 2IWQ A; 3U8T H; 1N99 A; 1U38 A; 3VB7 A; 2J2U A; 1TQ3 A; 6GCH F; 1QQU A; 1ETR H; 2ZQ1 A; 3L6P A; 1O2Y A; 4JZ1 A; 4NG9 H; 3A87 A; 3VFE A; 2JIL A; 2VIQ A; 2RRM A; 6LPR A; 1HAY B; 1Z6J H; 3MO6 A; 1N7E A; 1H9H E; 2R9P A; 9LPR A; 2HNT F; 4ZH8 A; 1O3E A; 1GBC A; 3QDO A; 2JET B; 1IQL A; 1LHD H; 2OD3 B; 4NMP A; 1GMC G; 3M3T A; 2YUY A; 1BCU H; 1C5W B; 5F8T A; 1CHO G; 2Y80 A; 7GCH F; 6EST A; 2DC2 A; 2H5C A; 2Y82 A; 3B9F H; 3ZZB A; 5HM9 A; 3TNT A; 2AWW A; 4MLF B; 5C2Z A; 1RGR A; 1ANB A; 3LU9 B; 1DOJ A; 5A8Z A; 3B8J A; 3HPT B; 2BD2 A; 1QJ7 B; 2TPI Z; 2VSP A; 4D76 A; 2AS9 A; 3SGA E; 1GDU A; 3QK1 A; 4AN7 A; 1YLD A; 2UZC A; 2M0U A; 2LPR A; 1FDP A; 1GWA A; 1S0Q A; 1ZMJ A; 4DOQ A; 1TRN A; 1P3E A; 5ILB A; 1O2O A; 5G4B A; 1WUN H; 1GJA B; 2EGO A; 3QWC H; 1EZX C; 4X0W U; 4Y11 E; 2ZHQ H; 3M3S A; 1DST A; 2NWN A; 3UPE A; 3PB1 E; 5C1X A; 1CGJ E; 1J8A A; 1GHA F; 4NA7 A; 3WY8 A; 3QJM A; 1BMM H; 1YPG H; 3M35 A; 2FP7 B; 5HFE A; 1HRT H; 3U8O H; 1MCV A; 4NSV A; 1HAP H; 3TH3 H; 4NXR A; 1SBW A; 2YOL A; 3BG4 C; 2KAI A; 1FLE E; 3RM0 H; 1O5A B; 1C5S A; 5EW1 H; 1XUI A; 1J17 T; 1Y3Y A; 1YCP H; 1SGT A; 4NFE A; 3S9C A; 2A7H A; 5HYO A; 2BLV A; 3QGN B; 5E2O A; 1BBR H; 2BLQ A; 2ZIQ H; 2A4O A; 2BY7 X; 3LDX H; 1WHD A; 1QHR B; 3SFJ A; 3RLY H; 1WFG A; 4TY6 A; 2W3K A; 1A61 H; 1GI6 A; 4OS1 A; 2AEI H; 4I33 A; 2FI5 E; 1DY9 A; 1YKT A; 1AFE H; 3W94 A; 2ZGH A; 1C1P A; 1EJA A; 5SGA E; 1CT4 E; 1TWX B; 2FYQ A; 3SU0 A; 3HGN A; 1H9L B; 3EGH C; 3PMB B; 1SOT A; 2Z9G A; 2C3S A; 2SGA A; 3SU2 A; 2ZCK P; 3MYW A; 3KDK A; 2XRC A; 3Q76 A; 5HF1 A; 1DAN H; 2PKT A; 5FPY A; 4B74 A; 3R0F A; 7GCH G; 5B42 A; 2QV1 A; 3WKM A; 4DII H; 2FS8 A; 1SGP E; 4MPX A; 3SJ9 A; 4BXW A; 2W26 A; 1K1M A; 4B1T A; 1TX8 A; 2VVV A; 3PDV A; 2UWO A; 2EHR A; 3NGH A; 2P3F H; 2CJI A; 5H4I B; 4ABH A; 1O39 A; 1C5T A; 2EEK A; 3PS4 A; 1GBT A; 2FX6 A; 1O2T A; 3QUM P; 3SJI A; 1DWC H; 3ID1 A; 2XCN A; 3P8G A; 1O38 A; 1V2L T; 1GL0 E; 2EXG A; 1X7A C; 5FBI A; 1P2M A; 1GHA G; 1TGT A; 1W10 U; 1SGD E; 1MCT A; 1FV9 A; 4D7G A; 3QJN A; 1Q31 A; 1C2M A; 5GPI B; 2V6N A; 1FXY A; 3EE0 B; 1C5L H; 1EOJ A; 5ABW A; 2DLU A; 