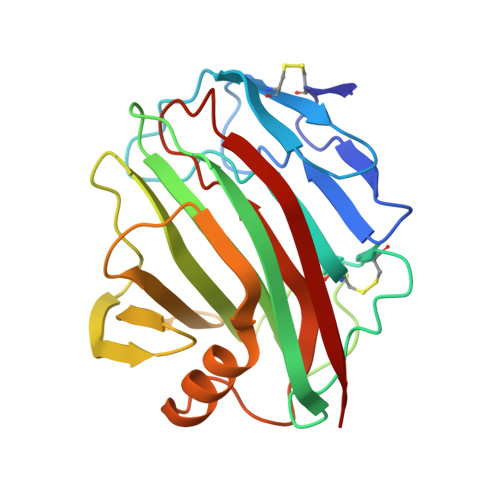
The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
62
|
sequence length |
226
|
structure length |
226
|
Chain Sequence |
TVELCGRWDARDVAGGRYRVINNVWGAETAQCIEVGLETGNFTITRADHDNGNNVAAYPAIYFGCHWGACTSNSGLPRRVQELSDVRTSWTLTPITTGRWNAAYDIWFSPVTNSGNGYSGGAELMIWLNWNGGVMPGGSRVATVELAGATWEVWYADWDWNYIAYRRTTPTTSVSELDLKAFIDDAVARGYIRPEWYLHAVETGFELWEGGAGLRSADFSVTVQKL
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
Dimerisation and an increase in active site aromatic groups as adaptations to high temperatures: X-ray solution scattering and substrate-bound crystal structures of Rhodothermus marinus endoglucanase Cel12A.
pubmed doi rcsb |
| molecule keywords |
ENDOGLUCANASE
|
| molecule tags |
Hydrolase
|
| source organism |
Rhodothermus marinus
|
| total genus |
62
|
| structure length |
226
|
| sequence length |
226
|
| chains with identical sequence |
B
|
| ec nomenclature |
ec
3.2.1.4: Cellulase. |
| pdb deposition date | 2005-07-13 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF01670 | Glyco_hydro_12 | Glycosyl hydrolase family 12 |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Beta | Sandwich | Jelly Rolls | Jelly Rolls |
#chains in the Genus database with same CATH superfamily 3AKP A; 2D98 A; 4XQ4 A; 3VZO A; 1C5H A; 2VG9 A; 3VL8 A; 3ZSE A; 3AKT A; 3WXP A; 4XQW A; 1REF A; 1OA4 A; 3AMP A; 2D97 A; 3AMM A; 3VZM A; 1UU5 A; 2DFC A; 1HV0 A; 3VLB B; 2JEN A; 3VZN A; 4HK8 A; 1BK1 A; 1H1A A; 3AMH A; 3M4F A; 3WP3 A; 1W2U A; 4IXL A; 3WP6 A; 4HK9 A; 2BWC A; 4S2D A; 3LB9 A; 3WQ7 A; 3WX5 A; 5K9Y A; 3MFA A; 3WQ1 A; 3VL9 A; 2NLR A; 4H7M A; 5EJ3 A; 1XYN A; 2BW8 A; 1XYO A; 1XNC A; 2JEM A; 4S2H A; 4HKL A; 1OLR A; 3AKR A; 4HKW A; 2BVV A; 1BVV A; 4S2F A; 1QH7 A; 3WT3 A; 3AKQ A; 3VZK A; 3AKS A; 4HKO A; 1H4G A; 2B42 B; 1BCX A; 2C1F A; 3HD8 B; 2DCJ A; 3WP5 A; 1T6G C; 2VUJ A; 3O7O A; 3VHO A; 3VZJ A; 1F5J A; 1YNA A; 3WR0 A; 4S2G A; 1H4H A; 3RI9 A; 1OLQ A; 1TE1 B; 4XQD A; 3VZL A; 2BWA A; 1XND A; 3MF6 A; 1C5I A; 1HIX A; 1M4W A; 3MFC A; 3MF9 A; 1AXK A; 2VUL A; 5HXV A; 3RI8 A; 1UU6 A; 1KS5 A; 1REE A; 1UKR A; 1IGO A; 4XPV A; 1XNK A; 2DCY A; 2F6B A; 2NQY A; 3VGI A; 2QZ2 A; 1H8V A; 3AMQ A; 3WQ0 A; 3WP4 A; 1XNB A; 1OA3 A; 1OA2 A; 5K7P A; 1PVX A; 3B5L B; 1ENX A; 3AMN A; 1RED A; 2DFB A; 1H0B A; 2DCZ A; 1XYP A; 2JIC A; 3B7M A; 1KS4 A; 3WY6 A; 1UU4 A; 2Z79 A; 3VHP A; 1QH6 A; 2B46 X; 3VHN A; 2DCK A; 3EXU A; 2B45 X; 3LGR A; 4NPR A; 2VGD A; 2QZ3 A; 1XXN A; 1HV1 A; 1NLR A; #chains in the Genus database with same CATH topology 1RMU 3; 2TEP A; 4CDU C; 5CFC B; 1VBA 3; 4LXL A; 2IFR A; 3ME2 A; 4QAW A; 1N1S A; 3M3C A; 4RHV 2; 5FYY A; 3H1W A; 4ATF A; 5C8C B; 1TME 1; 4XET A; 4JGY C; 2YC0 A; 5BNO B; 2FBE A; 4XUP A; 1QJF A; 1V0A A; 1EAH 3; 4B7K A; 5CVR A; 1IPJ A; 2K0G A; 4FFG A; 2Y34 A; 1I5P A; 2WWJ A; 4JC1 A; 1ASJ 1; 4D7T A; 4XIK A; 4J1X A; 3POB A; 4RGK A; 4IV1 A; 2QQU A; 5FMD A; 2XBO 2; 4WVZ A; 4X6R B; 1LHW A; 1BK1 A; 4RS5 C; 3OG2 A; 1BLZ A; 1MS3 A; 3FMG A; 3T2T A; 5SV8 A; 1SFP A; 1EYB A; 1O5U A; 1OPO A; 2RE9 A; 3I2I A; 2XJR A; 1MR5 A; 3PLQ A; 3WNM A; 2R04 2; 5D2A A; 4CDX B; 5CEH A; 5JZG A; 1PGL 1; 5I0D A; 4MHR A; 2J1T A; 2Y26 A; 3ZSJ A; 1V6O A; 4LUL A; 5EJ3 A; 5I0R A; 2WKK A; 1Q43 A; 2RS5 3; 5FZO A; 1HXS 1; 4HKL A; 4Z07 A; 3CJI A; 1Y7B A; 1ULO A; 1FPN 2; 3INA A; 2W39 A; 1VAM A; 3WT3 A; 1CES A; 3OOX A; 4R4Z A; 1H2L A; 2ZOE B; 1W2T A; 1UOG A; 5KC5 A; 2ZS6 B; 1NY7 1; 2EAL A; 4WZC A; 1JYC A; 3VBS B; 3WUC A; 2DVB A; 1LHN A; 3QD6 A; 3JXS A; 3WP5 A; 1GU3 A; 3MO4 A; 4P00 B; 2VMG A; 4KG8 A; 2BEO A; 3OEB A; 4QD2 A; 2VQ0 A; 2C24 A; 4XTP A; 2FDG A; 2XUE A; 1UNB A; 4MET A; 1UOB A; 4R9D A; 5FZ8 A; 1NZI A; 4XZU G; 5A3U A; 2JB8 A; 2WFF 3; 1HJG A; 2P34 A; 3J7N A; 1WCD J; 4AQ6 A; 4CDX C; 4DXS A; 5DG1 A; 5BUV A; 1RMU 1; 3KF3 A; 4MGQ A; 2RR1 3; 2XG3 A; 2UY9 A; 2AEZ A; 2A3Y A; 1DZT A; 3VZL A; 2HWC 1; 2A5Z A; 4JZR A; 2YU2 A; 5KWR A; 2ZKL A; 3QBQ A; 4PT6 A; 5A7N A; 4EZ4 A; 5DZG A; 1CPM A; 4I30 A; 1W05 A; 4IE7 A; 4YM2 A; 5EPA A; 3AX4 A; 3AZW A; 4P9J A; 4O7X A; 1PGS A; 5E8A A; 3LWC A; 1YFX A; 3N9P A; 4AQ2 A; 4BK4 A; 2RDR A; 4CCL A; 2PHT A; 4A4A A; 1GZT A; 1W2O A; 3WF1 A; 4Q0Q A; 2NPI A; 5F32 A; 3NO8 A; 5K8S A; 4LBN A; 2I2S A; 5GZE A; 1RUN A; 1DLL A; 1K5M A; 1OKO A; 2VOW A; 2WNV B; 5FY0 A; 4Q0P A; 3AMQ A; 1W8U A; 4QMA A; 3HOB A; 5FZG A; 5GZC A; 4I09 A; 2SLI A; 3KZ4 C; 4P08 A; 1DQ2 A; 1ZBE 2; 3AVR A; 1FNY A; 2FZ2 A; 1H30 A; 4PLO A; 2JIC A; 4K3U A; 1KDK A; 3DEC A; 4LUM A; 5GLZ A; 2R06 3; 4Q4X 1; 3VKM A; 3ZKU A; 4AF9 A; 4FPE A; 1C3H A; 1TMF 3; 3HVD A; 2JG9 A; 1NLS A; 1IQA A; 3AFK A; 4QPI B; 3IN9 A; 4C0H A; 4ONU A; 5LRS A; 1RGS A; 3EXU A; 4ZS2 A; 3WUD A; 2B45 X; 2W1S A; 4A2U A; 4IGP A; 3S7I A; 2QZ3 A; 1AL2 3; 5GAL A; 4BMB A; 1KQR A; 1POV 3; 1UL9 A; 2XHN A; 5T50 A; 1WBL A; 1JN2 P; 2VZQ A; 5LK5 A; 2RS3 3; 2ZHL A; 2E51 A; 4GXL A; 5FY5 A; 3WXP A; 1SEF A; 4UBH A; 2QQO A; 2WNV C; 1RYY A; 2J42 A; 2ZGT A; 5EHD A; 1W9W A; 4V2V A; 1COV 2; 4YRD A; 4PJY A; 3K75 B; 2Q8D A; 3TOJ A; 3VDC A; 1POV 0; 1NE6 A; 2Z4F A; 1QNW A; 3ACF A; 4RL7 A; 5T54 A; 1SCR A; 1PIV 3; 4YZ1 A; 1JYT A; 3O5S A; 5FAX A; 5DAV A; 5FP3 A; 4OMD A; 4NMI A; 3PL0 A; 1DQ6 A; 1OBN A; 2WFF 2; 4MOA A; 1V00 A; 3JCI A; 1DYK A; 2XPJ A; 3S30 A; 2NM1 A; 4QPI C; 1D4M 1; 2XKO A; 5FZ7 A; 3LB9 A; 4EN6 B; 2IXJ A; 2YPJ A; 5FKC A; 3LIH A; 2J1E A; 5EXO A; 5FYC A; 4CEY B; 2NLR A; 3GZE A; 3WF4 A; 2D6K A; 1XYO A; 5IVF A; 3AL6 A; 3OGR A; 1G6N A; 4CCK A; 3N9L A; 4I3G A; 4QQO A; 1AJO A; 4LBJ A; 5FMC A; 2WBK A; 3NL1 A; 2TUN A; 3K4D A; 4OZX A; 4RR3 A; 2FDI A; 1BKZ A; 3AKQ A; 4MVT A; 3SHR A; 3OEA A; 3VZK A; 1XE7 A; 3AKS A; 1ZRD A; 5ERL A; 5JOY A; 2PXJ A; 4MGS A; 1R64 A; 2D3P A; 3P3P A; 3AYD A; 3E5U A; 3LEM A; 1QXR A; 1AR6 3; 2N7G A; 5C70 A; 4DUX A; 4CDQ A; 3D8C A; 3I8T A; 4XJU A; 3MV1 1; 2LRP A; 4LK7 A; 4BQY A; 3BPZ A; 1N3Q A; 4DPN A; 1B35 A; 2DYC A; 2BNN A; 1MS1 A; 1KFU L; 2R06 2; 1AUY A; 1G9F A; 1GSL A; 1GWK A; 1U5I A; 3IB3 A; 4E4Z A; 4TLF A; 1YQ6 A; 3ZXL A; 2XQR A; 1TMF 2; 4DRV A; 3NJY A; 4X8P A; 1O59 A; 2VUD A; 3JU9 A; 3MF6 A; 2CGO A; 2RS3 1; 4XJA A; 2FQP A; 3EBR A; 4LO8 A; 2IXL A; 4Y5S A; 3RJ2 X; 2JKX A; 4Q4Y 3; 1RUD 3; 3ZFE B; 1MZE A; 3MF9 A; 3USU B; 2X05 A; 4Q4V 2; 1AL2 2; 3DKW A; 3IBM A; 3TEY A; 2ZEW A; 4GMP 1; 1WD3 A; 1VBD 1; 4CDU A; 2RJL A; 4IV3 B; 3EJK A; 1REE A; 3ZYG A; 1QY4 A; 1AR9 1; 3BU7 A; 4JGY A; 1GZW A; 3I3E A; 4AI8 A; 4KBB A; 4Y20 B; 1GP6 A; 4CXY A; 1UF2 C; 1C8M 2; 1W8H A; 1XU2 A; 1ZRE A; 2D3S A; 1PIV 2; 2NMO A; 5K7P A; 1Z78 A; 4HMZ A; 1SDD B; 3VBH B; 4DMI A; 1N1V A; 4CA9 A; 2PHX A; 1MZ6 A; 1HRV 2; 1X8E A; 5LCW L; 3OF1 A; 1LES A; 2DFB A; 4U36 A; 4Y24 A; 3ZIW A; 4K9S A; 3POW A; 3I4I A; 5CG8 A; 1WW6 A; 2ZGO A; 3VDD B; 5JPM B; 4QX3 A; 2VBB A; 4OUL A; 2PLV 3; 3TBD A; 1XU1 A; 3IPV A; 2ERF A; 3ZFE C; 4KGQ A; 2D40 A; 5L7O B; 1OQ1 A; 5J3U A; 3BAL A; 3GJB A; 2J22 A; 5FZE A; 3CDZ B; 4GA9 A; 2VZ3 A; 1SFN A; 3HQU A; 3VKF C; 2Q30 A; 1YFU A; 2DTW A; 4IV3 C; 3B94 A; 1OI6 A; 3B69 A; 4PQ9 A; 5CFC A; 2ZGU A; 4MLO A; 4OVE A; 1NCQ B; 5C8C A; 5K0U B; 3T0D A; 5JDB A; 1RUI 1; 1VMH A; 3RS6 A; 3VHR A; 4AE2 A; 3AVS A; 4BXS V; 2QBX A; 4XQW A; 5CU1 A; 4LLO A; 3SIT A; 5BNO A; 3HWJ A; 1GOF A; 1PM4 A; 1NQD A; 1R08 2; 4IER A; 3I3Q A; 5LK7 B; 4IET A; 4YM3 A; 2V4A A; 3POI A; 3QH7 A; 4CU8 A; 2EXI A; 2HWD 3; 1BXH A; 2X10 A; 3VBH C; 4UT5 A; 1UOF A; 2R6Q A; 4BCU A; 2JEN A; 2JG8 B; 2DBI A; 1EV1 3; 2CWM A; 3GHN A; 4N7U A; 3PU8 A; 4FI9 A; 3VZN A; 1JMU B; 3UKT A; 4FTB A; 2XQX A; 4XIL A; 3VDD C; 3M4F A; 1W0P A; 1AX0 A; 2CAU A; 3E5Q A; 1RUJ 3; 5F8P A; 1EUT A; 2Q5T A; 4XUR A; 4UFH A; 4IUG A; 1DGW X; 5L7O C; 3OGV A; 3MFA A; 2O0O A; 1Q56 A; 1POQ A; 3JZV A; 5IVY A; 5T3N A; 1QWR A; 3DKQ A; 5CIS A; 4Q4Y 2; 4OHW A; 1VBB 3; 3LIG A; 4H7M A; 5D8A A; 5ANN A; 2DDU A; 3THP A; 5FZF A; 1NCQ C; 3ZDV A; 3GAL A; 4L99 A; 2E6V A; 3OPZ A; 3M7A A; 2VVE A; 3OD4 A; 4LHS A; 4WZF A; 4PLN A; 1PO2 3; 3DUI A; 4HG6 B; 4KWL A; 2ZHM A; 2RMU 3; 1BBT 3; 3VBS A; 4BA6 A; 4JTN A; 3D0S A; 1CAX B; 1GWM A; 1NA1 C; 1BCX A; 1RJ7 A; 4HET A; 1JYX A; 2DTY A; 4QQL A; 4TWT A; 4AW7 A; 1JOB A; 4H14 A; 3CJX A; 2JG8 C; 2CAV A; 3GF6 A; 1XNT A; 3BKZ A; 4MGI E; 5FMO S; 3JBG 1; 3X29 B; 1W0N A; 1WLW A; 2VE1 A; 1PO1 3; 2MEV 1; 3OUH A; 4XAE A; 5FZH A; 3RVH A; 3ZN5 A; 3M3Q A; 3PHM A; 2YMZ A; 4DZG A; 1OLQ A; 1FNZ A; 2VH9 A; 3O14 A; 2Q8C A; 3WW1 A; 3TEZ A; 1W99 A; 4Q8K A; 1M4W A; 1WCU A; 3PVN A; 5LBE A; 5ACA 3; 5JOW A; 2WSU A; 3L9J T; 4GKY A; 3RI8 A; 4XFH A; 4Q5O A; 1KS5 A; 4BB3 A; 1DGR N; 4NIH A; 1AF9 A; 4GWN A; 3O1S A; 3MIF A; 2ZPX A; 1SCS A; 1AR7 2; 2BU9 A; 2DCY A; 2ZGQ A; 3KB5 A; 4O61 A; 3RJR A; 1QKQ A; 2NQY A; 1J84 A; 2WS9 3; 3HNM A; 4XYX A; 4JDM A; 3E6D A; 4OHZ A; 4BME A; 3ZXA C; 5IWF A; 3OGS A; 4RYD A; 1CAV B; 2YC2 A; 2ZEZ A; 4AVA A; 1EV1 2; 4Q4W 3; 1UZW A; 3VD3 A; 3FLT A; 1LEM A; 1RXG A; 1F2N A; 1QF3 A; 1ULC A; 3AMN A; 4GZ9 A; 4LKD A; 5BPX A; 3H8R A; 1DGW A; 2BBV A; 1GUI A; 3ZXJ A; 5A7O A; 3RMY A; 1RUJ 2; 2YBK A; 4QPI A; 4WV8 A; 2HH9 A; 1C28 A; 5FUI A; 1ZM1 A; 2Z42 A; 3J8F 2; 1F35 A; 3N9M A; 4OCT A; 3VBR B; 2WWU A; 3ECQ A; 1JYI A; 4AGG A; 1OXC A; 2DCK A; 3WCS A; 3ZYF A; 3PDQ A; 4P9X A; 3D82 A; 3N9N A; 1WW4 A; 3UGG A; 4CDW B; 5I0T A; 1Y9G A; 2VGD A; 3C7F A; 5LY2 A; 5B2G B; 1BBT 1; 1SLB A; 2ID4 A; 2WUH A; 1OD5 A; 3AKP A; 3C7G A; 3WSO A; 2VW1 A; 4AWD A; 2XL7 A; 3CEW A; 2YYO A; 2RDQ A; 1PO2 2; 4AGV A; 4YG6 A; 4LBK A; 4E2G A; 4KWJ A; 2RMU 2; 2W1Q A; 1O3T A; 2XR4 A; 4TZD A; 1O4Z A; 2D6L X; 3EMR A; 2F4P A; 4R8H A; 3ZXD A; 1OKQ A; 1L7L A; 5KXC A; 3I2G A; 4XF3 A; 3I7D A; 2P3Y A; 2BZD A; 4E4S A; 1R1A 2; 3WDX A; 2V72 A; 2HBU A; 4IE4 A; 1DGL A; 1WCS A; 2JDN A; 2W1U A; 2W94 A; 3WNL A; 1F15 A; 5F2W A; 1J3R A; 5TSW A; 1RTV A; 4BQ4 A; 3VBR C; 1C57 A; 5FWJ A; 4HK9 A; 2A6Z A; 2ZGS A; 4AVV A; 1GQP A; 2AGS A; 5B4X A; 4DXT A; 1FV2 A; 1JH5 A; 3JQX A; 4CEY A; 5IFT A; 2FDH A; 4CDW C; 5CYY A; 2GAL A; 4CEW B; 1HRI 3; 1O5L A; 1VRB A; 3BTA A; 3FE5 A; 3ILR A; 2XML A; 1F9K A; 3GNE A; 4FCA A; 5ACA 2; 2JEM A; 1PX4 A; 3RWN A; 3OBA A; 3C7H A; 3W9A A; 3AP9 A; 3BU0 A; 4MGZ E; 1D2S A; 5EZW A; 3ZYI B; 4CTE A; 4UFI A; 3H8X A; 1X7N A; 3UJO A; 3T2O A; 3AZV A; 2IUW A; 1Q8S A; 1Z7S 1; 1ZVF A; 4KGG A; 2IXI A; 4CP1 A; 4N53 C; 3VKL A; 2YGL A; 1OHF A; 4M3C B; 3ZFF B; 3PUH A; 1G9A A; 1AX2 A; 1WMX A; 2RM2 2; 2B42 B; 2VXR A; 3NNJ A; 2WWS A; 4HVQ A; 1YCI A; 2AAC A; 2BNM A; 4Q4W 2; 4URA A; 5CIR A; 3D5O A; 3E97 A; 1OUX A; 5DDJ 3; 1AYM 3; 1Y3T A; 1VLN A; 4LOR A; 1ULD A; 3NNM A; 3WF3 A; 2O8Q A; 2D6N A; 4AAP A; 1AYN 3; 1D4V B; 5J1X A; 4JAA A; 1S0E A; 2YGM A; 2V5D A; 3BB6 A; 4W50 A; 1ZBA 1; 2YY1 A; 2H0V A; 3LFI A; 1OOP B; 3NW4 A; 2ZJC A; 5J98 B; 5FTT C; 1N1Y A; 3FHA A; 3ZFE A; 4QGM A; 5DAX A; 4BM8 A; 4EN8 B; 4XCA A; 2YRO A; 1IPK A; 2JE8 A; 1W3V A; 1CON A; 2RS1 3; 3USU A; 1N47 A; 1AX1 A; 4BE3 A; 2VCM A; 1LEC A; 3H0O A; 2EAA A; 4PDW A; 2KR2 A; 3LEK A; 1UU6 A; 1HB3 A; 1K45 A; 5CKQ A; 1RUG 3; 3KWV A; 2WYS A; 4N43 B; 5FZB A; 3LDB A; 2BQP A; 4TYS A; 3ZFF C; 3HAG A; 1O4T A; 1LB2 A; 2VJX A; 5FJK A; 1CX1 A; 4OI2 A; 4Y20 A; 4QXH A; 4NWW A; 4D6R A; 1RUO A; 3GCW A; 3WQ0 A; 3WP4 A; 3RPQ A; 1FMD 1; 4ASL A; 3WNP A; 2PAE A; 4R4X A; 4OI1 A; 3KM5 A; 1UD1 A; 5T5P A; 4A3X A; 4CU9 A; 3B7M A; 1XYP A; 3M70 A; 2X5I B; 3VDD A; 5L73 A; 3CLP A; 1V9U 1; 1OOP C; 3KCX A; 3MIE A; 3OBT A; 4PF1 A; 1PVC 3; 2HEV F; 2ENR A; 5L7O A; 2VMC A; 3VHN A; 2OZJ A; 3D3A A; 4TLG A; 1HRI 2; 4XBN A; 3HPN A; 4YPJ A; 1A8D A; 1W8Z A; 1UXX X; 3KGL A; 1NCQ A; 5K0U A; 3PIJ A; 4WM8 B; 2D98 A; 2EIB A; 4N43 C; 2W47 A; 2XON A; 5GZF A; 3URF A; 3PTY A; 3D4K A; 2IVI B; 