5EPY A; 1Z1J A; 3RXQ A; 1SSX A; 1O2Z A; 2AMP A; 3AAS A; 3M5M A; 1NC6 A; 1C1R A; 1GBF A; 1KXE A; 3AW1 A; 3P8O A; 5F8Z A; 2KV8 A; 2OC8 A; 1C2D A; 1YPK H; 4BTT B; 4E3B A; 2ZKS A; 1EST A; 3SUG A; 4ZMA H; 1AY6 H; 2XCF A; 5EG4 A; 1HFD A; 5I7Z A; 1O2L A; 3HPK A; 1BMN H; 4GVC A; 4OEP A; 1K1I A; 3QN7 A; 2TGP Z; 2ULL A; 3NWU A; 1GGD B; 4CH8 B; 2JIK A; 1ELV A; 2BDG A; 3SOE A; 1YLC A; 2AIP A; 4TY7 A; 5DPA A; 1IHJ A; 2KAI B; 3CEN A; 5B6L A; 3BTK E; 3E3T A; 4JOP A; 4U01 A; 3A82 A; 1DXW A; 1P0S H; 2D90 A; 1PJP A; 1X8S A; 4E35 A; 1PQ5 A; 1B8Q A; 2P3T B; 1FC9 A; 1AFQ B; 1CBW B; 2BD9 A; 1OP0 A; 1NRQ H; 5EXL A; 1MKW K; 3DOR A; 3KZE A; 1HAX B; 1ZSK A; 3ZVC A; 1C1T A; 1D3T B; 1OWH A; 4NIV A; 1VCQ A; 2FOE A; 2QA9 E; 1A0L A; 1O2N A; 4NGA H; 2HOB A; 1Z86 A; 1NS3 A; 2BY5 X; 3LJO A; 2FNE A; 2M3M A; 2O8L A; 1LVM A; 2I1N A; 2BY9 X; 2DMZ A; 2GT7 A; 3KZD A; 2ZFT A; 1YBO A; 1KIG H; 4ZHM U; 3DUX H; 1OWI A; 4AOQ A; 1SLU B; 1EZQ A; 2CN0 H; 4JJ0 A; 3MU0 A; 2FIR H; 3CBZ A; 1Z7K A; 1MTV A; 1SGY E; 3RXH A; 1GJ8 B; 1AGJ A; 4DT7 B; 5JBA S; 1HAG E; 2JXO A; 4DJZ B; 1O3B A; 4K69 A; 5F67 A; 1YYY 1; 3H7O A; 2QGR A; 3SW2 B; 4K78 A; 1OYT H; 2V3H H; 3S7H B; 1TAL A; 2BQ7 B; 1C1S A; 1CHG A; 3LH1 A; 2A45 B; 4MNV A; 2QKT A; 1FC7 A; 2Z3C A; 2ZCL P; 3VB6 A; 3PV5 A; 1F0S A; 2KJK A; 2H2C A; 4YOG A; 1O3H A; 2L97 A; 1ELF A; 2IWP A; 4MG3 A; 3IIT A; 1LPG B; 1EX3 A; 1O2M A; 2BXU H; 1YPM H; 2JRE A; 2QIQ A; 4CBO A; 1MTN C; 1AB9 C; 3FP7 E; 1T4U H; 3HGP A; 4CRD A; 1MQ5 A; 2C8Y B; 1ACB E; 2ZA5 A; 3MNX A; 5EW2 H; 2BLW A; 2FOG A; 3S9B A; 1LKA A; 2TGT A; 1W0Y H; 1YGC H; 2GMT B; 4A1V A; 1ZOM A; 1TMU H; 1ZSL A; 1QUR H; 2C93 B; 1QL7 A; 4ZUH A; 5FAH A; 4GVD A; 4ZKN U; 1VCW A; 5E22 A; 1DDJ A; 2BOH B; 1SFQ B; 4IMZ A; 4D8N A; 3NXP A; 1EXF A; 2ZFS A; 2ZFQ H; 5E2P A; 3A7W A; 3FZZ A; 1VIT F; 1PPB H; 1ANE A; 3RM2 H; 4IGD A; 1VGC B; 4DGJ A; 5EOK A; 4BOH A; 2TRM A; 1WV7 H; 1FON A; 1RZX A; 1UFX A; 3R0H A; 1AFQ C; 1CBW C; 4A1T A; 3GSL A; 1Y3X A; 2F5Y A; 4RSP A; 1TGN A; 2BD5 A; 5DP8 A; 3DJ3 A; 2NU1 E; 3A8D A; 3PTN A; 5HFB A; 2PKS B; 2WUC A; 4CHA B; 1D3Q B; 1MU8 B; 1N7F A; 4FUC A; 