3WCR A; 2V09 A; 4IW3 A; 3AP6 A; 1JZ5 A; 1V6J A; 5GKA C; 1OF3 A; 4GAZ A; 4HL0 A; 3RZK A; 4XFA A; 3FWE A; 1KFX L; 2BOI A; 3ETQ A; 2Q4S A; 4LR4 A; 1LAJ A; 4LO4 B; 2JWP A; 4UWD A; 5D1I A; 2CDO A; 4KWU A; 4PLM A; 4URT A; 3WNK A; 2E26 A; 1AYN 2; 1GQG A; 3LEG A; 4NRO A; 3UL2 A; 1ENQ A; 1RUC 1; 3MPP G; 4OMC A; 1W6P B; 3HQX A; 1E5R A; 2WIN B; 4LO7 A; 1WD4 A; 2ZGP A; 2BWC A; 3WQ7 A; 2WN2 A; 3QOP A; 4FPY A; 5JVV A; 4K20 A; 2RS1 2; 2ZML A; 2W3J A; 1C1F A; 1CAU B; 4FPO B; 5DAQ A; 3WG2 A; 4CUC A; 1BMV 1; 3K77 A; 1DCS A; 1XNC A; 2PHU A; 4GH4 A; 5A1F A; 3H3L A; 1WGP A; 1K5M C; 1RUG 2; 2HWF 3; 3ZMN A; 1JW6 A; 2CSG A; 3DF0 A; 1QQP 2; 4GUL A; 1LHU A; 1CAX A; 1SLT A; 3FLP A; 3AC0 A; 4I0B A; 2YFU A; 4LBQ D; 5FYS A; 1S1A A; 1YFW A; 2E32 A; 3LOI A; 4W65 A; 2A7A A; 1NKG A; 2WZR 2; 3O7O A; 5JWP A; 3MYX A; 4N9H A; 1LUL A; 3LSD A; 2JG9 C; 1LGB A; 4HN0 A; 2UWC A; 4DRR A; 1D0H A; 3AZY A; 1YQ8 A; 5H9Q A; 1OUS A; 1AHS A; 1RIT A; 1PVC 2; 2V4V A; 1DQ0 A; 3KCY A; 1JBC A; 1GOG A; 4DY3 A; 1SMV A; 3O1U A; 1L3J A; 1CD3 G; 5ABJ B; 4DO0 A; 5LBB A; 4AXO A; 5BV6 A; 2FD8 A; 1OFN A; 2WNE A; 3VD9 A; 3BTX A; 5I0S A; 1OD3 A; 5E88 A; 4LBO A; 3I97 A; 5GLW A; 1PNG A; 1QJX 2; 4G4F A; 3GGL A; 4QPY A; 5FYV A; 4YSF A; 5GM0 A; 1IGO A; 2BJS A; 1GP4 A; 4JGZ B; 3HT1 A; 3R0R A; 1UCX A; 2DBN A; 2QCS B; 5C3S A; 4HOO A; 1UY0 A; 3I2D A; 4AYU A; 4GB3 2; 1CAV A; 3POJ A; 4IE5 A; 4YG3 A; 4AGR A; 1S0J A; 3HQR A; 2VOT A; 4QDR A; 4XUN A; 2BVB A; 5FLH A; 3ZYR A; 4D7Y A; 2FDK A; 2XLF A; 3I2H A; 2A75 A; 3H50 A; 1FOD 1; 2BFU L; 2DV9 A; 2R04 3; 4QGL A; 3PJQ A; 4P02 B; 4DQA A; 5FOB B; 5LRR A; 1GLH A; 3VBR A; 3WD5 A; 3VKO A; 4D94 A; 1URX A; 5F3G A; 4H2A A; 2HWF 1; 5JOZ A; 5ABJ C; 4NPL A; 1WWY A; 1WCK A; 3S5Q A; 5C3O A; 1W04 A; 1UQX A; 1Z0H A; 2GC0 A; 2ZCW A; 3RZJ A; 2GSY A; 3BN6 A; 5FO8 B; 1O3R A; 4QQP A; 1FPN 3; 4RR3 C; 2BRT A; 4ZPI A; 1Q8V A; 3VD7 A; 1O8P A; 1Y9Q A; 1F31 A; 5F9T A; 2F4L A; 2JDY A; 2OUJ A; 4JGZ C; 2GC1 A; 1V6M A; 5AFB A; 4EWE A; 5I91 A; 2CGN A; 2WT0 A; 1J3Q A; 2EIG A; 1A6C A; 4XE9 A; 4Q27 A; 2A73 B; 1OQE A; 4CDQ C; 2D97 A; 4NOT A; 5FV3 A; 3DYO A; 3O0V A; 3OPW A; 3ZM8 A; 2YC4 A; 4CXW A; 1TNF A; 3A0K A; 4XZP A; 4ZXE A; 2WQ4 A; 4CYD A; 2TBV A; 5F3I A; 1SPP A; 2PHL A; 2WRA A; 3N35 A; 4UFK A; 2E7A A; 2GNB A; 5LBC A; 4F8A A; 4QKF A; 4RHV 1; 3UT0 A; 1R09 2; 5CKM A; 1BHG A; 4Q29 A; 4YW3 A; 5IK4 A; 1UX6 A; 4J6G A; 3M3E A; 4FP2 A; 2VD7 A; 5GLU A; 4CEW A; 2B5H A; 3GBG A; 3THT A; 5JUV A; 4AG4 A; 1VBE 3; 3MID A; 1FAY A; 1VRH 1; 3UES A; 3THD A; 4QX2 A; 3MLK A; 1VIW B; 3CMG A; 3FHQ A; 3CKH A; 1B09 A; 1MEC 1; 2VY0 A; 1UAI A; 2LUW A; 5FO9 B; 3LEI A; 1QDC A; 4LKF A; 3UET A; 1BVP 1; 4UXA A; 2XBO 1; 3AJZ A; 1WME A; 2BVV A; 3TYJ A; 4Y5T A; 5FYB A; 1UZY A; 3RG0 A; 1GAN A; 4TTG A; 1RUF 2; 2R32 A; 3HBZ A; 4JDN A; 4M3C A; 3ZFF A; 2LW8 A; 3ZIX A; 1IS3 A; 3S0M A; 4HKO A; 1ZBE 3; 3KQ4 B; 4HN1 A; 4J1W A; 1RUE 3; 2VMH A; 2ZAA A; 4FPJ A; 5FYU A; 5FYM A; 1AJK A; 4XAB A; 2EVX A; 4OUM A; 4G9S B; 2ZMN A; 2K2S A; 3VHO A; 3U11 A; 1XC6 A; 1JXN A; 2YHJ A; 4KNC A; 4LBQ B; 3U14 A; 1ZYB A; 2J1S A; 5FUP A; 3CNA A; 5F2S A; 4C8D A; 1OOP A; 4KH9 A; 1UO9 A; 4CVU A; 1S0B A; 1YUD A; 1O6U A; 3QLQ A; 5CFC C; 5D8A B; 4IEU A; 5C8C C; 2VZS A; 2HEW F; 1TME 2; 2R30 A; 3KVB A; 1D2Q A; 5HXM A; 1OQD A; 3NJX A; 5BNO C; 3MLJ A; 5FP9 A; 5KHH A; 3POS A; 5F39 A; 2YV8 A; 4PF5 A; 1W0O A; 4N43 A; 1O3S A; 2ZX0 A; 1ASJ 2; 2CY6 A; 2G9G A; 2XUM A; 2WT2 A; 1COV 3; 2ILM A; 2Q1Z B; 2OW4 A; 3H1Y A; 4XF0 A; 3UK9 A; 1H6Y A; 2D44 A; 1BJQ A; 2Q8E A; 4ZXK A; 1ZZ9 A; 5CFD B; 1XNB A; 3JB8 A; 4HZF A; 1NXD 1; 5F6L B; 2QDN A; 4JRL A; 4K7J A; 3W59 A; 3S5A A; 1PX3 A; 2CYF A; 2A1X A; 5A3W A; 5IVC A; 4IL7 A; 5LA2 A; 1JHJ A; 2X5I A; 1H2K A; 5DUU A; 2ADE A; 4DIN B; 1VAV A; 1VBA 1; 1KEX A; 3T1M A; 2VW0 A; 1KS4 A; 4OLL A; 1LHV A; 3USS A; 1NZC A; 2BP6 A; 2BFU S; 5JI7 A; 1ENS A; 3WW4 A; 5A6Z A; 5ISL A; 1HXS 2; 2O8O A; 5KHI A; 3IMM A; 5D8A C; 3VHP A; 4A7Y A; 1DU3 D; 1EAH 1; 1QH6 A; 1VBE 2; 2P6H A; 1O91 A; 4LUK A; 1IS6 A; 3J7H A; 4I0W B; 3BUC A; 4WM8 A; 4LHN A; 4QKB A; 3WNN A; 1OLM A; 3C7O A; 3AP5 A; 3V56 A; 2FCT A; 1V6I A; 1VPH A; 3VBS C; 5FY9 A; 1DQ4 A; 2X09 A; 2C26 A; 2FMY A; 4CCO A; 3IDB B; 3MBW A; 3OUJ A; 3QAC A; 4DUW A; 1XOY A; 4Y4Y A; 1OA4 A; 5CZK A; 3AZX A; 2MPF A; 5CFD C; 2R07 2; 1CR7 A; 1RUE 2; 3D0J A; 2EAK A; 1M06 G; 3ILN A; 5SV5 A; 3WW3 A; 1ND3 B; 4CZS A; 4QB6 A; 1OT5 A; 1HV0 A; 2RS5 1; 3OUI A; 2HWC 2; 2WZ8 A; 1GNY A; 2KD7 A; 4YZ4 A; 5FUN A; 4FBF A; 2A6X A; 4C0B A; 3KLS A; 1W6P A; 1WUV A; 1NCR B; 3SCG A; 3WP3 A; 5BXX A; 5DUV A; 3CHX A; 2OYZ A; 2HYK A; 1G8W A; 3FFZ A; 1G9B A; 1KXG A; 2IXK A; 1DQ1 A; 3WX5 A; 5K9Y A; 4Q4V 3; 1CAU A; 2BBA A; 3ZXK A; 5IK5 A; 2ZEY A; 1C1L A; 1COV 1; 4R9A A; 4NIA A; 1W6M B; 3VL9 A; 4F83 A; 3NNF A; 4AOC A; 4WBB A; 3HJR A; 2CTV A; 3SLI A; 1DDL A; 2K46 A; 2BYV E; 3NAP B; 2VMD A; 4Y4R A; 5FRE A; 4ATE A; 3K4A A; 1UZZ A; 3ZDS A; 2WR9 A; 2YXS A; 1JU4 A; 3VBF B; 2WFF 1; 3GHM A; 3HT2 A; 5BNQ A; 4QM9 A; 1C8M 3; 2EID A; 2RR1 1; 2QQJ A; 4FFF A; 3AYC A; 4IE0 A; 5LY1 A; 4BXF A; 4Q4X 2; 2J1R A; 1ND3 C; 1HRV 3; 4NVP A; 2CDP A; 3WDU A; 4MXW B; 1W9T A; 5FXW A; 5GZD A; 1IKP A; 4EN0 A; 4A41 A; 4M4R A; 1LGN A; 1RC6 A; 3CQO A; 1QJZ A; 3E6C C; 2QQM A; 2WLQ A; 3SXW A; 4AED A; 2GN7 A; 1NXM A; 2UY8 A; 5DZF A; 1ODM A; 1YQC A; 3WR0 A; 4LT5 A; 4AVT A; 3PME A; 4HPK B; 5FXX A; 5FYZ A; 3Q8F A; 2K9Z A; 3IDA A; 2VBP A; 4HFS A; 5ABJ A; 1XND A; 5C33 A; 1T2X A; 1RJ8 A; 3DYM A; 3N7J A; 4XDO A; 2C0Z A; 2ORZ A; 4ZZ5 A; 3I2O A; 4LBM A; 1AXK A; 4YW6 A; 1VB2 A; 1VR3 A; 4A7Z A; 1KGY A; 1U5X A; 4CCD A; 3VBF C; 4BHP A; 1UY1 A; 1R08 3; 3ZYH A; 4JGZ A; 1UKR A; 2R06 1; 3POF A; 1UWW A; 3VBO A; 2GMM A; 2QVS B; 2P3I A; 3TAY A; 5B4N A; 1CN1 A; 1ND2 B; 1TMF 1; 1BYH A; 1H2N A; 2A3X A; 3VGI A; 1SLC A; 4NHX A; 1VMF A; 2P5B A; 3KV5 A; 3WDY A; 3PUI A; 1JI6 A; 1QXJ A; 5DG2 A; 1OA3 A; 1D4M 2; 4UYB A; 1GKB A; 1DHK B; 2H6C A; 1AL2 1; 2W2I A; 1XSR A; 1V6N A; 3MGU A; 5CB7 A; 1POV 1; 1H0B A; 4QDS A; 4CEY C; 3R05 A; 3OD0 A; 4RQP A; 2VC9 A; 4P9W A; 5K4L A; 4NHY A; 5C3Q A; 4JHT A; 2F17 A; 1WW5 A; 2WQZ C; 4IEV A; 1UU4 A; 2Z79 A; 1YEW A; 3ETP A; 5JR2 A; 3KQR A; 1T6B X; 1SG3 A; 3M3O A; 1NLQ A; 4PDW B; 3M3I A; 1PZ7 A; 4OQ9 A; 2O1Q A; 2OXB A; 2VZR A; 4NPR A; 2EA7 A; 1JS9 A; 3NQH A; 5JIA A; 5FXV A; 4E27 A; 5A33 B; 2B8M A; 1PIV 1; 2IC1 A; 3Q8E A; 1EPW A; 4NAO A; 4OUE A; 4WVM B; 5FZI A; 3ZN6 B; 1W6N B; 4AFB A; 2EIE A; 5AC9 3; 2HWB 3; 2VG9 A; 5EJZ B; 3VL8 A; 4A6S A; 4V2W A; 5FLJ A; 4A45 A; 1SBF A; 3ORJ A; 1ND2 C; 5F37 A; 4GAL A; 4AVB A; 1JU3 A; 1REF A; 3AK0 A; 5H9P A; 1L7Q A; 3W58 A; 3EB7 A; 3ACI A; 3GBF A; 3VZM A; 5BY5 A; 1S2K A; 3R4D B; 4FPK A; 2ET1 A; 2WO3 A; 1I5Z A; 1UU5 A; 4FQZ A; 1YJK A; 3FUO A; 2JKB A; 1XB9 A; 2XVK A; 1U5Y A; 1CZS A; 4DXZ A; 4V1P A; 4E2Q A; 2JD9 A; 3TNQ A; 5FZ0 A; 5IAV A; 1AXY A; 1VBD 2; 2LTN A; 1XRU A; 4WVV A; 4PDW C; 4YZ5 A; 4O9E A; 1S4C A; 3VDA A; 3H7J A; 4MSV A; 4AFA A; 3IDC B; 4LQR A; 5G05 L; 1AR6 1; 1ULE A; 1YIP A; 2VBD A; 1J83 A; 2VCA A; 2XH6 A; 2Z5W A; 4ZU5 A; 2A3W A; 5J37 A; 1AR7 3; 3H8O A; 1HJF A; 4LO5 B; 4BQ2 A; 5IAT A; 2J7M A; 1NYW A; 5A7Q A; 2ARB A; 5I8T A; 1LOF A; 3F2Z A; 2W5F A; 4Z2A A; 2E33 A; 1TEI A; 5L36 A; 1G9C A; 5F6K A; 2ZX2 A; 4Q4Y 1; 1RUD 1; 5KBF A; 4E4D X; 4LBQ A; 5FYI A; 1VBC 3; 3PUR A; 3MW2 A; 4JDO A; 4D6S A; 3J8F 3; 4FSJ A; 4NHL A; 1PM7 A; 3I3D A; 2GAU A; 3FBM A; 3T0A A; 3NV3 A; 3OTF A; 3WV6 A; 2QE3 A; 5E16 A; 4B7E A; 4ZS3 A; 5K0U C; 4LPL A; 2PAM A; 5I8Y A; 1QJE A; 4GH4 B; 3PUQ A; 2OX0 A; 2OPK A; 2PHW A; 5LB6 A; 1IZ3 A; 5AC9 2; 4MO3 M; 1OC1 A; 2HWB 2; 4AHX A; 1YQ5 A; 1W90 A; 4LK6 A; 4XOG A; 4AHY A; 3UJQ A; 3Q8B A; 1RHI 3; 4PI2 A; 1O8S A; 5J36 A; 2PLV 1; 4X6Q B; 1XSQ A; 5JLV A; 1E5S A; 1R1A 3; 4SLI A; 4F3J A; 2FZ1 A; 1O4Y A; 1F4A A; 4KBZ A; 4MGK E; 5CFD A; 1P8J A; 4X8N A; 3UGH A; 2PHF A; 4JQT A; 4XFB A; 2GN3 A; 2DUQ A; 2YB7 A; 4ERO A; 2E31 A; 4BWO A; 4QB1 A; 4MH0 E; 4KU7 A; 3LDK A; 1VAK A; 2WNU B; 4YFZ A; 3HMY A; 1TG7 A; 2DCZ A; 3NVC A; 4BOW A; 5CAZ A; 3BTY A; 1W6Q A; 2W68 A; 5FCC A; 3VKN A; 3VUO A; 3ZFG B; 3BYW A; 3KV4 A; 1SLA A; 4IEP A; 4QKD A; 4HSJ A; 4I3P A; 5AFE A; 1D0G A; 1DZR A; 2P3J A; 4GH4 C; 3SC7 X; 4EWA A; 1EPZ A; 1WLC A; 4GQ7 A; 4HXC A; 2HWD 1; 2HBT A; 3J7M A; 4BK5 A; 2IFW A; 3WDW A; 1EV1 1; 5DZE A; 2FDF A; 4DS0 A; 1LGC A; 1Y9M A; 2ATF A; 1SBD A; 1DQ5 A; 4ET7 A; 1WA6 X; 2JD4 A; 2WAA A; 2RM2 3; 2GZW A; 3AKT A; 4AVS A; 5F3C A; 1RUJ 1; 5FPZ A; 2AYH A; 3JD7 3; 3ENR A; 2BOJ A; 2WC2 A; 1IKQ A; 1SLI A; 2XL9 A; 1RUH 3; 3AMP A; 5EJ1 B; 2MEV 2; 1ND3 A; 3DV8 A; 2DWR A; 4JQR A; 4JKM A; 1VBB 1; 5EWS A; 5T55 A; 1Y43 B; 1BEV 3; 4NJ4 A; 1ZZ6 A; 2ZTN A; 1BQP A; 1CX4 A; 2WNU C; 3U10 A; 4A42 A; 4UW6 A; 1VBC 2; 3HN1 A; 1NCR A; 1YYN A; 3OCP A; 5IK8 A; 2PFW A; 3AMH A; 3IMN A; 3ZFG C; 5LA0 A; 2ZGR A; 1UZ0 A; 1PO2 1; 4IES A; 3WP6 A; 4EQV A; 5FBJ A; 4BQ1 A; 2RMU 1; 1W6M A; 5FPX A; 3FLR A; 3HNB M; 5A70 A; 1NE4 A; 2VZT A; 3HEI A; 4IDZ A; 2AC1 A; 3NAP A; 2YBP A; 5F3E A; 1HLC A; 3B01 A; 3SCH A; 3E6B A; 2JIG A; 3VBF A; 1HQN A; 4KI5 M; 5DAP A; 1PO1 1; 4FTS A; 1RHI 2; 3ZX7 A; 2Y6G A; 2E9Q A; 3T3Y A; 3UKV A; 3RQ0 A; 4OYU A; 3U4S A; 1H8T A; 3W5N A; 2BNO A; 5I0U A; 1QDO A; 4NHM A; 2R0H A; 4JX0 A; 4Q0V A; 3RZH A; 4I0A A; 4YGC A; 4MXW A; 3MUZ 1; 4CPB D; 3NKT A; 1SPP B; 5EV7 A; 4LLF A; 5ACA 1; 1XBF A; 3CZJ A; 3RYR A; 2XXZ A; 4KQ7 A; 3T1L A; 4RN5 A; 2WII B; 3DPR B; 3VZJ A; 2VAU A; 3RYK A; 3V0B B; 1K5J A; 4P9G A; 4XFG A; 4XF9 A; 3T08 A; 4HPK A; 1V6L A; 1J1L A; 4PCR A; 2EXK A; 4DEQ A; 4FPG A; 2WS9 1; 3ALQ A; 3Q9O A; 5M0T A; 2ORX A; 2JE7 A; 2VVD A; 4C90 A; 5FZ9 A; 1HIX A; 2JDK A; 3N9Q A; 1I6X A; 3PIG A; 4Q4W 1; 1ZZC A; 2DCT A; 5KXD A; 1MVE A; 3I2F A; 2G1M A; 1JTZ X; 1SHW B; 1ND2 A; 1XNK A; 1AL0 G; 1MAC A; 3WDT A; 2Y33 A; 3JD7 2; 2VZU A; 3ZVX A; 3TN9 3; 2NZ9 A; 4LHL A; 4AK3 A; 1RUH 2; 3A22 A; 4FOY A; 4HT1 T; 3RZL A; 4NIG A; 1HQL A; 2NYY A; 1BEV 2; 1OH3 A; 2C4X A; 4XUQ A; 3DPR C; 3KGZ A; 2EXH A; 1E5H A; 3DYP A; 1GNZ A; 1I3H A; 5H9R A; 4CXX A; 4CEW C; 5A3O A; 2ZMK A; 4ARX A; 5JMC A; 4XEZ A; 4NAM A; 1EY2 A; 1GNH A; 4XW3 A; 1L7R A; 2Y6F A; 4XVH A; 1ZRC A; 1UPS A; 3HN3 A; 1PGW 1; 3O1M A; 4XUT A; 5A33 A; 5CNA A; 2P17 A; 2D7F A; 3B9C A; 4WVM A; 3EHK A; 3ZO0 B; 4YM1 A; 3PNA A; 5FY8 A; 1W6N A; 3ZN6 A; 2X53 S; 3B9I A; 1QQP 3; 2QQL A; 4FOQ A; 1SQ4 A; 4MXV A; 3I2K A; 2PA7 A; 3GZT B; 4LIV A; 4GJY A; 3DN7 A; 3ST7 A; 1Q8O A; 3LFM A; 1NT0 A; 4CUB A; 2Q8O A; 4XXP A; 1HB2 A; 3IAQ A; 4Y1Z A; 3O0W A; 4FTE A; 1W39 A; 2FMD A; 2BUK A; 2RHP A; 3III A; 4H7L A; 4MX3 A; 4QX8 A; 4YWA A; 5FLI A; 5FZ4 A; 4AFD A; 2WZR 3; 2VQU A; 3LDR A; 4WM7 A; 3TEW A; 1VJ2 A; 1HRI 1; 2VNG A; 2R1B A; 5FDB A; 1ZBA 2; 4L2N A; 3PVO A; 1R1Z A; 2DFC A; 1WA4 A; 4Y22 B; 1A34 A; 4XMA A; 2XJQ A; 1S0I A; 3F95 A; 4LA3 A; 2OA2 A; 3KCC A; 5FZK A; 1H1A A; 4LOT A; 1S2E A; 5BNN B; 1O3Q A; 4N90 A; 2HWE 3; 5JSP A; 5IK7 A; 2CV6 A; 2ZGM A; 1ZZB A; 5IHR A; 2D43 A; 4I01 A; 1UMZ A; 1JYV A; 2GU9 A; 5KXB A; 4XFC A; 3M1H A; 3FN9 A; 1QJX 3; 5F5A A; 1JZ4 A; 1UN1 A; 2BW8 A; 3U4X A; 1W2A X; 3N7K A; 3V0A B; 4D0Q A; 4AL9 A; 3K4Z A; 5DDJ 1; 1AYM 1; 2WFP A; 3HBK A; 3L2Y A; 2VMI A; 1FMD 2; 1AYN 1; 2ZGL A; 1H1I A; 1S2B A; 4S2F A; 5DUW A; 4GB3 3; 3TN9 2; 4CU6 A; 2OII A; 4GWM A; 4A7K A; 3NRU A; 4XAC A; 5KHJ A; 3P1I A; 4OHV A; 1GZC A; 2X5I C; 1V9U 2; 2RS1 1; 2A70 A; 3NMB A; 3ILF A; 2VZP A; 1T6G C; 4P5H A; 2VM9 A; 4DFS A; 1GOH A; 2ZBJ A; 1QNY A; 4LKS A; 4R52 A; 4CP9 B; 3KF5 A; 3AFG A; 5BNN C; 4CCM A; 1RUG 1; 3CL1 A; 5JSO A; 3ZPY A; 1ALY A; 1F5J A; 1ULP A; 5F5C A; 1QQP 1; 5L9V A; 2LS6 A; 1UIJ A; 1AR8 3; 4WM8 C; 4G3Y C; 3RI9 A; 2FCV