2KOM A; 1A5H A; 1AE8 H; 3SV6 A; 1NRP H; 1RIW B; 3GY6 A; 1E0F D; 3NCV A; 3UNS A; 1W7X H; 1BTH H; 5FHT A; 3ID2 A; 2FEQ H; 4O9V A; 1C2K A; 4XBB A; 4A92 A; 8EST E; 3AVZ A; 1N7T A; 1ZUB A; 1FAK H; 1G32 B; 1HJ8 A; 2TIO A; 2ZDL A; 2I04 A; 2PDZ A; 5FXL A; 2SGD E; 1OXG A; 4MTB A; 1GHV H; 1Y5A T; 1TON A; 2F83 A; 1XUG A; 3ZZC A; 2XBW A; 4AB8 A; 2PZD A; 1SFI A; 2PSX A; 1NFW A; 3LC3 A; 1FQ3 A; 1V2T T; 4DG4 A; 4KEL A; 1LQD B; 5AVD A; 3N7O A; 4Q2K A; 3ZV9 A; 1QB1 A; 3V7T A; 1ZZZ A; 1G2L A; 1VB7 A; 1GBL A; 1MAY A; 1PQA A; 4UFF H; 4VGC B; 4X1R U; 4O06 A; 3VB4 A; 3GYM A; 2DKR A; 1O5E H; 2WPK S; 3PWB A; 4MPU A; 4AOR A; 3H5C B; 1EAW A; 4BXS A; 3GY2 A; 1HGT H; 1SI5 H; 1YPJ H; 4SGB E; 2X7Z A; 3EYD A; 2EI7 A; 2M0Z A; 2QXI A; 3TH4 H; 2VIO A; 1UEP A; 3I18 A; 1T32 A; 4LZ4 B; 2JH0 D; 2VGC C; 2ZU4 A; 3O8D A; 1KY9 A; 1X6D A; 3L4F D; 3LGV A; 1OKX A; 2G4U A; 4R2Z A; 1WAY B; 4D7F A; 1P9S A; 3QIK A; 3PRO A; 2JH6 D; 2FS9 A; 1QCP A; 1L0Z A; 4F4O C; 4U2W A; 5B6O A; 4Y8Y A; 2EGN A; 1P10 A; 2QG1 A; 1V2W T; 1TPA E; 4UFD H; 3EA7 A; 2PR3 A; 1SHY A; 4H6T A; 4JOR A; 1TQ0 B; 1H4W A; 1GBH A; 1H8I H; 1FNI A; 4B76 A; 2C8Z B; 2KRG A; 1ZRB A; 2GMT C; 3E0N B; 3U8R H; 4Y7B A; 1TOM H; 2BTC E; 3Q00 A; 3M5L A; 2P94 A; 1TA6 A; 3KEE A; 1D6W A; 1FUJ A; 1MC7 A; 2W7S A; 3LT3 A; 1P06 A; 2ILN A; 3M7Q A; 2BZ6 H; 5DK1 A; 1SR5 C; 4NSY A; 2FCF A; 2A5I A; 4PRO A; 1VGC C; 1RFN A; 1GHB G; 3U69 H; 2Z7F E; 1S84 A; 4LXB H; 1ETT H; 2HLC A; 1G9O A; 1SGF A; 2FO9 A; 3T2N A; 1PPF E; 2I4S A; 3T27 A; 1A2C H; 1KXD A; 4WXV A; 2GZV A; 5F6M A; 1TA2 A; 1EAU A; 1TOC B; 5DTH A; 1K22 H; 2R3Y A; 2PKS C; 2KPK A; 3KDG A; 2KJD A; 4ABJ A; 2QY0 B; 3GCO A; 2BDA A; 1RIW C; 4Y76 A; 2ZTX A; 2O2T A; 2Q6G A; 1D4P B; 3GCH B; 1RS0 A; 2FN5 A; 2VZ5 A; 3GCT G; 1LKB A; 2V35 A; 3P6Z B; 3OU0 A; 3JZ2 B; 3UUZ A; 5LPF A; 1KLJ H; 3SBK A; 5DJ7 A; 5A09 A; 1W13 U; 1IAU A; 1P3C A; 4JJ3 A; 4Z33 A; 1UCY K; 4AFZ A; 1WI4 A; 1HV7 A; 4DW2 U; 3SJ8 A; 1V6D A; 2CMY A; 2BQW B; 3ITI A; 1XM1 A; 1WG6 A; 3SJK A; 5I46 H; 1VJ9 U; 3VB3 A; 4H11 A; 2F9O A; 4NMS A; 1LQE A; 3BF6 H; 4VGC C; 1K1J A; 4IW4 E; 1JXP A; 1THP B; 1V6B A; 3EQ0 