A; 3KV6 A; 2BWA A; 1NX9 A; 4K8F A; 4COV A; 2OUH A; 4M0H A; 3AXE A; 3RGB A; 4QXB A; 5FJH A; 4M3C F; 2VMF A; 5LAT A; 4P9I A; 5HXV A; 5EYX A; 3MLL A; 5DQ0 A; 1SZB A; 3K46 A; 3BPS A; 3O1R A; 1LU2 A; 4A6O A; 1HN0 A; 3ACH A; 3GYD A; 5A3M A; 1PNF A; 2R1D A; 2XJP A; 2XWJ B; 3VD4 A; 2VPV A; 4YGD A; 5T52 A; 1PVC 1; 2XJU A; 1HDK A; 3LCP A; 3WHU A; 1OMI A; 1RUC 2; 2PAK A; 2XDV A; 2RG4 A; 4O9G A; 3ATG A; 5KR7 A; 4EV0 A; 2WNU A; 3MPB A; 4AED B; 2Y0O A; 1OA2 A; 1R09 3; 1YFY A; 5FRA A; 1LOD A; 4KMQ A; 4R9C A; 3ZYB A; 1JZ6 A; 1WKY A; 4COU A; 1RED A; 2G4I A; 2QJV A; 1BMV 2; 2C5D A; 3ZFG A; 2HWE 2; 3POG A; 5A1A A; 4LA2 A; 3T0B A; 1G9D A; 3UKN A; 4YYO A; 3OJB A; 2EIC A; 2UYA A; 2VCC A; 4QX7 A; 1F5F A; 2QQN A; 1XPW A; 3KT7 A; 3WF0 A; 4NWV A; 5IUQ A; 1UYZ A; 1NUK A; 4ISR A; 2ZZJ A; 1RUF 3; 2JDA A; 1HV1 A; 1A8M A; 3VBO B; 4BJ0 A; 4EN9 B; 3OYW B; 4XQ4 A; 5JXH A; 1QJU 3; 3O1O A; 4PIX A; 2UU8 A; 2Y6H A; 4QHZ A; 2R16 A; 3GCX A; 4Y5Z 0; 2XJT A; 1RVF 3; 2WAO A; 4LE4 A; 3SCF A; 1QJY 2; 3K48 A; 3B93 A; 4CCN A; 5A3L A; 2KXL A; 4AFC A; 3WQB A; 5FPB A; 2P6C A; 2P2K A; 3LA7 A; 1U0A A; 4QT6 A; 5FKB A; 4AED C; 5CBL A; 4MAD A; 3THC A; 5J96 B; 3GDB A; 3VB9 A; 4D6Q A; 3OGG A; 3VLB B; 3A23 A; 3I3B A; 3ON7 A; 4NN0 A; 4OHY A; 4RQP B; 3J7L A; 1MS8 A; 5FPV A; 5A7P A; 3BGA A; 3HNY M; 4K6B A; 4QXC A; 1ZA4 A; 3C8X A; 1MQT B; 1OSG A; 3LQC A; 3ZMO A; 1TME 3; 2XHK A; 4OQ8 A; 1AR8 2; 1H2M A; 2WZE A; 4IEO A; 4HON A; 1YXW A; 4IEY A; 3JQW A; 2ZX4 A; 5FZD A; 4QX0 A; 3NKG A; 2ZAH A; 4M0N A; 3RZZ A; 1ASJ 3; 1HQW A; 1IQD C; 1AXZ A; 2VOK A; 4M01 A; 3VBO C; 1N3O A; 1W8O A; 4SBV A; 2CU5 A; 3JB4 C; 4S2H A; 2JDH A; 2VNR A; 4FAG A; 1HX6 A; 4ZZ8 A; 1OUR A; 5HV0 A; 1YIF A; 2FDJ A; 3KVA A; 5L9R A; 4LOS A; 3EQE A; 1LU1 A; 1FOD 2; 3A7Q A; 3PMS A; 5FZ1 A; 5LBF A; 3K51 A; 4P9Y A; 1GV9 A; 4KWK A; 3LOB A; 3AYA A; 2ZHN A; 4JKK A; 5T5O A; 3BTZ A; 4TN7 A; 2YBS A; 4RQP C; 4BIS A; 3DPR A; 3V0B A; 1X36 A; 2WA3 A; 1NOV A; 1HXS 3; 4FOV A; 3AP7 A; 4O65 A; 1MQT C; 2QQV A; 3N36 A; 4UFL A; 2A6W A; 3V64 A; 2PHR A; 2MNG A; 4XQD A; 1KJR A; 1VAL A; 3WDV A; 4AVC A; 4Q2F A; 3DXT A; 2GC3 A; 2VQA A; 3FL7 A; 2WA4 A; 1WMD A; 2PTM A; 2RJK A; 2ZAB A; 2YG0 A; 3CO2 A; 1XNA A; 3WW2 A; 1QJU 2; 2HJI A; 3LA3 A; 4CE8 A; 2R04 1; 1OH4 A; 2WL1 A; 2PQS A; 1HN1 A; 3WNX A; 3I2J A; 4DY5 A; 2R07 3; 1MZ5 A; 1RVF 2; 1ZA7 A; 1X9T A; 2VNV A; 2WO7 A; 1PHM A; 2VVF A; 2WQ8 A; 2GNM A; 1CGP A; 1WW7 A; 2HWC 3; 2BDR A; 5A7W B; 4BPZ A; 5IVB A; 1YLL A; 1JYW A; 2Y64 A; 1FPN 1; 2ES3 A; 4IEQ A; 3ZN4 A; 5FXZ A; 4E4H A; 2W9X A; 1TVG A; 1VRH 2; 1J1T A; 1ENX A; 2I74 A; 2VUC A; 2BGP A; 1ULG A; 3AP4 A; 5E89 A; 5GZG A; 1MEC 2; 4QXK A; 4XF1 A; 4HSL A; 4QKN A; 4XIO A; 1X9P A; 4Z82 A; 3ELN A; 2J1U A; 2V5Y A; 1WLD A; 3J4U H; 5JPN B; 4K40 A; 5DMY A; 2GME A; 2QQK A; 2DVF A; 3O1P A; 5IYY A; 1VP6 A; 3FHI B; 4N9I A; 4XC9 A; 5FBZ A; 1S0G A; 1JHN A; 5KC6 A; 1QHD A; 1LOF C; 2JE9 A; 4Q26 A; 2A71 A; 3C7E A; 1UOJ A; 3BIW E; 4OI4 A; 3VZO A; 5J48 A; 3MEP A; 2J1V A; 4LE3 A; 4NR1 A; 4PIZ A; 4Q4X 3; 4YZ3 A; 1DP0 A; 2ZGN A; 4UF0 B; 3MZH A; 4W8J A; 4ZY9 A; 3QH6 A; 4L9B A; 5JAX A; 1PGL 2; 4OZO A; 3ZLI A; 4XHB A; 1DF0 A; 1VBE 1; 2QDR A; 3AMM A; 1DYP A; 2BV4 A; 4IGO A; 1GIC A; 4BIO A; 4Y22 A; 3GXU A; 5BNP B; 4BQ5 A; 4EWD A; 5IFP A; 3D6E A; 3H3Z A; 4B29 A; 3LAG A; 4QX1 A; 5LEK A; 4BDV B; 3A4U A; 1A78 A; 1W03 A; 1W6Q B; 4I02 A; 1Q8Q A; 4W6X A; 5BNN A; 5T5J A; 2JDP A; 5C3P A; 1APN A; 2XVG A; 2DU1 A; 2VTF A; 1ZNP A; 3FX3 A; 2Y6J A; 1ZBE 1; 3VBU B; 4Y1U A; 4AEJ A; 3SH5 A; 4YVW B; 1NY7 2; 2Z2S B; 3WQ1 A; 2R07 1; 4HBN A; 4CSW A; 1RUE 1; 3DGT A; 2W91 A; 3SNM A; 2JXA A; 2I07 B; 3R4U A; 4JVA A; 3AB0 A; 4UBG A; 2W08 A; 1SDW A; 5ANQ A; 5FPL A; 1W6O A; 5F1R A; 5HPO A; 3V0A A; 1ACC A; 2VO5 A; 4XJ8 A; 3LA2 A; 4IE6 A; 1JOD A; 3DCQ A; 4PI0 A; 4A44 A; 3HIF A; 4QWN A; 5FP7 A; 5LA9 A; 4E8D A; 2YFZ A; 2E7T A; 4ZU4 A; 5JXG A; 1SAC A; 2GH8 A; 1RMU 2; 1D4M 3; 4XTM A; 1VBA 2; 2Q1F A; 5FK7 A; 1GR3 A; 1LTE A; 1C8N A; 5BNP C; 3O0X A; 2JDU A; 4UOJ A; 3WZ1 A; 4CCE A; 2C1F A; 5F7U A; 2DCJ A; 4ZHN A; 3NNL A; 1JZ8 A; 3SIS A; 2OKX A; 2JDZ A; 2VUJ A; 3N9O A; 1EAH 2; 2JEC A; 4ARY A; 1BK0 A; 1UZV A; 2VOV A; 4BZ4 A; 4CP9 A; 3WG3 A; 2BPA 2; 5C4W B; 4CPB A; 3H9A A; 4W6Y A; 1PMH X; 2DUP A; 3K2O A; 3VBU C; 3T4H B; 3M2M A; 5JMO A; 3I3M A; 2UUR A; 1QOO A; 3ACG A; 4YVW C; 1MPX A; 4GWI A; 2PEL A; 3NJZ A; 2ARC A; 2ZZQ A; 4MV2 A; 3IM0 A; 5FP4 A; 2AR6 A; 3Q8C A; 5C7G A; 5JXJ A; 4CP2 A; 1AZD A; 5LZ6 A; 1PZ8 A; 2HYX A; 2WNO A; 4QB2 A; 2W87 A; 1JZ2 A; 2Y38 A; 4XCB A; 4LJH A; 4DIQ A; 3B3Q E; 1NQJ A; 1H8T B; 3ABZ A; 2XLA A; 3U1X A; 4AEK A; 3N7M A; 5JXI A; 4BH9 A; 1JZ7 A; 2RS5 2; 5HV4 A; 3J26 N; 1S55 A; 4H55 A; 5DAW A; 2VOX A; 3ZSL A; 2ADD A; 4BYY A; 3KHB A; 5C5U A; 1OVS A; 4K1Y A; 5A1L A; 5EQU A; 5A32 B; 4UI9 L; 3FFQ A; 1FXZ A; 1NCR C; 5GLT A; 1FT9 A; 2OQ7 A; 4ISQ A; 2IZW A; 4D69 A; 2UWA A; 1TQ5 A; 5IT6 A; 5C4W C; 1JOJ A; 2D6P A; 1MS9 A; 2C9A A; 3H42 B; 4LNV A; 4EZH A; 1X33 A; 3OSD A; 4GMP 3; 1VBD 3; 4RD7 A; 1KJL A; 4E8C A; 1YWK A; 3AYE A; 3AJY A; 3J9S A; 5APA A; 4FPH A; 5AVA A; 1AR9 3; 3AM2 A; 1KIT A; 4QQH A; 2D5F A; 3NAP C; 1ENR A; 2D3R A; 2X8K A; 3PJY A; 1SLL A; 1K12 A; 3RMX A; 1LED A; 4XAA A; 4GMP 0; 2KA3 A; 4DXR A; 2B9V A; 2Q0A A; 2RR1 2; 3A0N A; 5L9B A; 3OYW A; 5C6C A; 1I1E A; 3SH4 A; 1W8T A; 3O1T A; 1H8T C; 4UW5 A; 2QFE A; 2CWS A; 4B1L A; 3DL3 A; 1MS5 A; 3SQO A; 2GP3 A; 4QPW A; 3S57 A; 2R49 A; 2WN3 A; 3BVC A; 2SBA A; 2ZA9 A; 3NST A; 1JUI A; 2NN8 A; 1Q8P A; 4LBL A; 4Q4V 1; 1QOS A; 4R9B A; 5CG9 A; 2XVL A; 4NII A; 1GMM A; 2WYF A; 4MHU A; 2ZGK A; 2L9L A; 1UPI A; 3CZU A; 3N4M A; 5FK8 A; 1TZO A; 1UYX A; 4FPL A; 3WOG A; 3OBR A; 1J59 A; 2YZ4 A; 3LKJ A; 3OPT A; 4OI0 A; 4B4R A; 1MQT A; 5FMO L; 4FOW A; 1RUI 3; 1STM A; 3VDB A; 4NPM A; 1W9Y A; 3SEP A; 