H; 4CRF A; 4Y8Z A; 2XBY A; 3SO3 A; 3S69 A; 3D49 H; 2VPH A; 3RUO A; 5A0A E; 4JOK A; 4GAW A; 1GI7 B; 1JBU H; 4NMQ A; 3UNR A; 4X8S H; 4A7I B; 1G3C A; 1AO5 A; 1O3F A; 2QXG A; 1WBG B; 1A3B H; 3I77 A; 2XBV A; 4HOP B; 2L4T A; 3E0P B; 2EDP A; 1OZI A; 1GVK B; 1IHT H; 1O34 A; 1UIT A; 3DFL A; 3FP8 E; 1DWB H; 2OS6 A; 4K60 A; 3SN8 A; 4UON A; 2SNW A; 3RXL A; 3E91 A; 1P8V C; 3SNC A; 1XUH A; 4XHV A; 4JL7 A; 3K6Y A; 4BAK B; 1ESB A; 2XC4 A; 4FLN A; 1DUE A; 1TQ7 B; 1CU1 A; 2SGQ E; 3RXS A; 4ISH H; 4INH A; 5T6G A; 2KG2 A; 2FOC A; 1EOL A; 3BTW E; 4OAJ A; 2BB4 A; 1NFX A; 1OSS A; 2BD8 A; 3RC6 A; 1IQN A; 2HGT H; 2P95 A; 4YO9 A; 4CRE A; 3B76 A; 5L30 H; 2BQ6 B; 1G3E A; 3FAO A; 1G37 A; 2SFA A; 1V2K T; 1IQM A; 4YLQ H; 4AX9 H; 2XBX A; 3DFJ A; 4TYD A; 1JMO H; 4O03 A; 5DP6 A; 1W9O A; 1OBZ A; 4NIW A; 9EST A; 3ATW A; 3P8N A; 1C4V 2; 1EUF A; 1PYT D; 3O8B A; 1SLX B; 1C5P A; 1B5G H; 2FMJ A; 3LGT A; 4JOE A; 2H5D A; 2OSG A; 3KCG H; 1SMF E; 1GMH G; 5AFZ H; 3FZD A; 3GCH C; 2A5A A; 3GYL B; 3TLO A; 4ZHL U; 4XSK U; 1WH1 A; 1FPC H; 2IPH A; 4Q7Y A; 4ABI A; 2ZTZ A; 2M0V A; 1ZPZ A; 1ZML A; 2FYR A; 4JZF H; 2PWX A; 2ODP A; 2PHB A; 2J95 A; 1UTP A; 2SGE E; 2H2B A; 3SV9 A; 1S85 A; 2FE5 A; 2OXS A; 1E35 B; 2Z3D A; 3ENS B; 4GUX A; 1A7S A; 1Y3W A; 3E1X B; 3OSY A; 1O2S A; 1UEW A; 2GVF A; 1D6R A; 2Y7Z A; 5T6F A; 5A0B A; 1NZQ H; 1VZQ H; 5IDK A; 3T62 A; 3M36 A; 5F8X A; 2D92 A; 1A1R A; 1FY4 A; 2RCE A; 3F68 H; 2O8W A; 1WI2 A; 1E36 B; 1TIO A; 2UUY A; 2ANK H; 1O2P A; 1NU7 B; 1MTU A; 2C8W B; 3MMG A; 3T25 A; 2BXT H; 1GDN A; 2FWW A; 4E7R G; 1TNI A; 1C4U 2; 3GIC B; 4R0I A; 2HNT C; 1FIZ A; 5GXP A; 1K2I 1; 4DIH H; 2AMQ A; 5E0J A; 4Q3H A; 4X1N U; 1C9P A; 2O8U A; 2ZZU H; 1O3I A; 1DT2 A; 1UVU H; 2CF8 H; 1CGH A; 3UQO A; 2LNC A; 2PV9 B; 4MDS A; 4K8B A; 2QCY A; 2V3O H; 2XWB I; 1TRM A; 4ZKR U; 4A1X A; 4Z6A H; 2VU8 E; 2VIN A; 2AGE X; 2DB5 A; 3M61 U; 1GVL A; 3SHW A; 1GM1 A; 1F5R A; 4AGJ A; 1GCT B; 2B5T B; 4K72 A; 2OUA A; 4K3J A; 1O30 A; 2XWA A; 2BOK A; 1ZYO A; 2C4F H; 1P01 A; 1BMA A; 1GMD F; 2W7U A; 2WIN I; 3TJU A; 1BQY A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...