1X8M A; 1C8M 1; 2AZ5 A; 4BLJ A; 3H1M A; 1SBE A; 4YW2 A; 4S2D A; 2VME A; 2HKX A; 2ZX3 A; 3H8U A; 2RJ2 A; 2DVD A; 2YDE A; 4ASK A; 4EN7 B; 1HRV 1; 1LOB A; 1HW5 A; 3AZZ A; 5IVE A; 3S7E A; 2EF6 A; 3OBZ A; 4XFF A; 4AQS A; 1UW8 A; 2XKP A; 3UNP A; 2VNO A; 2VZV A; 1XQN A; 4QU1 A; 1V6K A; 2Y8K A; 1G86 A; 3U37 A; 2G9F A; 3L2H A; 3AKR A; 1PK6 B; 3Q8A A; 5JR7 B; 3UV9 A; 5A3P A; 1GWL A; 3ASI A; 4QGN A; 4WM7 B; 2QQP A; 3RZM A; 3WLU A; 3MIH A; 2WT1 A; 2RS3 2; 3MUY 1; 4JTO A; 1CAW B; 2VTX A; 5FIX A; 5C9A B; 3RDI A; 3HD8 B; 4LO6 B; 4Z2W A; 2UWB A; 3G7D A; 3RYP A; 5FQ0 A; 3LPG A; 4C1W A; 4JV4 A; 3RZG A; 4Q1P A; 1R08 1; 1YJL A; 3U75 A; 4QHO A; 1AR9 2; 5J09 A; 2D6O X; 4FP3 A; 1N1T A; 2AEN A; 2G19 A; 4QQ2 A; 4S2G A; 2EXJ A; 3W5M A; 1AVB A; 1D7P M; 1U12 A; 1X82 A; 1UY3 A; 2VZO A; 1TE1 B; 2AH2 A; 4Z8B A; 4K1Z A; 4RS5 A; 3O12 A; 4XF4 A; 4NUB A; 4NE9 A; 2IWG B; 3MFC A; 1FAT A; 1LOC A; 3T2P A; 4Y1V A; 5IVJ A; 5A7S A; 4Y33 A; 2LAL A; 1LOE A; 1PK6 C; 3JBG 3; 4L0P A; 4Q0S A; 4DEV A; 2BER A; 2ETE A; 4HVR A; 4WM7 C; 4GD4 A; 4AZZ A; 1TNR A; 4I2O A; 1UY2 A; 4XPV A; 2MEV 3; 2MHF A; 3NJV A; 1UY4 A; 3IIA A; 2PN5 A; 4D8M A; 5A7W A; 5C9A C; 4FO9 A; 2ARA A; 3SEE A; 3O1V A; 2RDS A; 4OHX A; 4UW4 A; 2VUA A; 1UX7 A; 2GNT A; 3FUQ A; 1W9S A; 3VV1 A; 1LNS A; 1DIW A; 4B3N A; 3NV4 A; 5AC9 1; 4XDP A; 4XJZ A; 3W20 A; 3MW4 A; 2HWB 1; 4EJR A; 4P5Y A; 3IKW A; 1NA1 B; 1RUI 2; 1AR6 2; 1CIY A; 1JZ3 A; 5KHK A; 4JKL A; 3RFR A; 4L92 A; 2PYT A; 2BTV C; 1QMJ A; 1LHO A; 1ZRI A; 4PIY A; 2Y60 A; 5FWE A; 1RIN A; 3RSJ A; 3CF6 E; 3PUS A; 3LZZ A; 4CDX A; 4FT8 A; 2W52 A; 1HB1 A; 5FFO C; 2VEC A; 4QU2 A; 2O14 A; 2GP5 A; 1W8W A; 3PU3 A; 5H9S A; 4COW A; 4B8E A; 4Z1V A; 3CBN A; 4DB5 A; 2XLG A; 3H3U A; 3KSC A; 1DGR A; 3GZA A; 1RUD 2; 5IVV A; 4UF0 A; 1W9F A; 1QGL A; 1UYP A; 2C6S A; 4UFJ A; 4BLI A; 4YZ2 A; 1W2N A; 2XJS A; 4CUG A; 3U0Z A; 5FYT A; 1H1M A; 2Q1M A; 5BNP A; 4YW1 A; 4IQP A; 1OF4 A; 3UYJ A; 2C9K A; 3POE A; 1GZ9 A; 1K3I A; 1ODN A; 2IXH A; 4CP0 A; 4FFI A; 1AR7 1; 1I9R A; 3OGJ A; 4NID A; 1CQ9 A; 1W28 A; 5FOJ B; 5HI0 A; 4MG2 A; 2HEY F; 3B02 A; 3BCW A; 4OUS A; 4M4P A; 4RFR B; 1PMJ X; 2ZEX A; 3VBU A; 5EP9 A; 4YVW A; 3NNG A; 3UGF A; 1C4R A; 4ZU7 A; 3ZR5 A; 2GMP A; 5FP8 A; 4YG0 A; 1KD7 A; 2PLV 2; 2QUO A; 1XYN A; 1UXZ A; 4M3C E; 3SH3 A; 3WF2 A; 1WMF A; 3JBG 2; 1VBC 1; 1S0F A; 4QX5 A; 4B1M A; 1YHF A; 2B4K A; 3J8F 1; 2PQQ A; 4FFH A; 1LKN A; 4XFI A; 2ZKN A; 2GDF A; 2E53 A; 5D21 A; 1RL3 A; 1GFF 2; 2X29 A; 1DYO A; 2QKD A; 5FY4 A; 4COZ A; 3NWK A; 4BQ3 A; 4LEJ A; 4ORF A; 3ZPO A; 4AQT A; 3I49 A; 5FZ3 A; 4RGA A; 2DVG A; 2WNV A; 2D5H A; 5C4W A; 4D7S A; 1YRZ A; 2B7Y A; 3SMH A; 3TWI A; 2UYB A; 4NRP A; 4HLT A; 2GC2 A; 1WLT A; 2HWD 2; 4QPG B; 4AEM A; 1U5Z A; 4CRQ A; 4WVW A; 1RHI 1; 5FZ6 A; 1UYY A; 4A3Z A; 1EUU A; 1GP5 A; 1W3X A; 3GM8 A; 4RFT 0; 3OB8 A; 1H6X A; 1R1A 1; 1H4H A; 1X50 A; 1Z7S 3; 5LEJ A; 4Z92 B; 3OY8 B; 2P4E A; 2BGO A; 2V73 A; 1UMH A; 2WDB A; 2GH2 A; 2NMN A; 2IVJ A; 1SFY A; 2W95 A; 2WLP A; 3KS7 A; 4QDQ A; 4QM8 A; 1CZT A; 5J7M A; 1IS5 A; 3BI3 A; 4Y1X A; 5EYY A; 2FCU A; 4B4P A; 1CVN A; 1GYK A; 2CNA A; 4P9L A; 4CUA A; 1CPN A; 4N7I A; 3C2U A; 4DUV A; 1EP0 A; 1VBB 2; 1W8F A; 3MHZ A; 1QEX A; 3LPF A; 4U2A A; 2P3K A; 5A32 A; 1ZBA 3; 4AI1 A; 2F6B A; 2P1B A; 4MUV A; 4AI3 A; 2E7Q A; 2IXC A; 3I59 B; 3S6P A; 1CJD A; 2YU1 A; 1H8V A; 2AUY A; 3M8L A; 3MIG A; 4E2S A; 4BU2 A; 3ZSK A; 1QXP A; 5C8W A; 2RDN A; 3N7L A; 5HBA A; 2DF7 A; 1BBT 2; 4RGG A; 1PVX A; 4YVS B; 5A6X A; 1ZRF A; 4QPG C; 1Y4W A; 4N53 B; 3HR9 A; 1S0D A; 1E57 A; 4GIQ A; 5DAB A; 3KV9 A; 2RM2 1; 4ES9 A; 4EYU A; 1DLC A; 5I93 A; 4D3W A; 1W06 A; 1FYU A; 2XWB B; 3JD7 1; 3NV2 A; 3ROU A; 1E5I A; 1OLM E; 1KDM A; 1RUH 1; 2B46 X; 1LCL A; 1PO1 2; 2QQW A; 4LMF A; 3P5B A; 5A6Y A; 5IW0 A; 5FPU A; 1LOA A; 5LW9 A; 4PHZ A; 3LGR A; 1BEV 1; 1FMD 3; 2WZP P; 3RWK X; 2WO1 A; 5C5T A; 1WZX A; 4N8M A; 1XXN A; 3GJA A; 2Y6L A; 4CCJ A; 1NLR A; 3PTR B; 1DG6 A; 4UFM A; 4YGB A; 1S0C A; 4RA0 A; 2W46 A; 1FI2 A; 5JSR A; 4XJW A; 5IJR A; 4COY A; 1V9U 3; 2NP0 A; 1ZZ8 A; 3VD5 A; 5F5Q A; 1IG3 A; 4FPF A; 4UW3 A; 2ZHK A; 2XOM A; 3TRS B; 1QU0 A; 4CCC A; 5CA6 A; 2DVA A; 2W92 A; 3CIH A; 4AFM A; 1MS4 A; 1CWP A; 3ZKY A; 5FZM A; 4YVS C; 2H6B A; 3EO6 A; 1V70 A; 2RV9 A; 1LOG A; 2WS9 2; 3TB0 A; 3WG1 A; 3WEZ A; 1OVP A; 2PMW B; 2OQ6 A; 1HKD A; 4HK8 A; 5FMB A; 4PBO A; 1MS0 A; 3T30 A; 4IV1 B; 2A6Y A; 3EYP A; 2CHH A; 3AXD A; 2QQI A; 4IXL A; 2DUR A; 1Z7S 2; 3FC0 A; 1IS4 A; 3N3H A; 5E6H A; 4LIU A; 4Y1Y A; 4F7Z A; 2CL2 A; 2JB4 A; 1YO8 A; 4AI2 A; 4AR2 A; 5FO7 B; 4Q0U A; 5JZG B; 4LTA A; 3ZXF A; 2ARE A; 3KT1 A; 3P3N A; 3FBY A; 1LJ7 A; 5HCF A; 4K8R B; 1PK6 A; 1OLR A; 2RVA A; 5FZO B; 1BVV A; 3CJI B; 1RUC 3; 5DUX A; 1Q5O A; 4C91 A; 3ZXE A; 3H7Y A; 4IV3 A; 3ZXG A; 5C9A A; 1CAW A; 3LE0 A; 4KG0 A; 4ZKT B; 1H4G A; 2P37 A; 2I45 A; 3B00 A; 4AI0 A; 2H0B A; 4P14 A; 4L11 A; 5JOX A; 4JHZ A; 3ES4 A; 1PGW 2; 2LRO A; 1YI9 A; 4IV1 C; 4NO4 A; 5I8S A; 5A80 A; 3WVJ A; 3VBH A; 1DFQ A; 1YNA A; 2WJS A; 4Q8L A; 3FZ3 A; 4RSU A; 4Y3O A; 3TN9 1; 2FHR A; 5EIY B; 3SKJ E; 4YAR A; 1PMI A; 3CU7 A; 1ONA A; 3KT4 A; 5JZG C; 2OPW A; 1W6O B; 2VCB A; 3R4S A; 5DXD A; 1RB8 G; 3PUA A; 4MZU A; 4TXW A; 1OPM A; 2OVU A; 2VUL A; 5JIU A; 1J3P A; 3IZO A; 5LAS A; 1QJY 3; 3CJI C; 1HB4 A; 3MDP A; 2WO2 A; 1NG0 A; 1IG0 A; 4LIT A; 4ITC A; 3LF7 A; 3GBW A; 2A6V A; 4YFW A; 5G04 L; 3E1F 1; 3ZYJ B; 4ASM B; 4GJZ A; 3GZU C; 4YW4 A; 4CU7 A; 5KXE A; 2VTX J; 2QZ2 A; 1K5M B; 1DBN A; 1MVQ A; 2Z69 A; 4YNI A; 1IPS A; 2HLE A; 1IHM A; 1GBG A; 3WNO A; 4GKX A; 1LEN A; 2JX9 A; 4C1X A; 4EE2 A; 1NA1 A; 5GJ6 A; 1YC6 1; 3QCW A; 2VL4 A; 3MIB A; 1W8N A; 5FZA A; 4W6W A; 3MV0 1; 1IOA A; 4YW7 A; 2JG8 A; 4B4Q A; 4BIF A; 4XMI A; 2ET7 A; 4HVO A; 1MZF A; 4OFG A; 5DDJ 2; 1AYM 2; 3APB A; 3BOP A; 4FXK B; 3RNS A; 3VN4 A; 1LRH A; 2B39 A; 3ES1 A; 2R7E B; 2WZR 1; 3T2Q A; 2JG9 B; 5E1R A; 5HCH A; 3U07 A; 1XJA A; 3IDE A; 3DXU A; 1YI7 A; 3WHT A; 4JCK A; 2VZ1 A; 1UMI A; 5CKN A; 2D6M A; 4L8Q A; 3MI8 A; 1NAE A; 1FOD 3; 3ACL A; 1C5H A; 3K3O A; 4XBQ A; 2LM3 A; 4DOU A; 1ULF A; 2ZXQ A; 2PHD A; 5T5L A; 2DUO A; 1N3P A; 4JRA A; 2HWE 1; 3ZOI A; 3ZSE A; 4YLZ A; 4AI9 A; 2YUE A; 5IAX A; 1UKG A; 2XJV A; 4XHX A; 1O7F A; 4Y26 A; 1A3K A; 1PZ9 A; 1QJX 1; 4FQ4 A; 4GWJ A; 2VU9 A; 3P6B A; 3AL5 A; 1CJP A; 4HAN A; 4L97 A; 4LKE A; 4AHZ A; 4XUO A; 4PBP A; 5C71 A; 2OT7 A; 3WIN E; 3LSE A; 2AEY A; 5EFU A; 1ZZ7 A; 4LHK A; 4GB3 1; 3WG4 A; 1W2U A; 4DNY A; 1QJY 1; 1VE0 A; 1KRI A; 2DU0 A; 2ZAC A; 3PVB B; 1JYN A; 4XDQ A; 3MW3 A; 1FX5 A; 4IGQ A; 3SKV A; 3BIE A; 3ZR6 A; 1H9W A; 3NV1 A; 1WBF A; 3G6J B; 1QIQ A; 2VQT A; 4HKW A; 4ES8 A; 4FVV A; 4IEW A; 4RHV 3; 3TNP B; 2JDM A; 2QNK A; 1QH7 A; 3U78 A; 2XGX A; 1RXF A; 3DEM A; 1AR8 1; 2ZX1 A; 2BGC A; 2WSV A; 4IEZ A; 4RR3 B; 5GLV A; 3ZSM A; 1YQ2 A; 2ARX A; 5L19 A; 2Y0I A; 1F4H A; 1OFS A; 1VRH 3; 3BOW A; 3ITQ A; 1FV3 A; 3K3N A; 4NRQ A; 4YGE A; 1J58 A; 5HCA A; 5LWB A; 3C3V A; 3MIC A; 4NRM A; 1MEC 3; 2TNF A; 3W21 A; 3ZYP A; 4BQW A; 4BQX A; 4QPG A; 2WAB A; 4YM0 A; 1CZV A; 4CDQ B; 2XBO 3; 4K21 A; 5DPN A; 4M00 A; 3A21 A; 5JCT A; 1B35 B; 2XB6 C; 4XXA A; 3OY8 A; 4FXG B; 1QOT A; 2QTW B; 4YW5 A; 3E5X A; 4W4Z A; 1VB4 A; 1GQH A; 2JJ6 A; 5DN2 A; 5FPA A; 4FAH A; 1XE8 A; 2W1W A; 3POY A; 3KD4 A; 1C5I A; 3IWZ A; 4NHK A; 5KHG A; 2W0X A; 1ZX5 A; 3T4V A; 5FY1 A; 5FZL A; 5A3N A; 3T09 A; 2OS2 A; 1R09 1; 4IND A; 4AAX A; 4ZON A; 3LD8 A; 3V65 A; 2Y6K A; 1H9P A; 5HCB A; 3RRD A; 4CDW A; 2YPD A; 3IT8 A; 4FPC A; 4CDU B; 3JUU A; 1US3 A; 3PVE A; 1BZW A; 3R6S A; 5A6Q A; 5BYN A; 2JIJ A; 2KM2 A; 3I59 A; 4TSV A; 1F8V A; 2HWF 2; 1G7Y A; 1ST8 A; 1Q3E A; 4JGY B; 4QHB A; 1GZW B; 5CAD A; 5FQ0 B; 2CGP A; 4MGY E; 1CIW A; 4MGT A; 2VW2 A; 4XBL A; 4J25 A; 4YVS A; 1US2 A; 4IEX A; 4N53 A; 2VXJ A; 2J1A A; 1RUF 1; 2G9Z A; 3TEX A; 3I54 A; 1JUH A; 5F5I A; 3B5L B; 5JI9 A; 2E63 A; 1QJU 1; 4H3W A; 4AHW A; 1VMJ A; 1WCQ A; 2R0F A; 1B35 C; 1LR5 A; 4UAP A; 4XJ9 A; 5LA1 A; 5FY7 A; 2OZ6 A; 2GND A; 4RS5 B; 1RVF 1; 3IAP A; 3WY6 A; 5A3T A; 5FYH A; 3KMH A; 5AVP A; 5FZC A; 1XE0 A; 2B9U A; 3FJS A; 3IPV B; 4KL1 A; 1RIR A; 1ZRR A; 2VR4 A; 1JOP A; 3BOD A; 3KHC A; 4E9K A; 4MYE A; 1DZQ A; 1K42 A; 5C3R A; 4PQQ A; 2D93 A; 4Q1R A; 5FR3 A; 4KU8 A; 1UIK A; #chains in the Genus database with same CATH homology 1RMU 3; 2TEP A; 4CDU C; 5CFC B; 1VBA 3; 2IFR A; 3ME2 A; 4RHV 2; 1N1S A; 3M3C A; 5C8C B; 3H1W A; 4ATF A; 1TME 1; 4XET A; 4JGY C; 2YC0 A; 5BNO B; 2FBE A; 4B7K A; 1EAH 3; 5CVR A; 1IPJ A; 2K0G A; 4JC1 A; 1ASJ 1; 4D7T A; 4XIK A; 4J1X A; 4IV1 A; 2XBO 2; 4WVZ A; 4X6R B; 1LHW A; 1BK1 A; 4RS5 C; 1MS3 A; 3FMG A; 3T2T A; 5SV8 A; 1EYB A; 1O5U A; 1OPO A; 2RE9 A; 3PLQ A; 2XJR A; 1MR5 A; 4CDX B; 1PGL 1; 2R04 2; 5JZG A; 2Y26 A; 3ZSJ A; 1V6O A; 4LUL A; 5EJ3 A; 5I0R A; 2WKK A; 1Q43 A; 2RS5 3; 1HXS 1; 4HKL A; 4Z07 A; 3CJI A; 1Y7B A; 1FPN 2; 3INA A; 2W39 A; 1VAM A; 3WT3 A; 1CES A; 4R4Z A; 1H2L A; 2ZOE B; 5KC5 A; 2ZS6 B; 1NY7 1; 2EAL A; 4WZC A; 1JYC A; 3VBS B; 3WUC A; 2DVB A; 1LHN A; 3QD6 A; 3WP5 A; 2VMG A; 4KG8 A; 2BEO A; 4QD2 A; 2VQ0 A; 4XTP A; 4MET A; 4R9D A; 2WFF 3; 4AQ6 A; 2P34 A; 4CDX C; 1WCD J; 5DG1 A; 5BUV A; 1RMU 1; 2RR1 3; 2XG3 A; 2UY9 A; 3VZL A; 2A3Y A; 1DZT A; 2HWC 1; 2A5Z A; 5KWR A; 2ZKL A; 3QBQ A; 5DZG A; 1CPM A; 4I30 A; 4YM2 A; 3AX4 A; 3AZW A; 4P9J A; 1PGS A; 5E8A A; 3LWC A; 1YFX A; 4AQ2 A; 2PHT A; 4Q0Q A; 2NPI A; 3NO8 A; 5K8S A; 4LBN A; 2I2S A; 5GZE A; 1RUN A; 1DLL A; 1K5M A; 2VOW A; 2WNV B; 4Q0P A; 3AMQ A; 4QMA A; 5GZC A; 4I09 A; 2SLI A; 3KZ4 C; 1DQ2 A; 1ZBE 2; 1FNY A; 2FZ2 A; 1H30 A; 4PLO A; 2JIC A; 4K3U A; 1KDK A; 4LUM A; 5GLZ A; 2R06 3; 4Q4X 1; 3VKM A; 4AF9 A; 4FPE A; 1C3H A; 1TMF 3; 2JG9 A; 1NLS A; 1IQA A; 3AFK A; 4QPI B; 3IN9 A; 4C0H A; 4ONU A; 5LRS A; 1RGS A; 3EXU A; 3WUD A; 2B45 X; 4A2U A; 3S7I A; 2QZ3 A; 1AL2 3; 5GAL A; 4BMB A; 1KQR A; 1POV 3; 1UL9 A; 5T50 A; 1WBL A; 1JN2 P; 5LK5 A; 2RS3 3; 2ZHL A; 2E51 A; 4GXL A; 3WXP A; 1SEF A; 4UBH A; 2WNV C; 2ZGT A; 1COV 2; 4YRD A; 4PJY A; 3TOJ A; 1POV 0; 1NE6 A; 1QNW A; 4RL7 A; 5T54 A; 4YZ1 A; 1SCR A; 1PIV 3; 3O5S A; 5FAX A; 1DQ6 A; 2WFF 2; 1V00 A; 3JCI A; 1DYK A; 2XPJ A; 3S30 A; 2NM1 A; 4QPI C; 1D4M 1; 2XKO A; 3LB9 A; 4EN6 B; 2IXJ A; 2YPJ A; 5EXO A; 4CEY B; 2NLR A; 2D6K A; 1XYO A; 1G6N A; 4I3G A; 4QQO A; 1AJO A; 4LBJ A; 3NL1 A; 2TUN A; 4OZX A; 4RR3 A; 1BKZ A; 3AKQ A; 4MVT A; 3SHR A; 3VZK A; 1XE7 A; 3AKS A; 1ZRD A; 5JOY A; 2D3P A; 3P3P A; 3AYD A; 3E5U A; 1QXR A; 1AR6 3; 2N7G A; 4CDQ A; 3D8C A; 3I8T A; 4XJU A; 3BPZ A; 1N3Q A; 4DPN A; 1B35 A; 2DYC A; 1MS1 A; 1KFU L; 2BNN A; 2R06 2; 1AUY A; 1G9F A; 1GSL A; 1U5I A; 4E4Z A; 4TLF A; 3ZXL A; 1TMF 2; 4DRV A; 4X8P A; 3JU9 A; 3MF6 A; 4XJA A; 2CGO A; 2RS3 1; 2FQP A; 3EBR A; 4LO8 A; 2IXL A; 3ZFE B; 3RJ2 X; 4Q4Y 3; 1RUD 3; 1MZE A; 3MF9 A; 3USU B; 4Q4V 2; 1AL2 2; 3DKW A; 3IBM A; 4GMP 1; 1WD3 A; 1VBD 1; 4CDU A; 2RJL A; 4IV3 B; 3EJK A; 1REE A; 3ZYG A; 1QY4 A; 1AR9 1; 3BU7 A; 4JGY A; 1GZW A; 4AI8 A; 4KBB A; 4Y20 B; 1UF2 C; 1C8M 2; 1XU2 A; 1ZRE A; 2D3S A; 1PIV 2; 2NMO A; 5K7P A; 1Z78 A; 4HMZ A; 3VBH B; 4DMI A; 1N1V A; 2PHX A; 1MZ6 A; 1HRV 2; 1X8E A; 3OF1 A; 1LES A; 2DFB A; 4U36 A; 4Y24 A; 3ZIW A; 4K9S A; 3POW A; 3I4I A; 5CG8 A; 1WW6 A; 2ZGO A; 3VDD B; 5JPM B; 4OUL A; 2PLV 3; 3TBD A; 1XU1 A; 3IPV A; 2ERF A; 3ZFE C; 4KGQ A; 2D40 A; 5L7O B; 1OQ1 A; 5J3U A; 3BAL A; 4GA9 A; 3VKF C; 1SFN A; 2Q30 A; 1YFU A; 2DTW A; 4IV3 C; 3B94 A; 1OI6 A; 3B69 A; 4PQ9 A; 5CFC A; 2ZGU A; 4MLO A; 4OVE A; 1NCQ B; 5C8C A; 5K0U B; 5JDB A; 1RUI 1; 3RS6 A; 3VHR A; 4AE2 A; 5BNO A; 4XQW A; 3SIT A; 5CU1 A; 4LLO A; 3HWJ A; 1NQD A; 1R08 2; 4IER A; 5LK7 B; 4IET A; 4YM3 A; 3QH7 A; 2EXI A; 2HWD 3; 1BXH A; 3VBH C; 2R6Q A; 4BCU A; 2JEN A; 2JG8 B; 1EV1 3; 2CWM A; 3GHN A; 4N7U A; 3VZN A; 3UKT A; 4FTB A; 4XIL A; 3VDD C; 3M4F A; 1W0P A; 1AX0 A; 2CAU A; 3E5Q A; 1RUJ 3; 2Q5T A; 1DGW X; 5L7O C; 3MFA A; 2O0O A; 1Q56 A; 3JZV A; 5T3N A; 1QWR A; 4Q4Y 2; 4OHW A; 1VBB 3; 4H7M A; 5D8A A; 3THP A; 1NCQ C; 3GAL A; 4L99 A; 2E6V A; 3OPZ A; 3M7A A; 2VVE A; 3OD4 A; 4WZF A; 4PLN A; 1PO2 3; 3DUI A; 4KWL A; 2ZHM A; 2RMU 3; 1BBT 3; 3VBS A; 4BA6 A; 4JTN A; 3D0S A; 1CAX B; 1NA1 C; 1RJ7 A; 1BCX A; 4HET A; 2DTY A; 4QQL A; 4TWT A; 4AW7 A; 4H14 A; 3CJX A; 2JG8 C; 2CAV A; 3GF6 A; 4MGI E; 5FMO S; 3JBG 1; 3X29 B; 1WLW A; 1PO1 3; 2MEV 1; 3ZN5 A; 3M3Q A; 3PHM A; 2YMZ A; 4DZG A; 1OLQ A; 1FNZ A; 2VH9 A; 3O14 A; 3WW1 A; 4Q8K A; 1M4W A; 3PVN A; 5ACA 3; 5JOW A; 2WSU A; 3L9J T; 4GKY A; 3RI8 A; 4XFH A; 1KS5 A; 1DGR N; 1AF9 A; 4GWN A; 3MIF A; 2ZPX A; 1SCS A; 1AR7 2; 2DCY A; 2ZGQ A; 3KB5 A; 3RJR A; 1QKQ A; 2NQY A; 2WS9 3; 4XYX A; 4JDM A; 3E6D A; 4OHZ A; 4BME A; 3ZXA C; 1CAV B; 4AVA A; 1EV1 2; 4Q4W 3; 3FLT A; 1LEM A; 1F2N A; 1QF3 A; 1ULC A; 3AMN A; 5BPX A; 2BBV A; 1DGW A; 3ZXJ A; 3RMY A; 1RUJ 2; 4QPI A; 4WV8 A; 1C28 A; 3VBR B; 1ZM1 A; 2Z42 A; 3J8F 2; 3ECQ A; 1JYI A; 4AGG A; 2DCK A; 3WCS A; 4P9X A; 3D82 A; 4CDW B; 1WW4 A; 5I0T A; 2VGD A; 5B2G B; 1BBT 1; 1SLB A; 1OD5 A; 3AKP A; 2VW1 A; 4AWD A; 2XL7 A; 3CEW A; 2YYO A; 1PO2 2; 4AGV A; 4YG6 A; 4LBK A; 4E2G A; 4KWJ A; 2RMU 2; 1O3T A; 4TZD A; 1O4Z A; 2D6L X; 2F4P A; 4R8H A; 3ZXD A; 1OKQ A; 5KXC A; 4XF3 A; 3I7D A; 4E4S A; 1R1A 2; 3WDX A; 1DGL A; 1WCS A; 1J3R A; 5TSW A; 1RTV A; 3VBR C; 1C57 A; 4HK9 A; 2A6Z A; 2ZGS A; 4AVV A; 2AGS A; 1FV2 A; 1JH5 A; 3JQX A; 4CEY A; 4CDW C; 2GAL A; 4CEW B; 1HRI 3; 1O5L A; 3BTA A; 3FE5 A; 3ILR A; 1F9K A; 3GNE A; 4FCA A; 5ACA 2; 2JEM A; 3RWN A; 3W9A A; 3AP9 A; 4MGZ E; 1D2S A; 5EZW A; 3ZYI B; 4CTE A; 1X7N A; 3UJO A; 3AZV A; 1Q8S A; 1Z7S 1; 1ZVF A; 4KGG A; 2IXI A; 4CP1 A; 4N53 C; 3VKL A; 2YGL A; 1OHF A; 4M3C B; 3ZFF B; 1G9A A; 1AX2 A; 2RM2 2; 2B42 B; 2VXR A; 2WWS A; 4HVQ A; 1YCI A; 2BNM A; 4Q4W 2; 5CIR A; 3D5O A; 3E97 A; 5DDJ 3; 1AYM 3; 1Y3T A; 1VLN A; 1ULD A; 2O8Q A; 2D6N A; 1AYN 3; 1D4V B; 4JAA A; 1S0E A; 2YGM A; 3BB6 A; 1ZBA 1; 2YY1 A; 2H0V A; 3NW4 A; 1OOP B; 2ZJC A; 5J98 B; 5FTT C; 1N1Y A; 4BM8 A; 3ZFE A; 4QGM A; 4EN8 B; 2YRO A; 1IPK A; 3USU A; 1CON A; 2RS1 3; 1N47 A; 1AX1 A; 4BE3 A; 1LEC A; 3H0O A; 2EAA A; 4PDW A; 1UU6 A; 1RUG 3; 4N43 B; 2BQP A; 4TYS A; 3ZFF C; 3HAG A; 1O4T A; 1LB2 A; 4OI2 A; 4Y20 A; 4NWW A; 1RUO A; 3WQ0 A; 3WP4 A; 3RPQ A; 1FMD 1; 4ASL A; 2PAE A; 4R4X A; 4OI1 A; 3KM5 A; 1UD1 A; 5T5P A; 4A3X A; 3B7M A; 1XYP A; 3M70 A; 2X5I B; 3VDD A; 5L73 A; 3CLP A; 1V9U 1; 1OOP C; 3KCX A; 3MIE A; 3OBT A; 1PVC 3; 2HEV F; 2ENR A; 5L7O A; 3VHN A; 2OZJ A; 1HRI 2; 4XBN A; 1A8D A; 3KGL A; 1NCQ A; 5K0U A; 4WM8 B; 2D98 A; 5GZF A; 4N43 C; 3URF A; 3PTY A; 3D4K A; 3WCR A; 2V09 A; 3AP6 A; 1V6J A; 5GKA C; 4HL0 A; 4XFA A; 3FWE A; 1KFX L; 3ETQ A; 2Q4S A; 4LR4 A; 4LO4 B; 5D1I A; 4PLM A; 4URT A; 1AYN 2; 1GQG A; 3UL2 A; 1ENQ A; 1RUC 1; 3MPP G; 1W6P B; 3HQX A; 2WIN B; 4LO7 A; 1WD4 A; 2ZGP A; 2BWC A; 3WQ7 A; 3QOP A; 4FPY A; 5JVV A; 4K20 A; 2RS1 2; 2ZML A; 1C1F A; 1CAU B; 4FPO B; 3WG2 A; 1BMV 1; 1XNC A; 2PHU A; 4GH4 A; 1WGP A; 1K5M C; 1RUG 2; 2HWF 3; 3ZMN A; 1JW6 A; 3DF0 A; 1QQP 2; 4GUL A; 1LHU A; 1CAX A; 1SLT A; 3FLP A; 4I0B A; 4LBQ D; 1S1A A; 1YFW A; 3LOI A; 4W65 A; 2A7A A; 2WZR 2; 3O7O A; 5JWP A; 3MYX A; 4N9H A; 1LUL A; 3LSD A; 2JG9 C; 1LGB A; 4HN0 A; 2UWC A; 4DRR A; 1D0H A; 3AZY A; 5H9Q A; 1AHS A; 1RIT A; 1PVC 2; 1DQ0 A; 3KCY A; 1JBC A; 4DY3 A; 1SMV A; 1L3J A; 1CD3 G; 5ABJ B; 4AXO A; 5BV6 A; 1OFN A; 2WNE A; 5I0S A; 5E88 A; 4LBO A; 5GLW A; 1PNG A; 1QJX 2; 4G4F A; 4QPY A; 4YSF A; 5GM0 A; 1IGO A; 4JGZ B; 3HT1 A; 3R0R A; 1UCX A; 2QCS B; 3I2D A; 4AYU A; 4GB3 2; 1CAV A; 4AGR A; 4YG3 A; 1S0J A; 2BVB A; 5FLH A; 3ZYR A; 4D7Y A; 2XLF A; 2A75 A; 3H50 A; 1FOD 1; 2BFU L; 2DV9 A; 2R04 3; 4QGL A; 3PJQ A; 4DQA A; 5FOB B; 5LRR A; 1GLH A; 3VBR A; 3WD5 A; 3VKO A; 4D94 A; 1URX A; 2HWF 1; 5JOZ A; 5ABJ C; 1WCK A; 1Z0H A; 2GC0 A; 2ZCW A; 2GSY A; 5FO8 B; 1O3R A; 4QQP A; 1FPN 3; 4RR3 C; 1Q8V A; 1Y9Q A; 5F9T A; 1F31 A; 2OUJ A; 4JGZ C; 2GC1 A; 1V6M A; 5AFB A; 4EWE A; 5I91 A; 2CGN A; 2WT0 A; 1J3Q A; 2EIG A; 1A6C A; 4XE9 A; 4Q27 A; 2A73 B; 1OQE A; 4CDQ C; 2D97 A; 4NOT A; 3O0V A; 1TNF A; 3A0K A; 4XZP A; 2WQ4 A; 4CYD A; 2TBV A; 2PHL A; 3N35 A; 2E7A A; 2GNB A; 4F8A A; 4RHV 1; 1R09 2; 4Q29 A; 5IK4 A; 4YW3 A; 1UX6 A; 4J6G A; 3M3E A; 4FP2 A; 5GLU A; 4CEW A; 2B5H A; 3GBG A; 3THT A; 4AG4 A; 1VBE 3; 3MID A; 1FAY A; 1VRH 1; 3MLK A; 1VIW B; 1B09 A; 1MEC 1; 2VY0 A; 1UAI A; 2LUW A; 5FO9 B; 1QDC A; 1BVP 1; 4UXA A; 2XBO 1; 3AJZ A; 1WME A; 2BVV A; 3TYJ A; 3RG0 A; 1UZY A; 1GAN A; 1RUF 2; 2R32 A; 3HBZ A; 4JDN A; 4M3C A; 3ZFF A; 3ZIX A; 3S0M A; 1IS3 A; 4HKO A; 1ZBE 3; 4HN1 A; 4J1W A; 1RUE 3; 2VMH A; 2ZAA A; 4FPJ A; 1AJK A; 2EVX A; 4OUM A; 4G9S B; 2ZMN A; 2K2S A; 3VHO A; 3U11 A; 1JXN A; 2YHJ A; 4KNC A; 4LBQ B; 1ZYB A; 3CNA A; 1OOP A; 4KH9 A; 1S0B A; 1YUD A; 3QLQ A; 5CFC C; 5D8A B; 4IEU A; 5C8C C; 2HEW F; 1TME 2; 2R30 A; 1D2Q A; 1OQD A; 5BNO C; 3MLJ A; 5KHH A; 3POS A; 2YV8 A; 4PF5 A; 1W0O A; 4N43 A; 1O3S A; 2ZX0 A; 1ASJ 2; 2CY6 A; 2G9G A; 2XUM A; 2WT2 A; 1COV 3; 2ILM A; 2Q1Z B; 2OW4 A; 3H1Y A; 4XF0 A; 3UK9 A; 2D44 A; 1BJQ A; 4ZXK A; 1ZZ9 A; 5CFD B; 1XNB A; 3JB8 A; 4HZF A; 1NXD 1; 5F6L B; 2QDN A; 4JRL A; 4K7J A; 3W59 A; 2CYF A; 4IL7 A; 5DUU A; 2X5I A; 1H2K A; 4DIN B; 1VAV A; 1VBA 1; 3T1M A; 2VW0 A; 1KS4 A; 4OLL A; 1LHV A; 3USS A; 1NZC A; 3WW4 A; 2BFU S; 5JI7 A; 1ENS A; 1HXS 2; 2O8O A; 5KHI A; 5D8A C; 3VHP A; 4A7Y A; 1DU3 D; 1EAH 1; 1QH6 A; 1VBE 2; 1O91 A; 4LUK A; 1IS6 A; 4I0W B; 4WM8 A; 4LHN A; 3AP5 A; 3V56 A; 3VBS C; 1V6I A; 1DQ4 A; 2FMY A; 3IDB B; 3QAC A; 4Y4Y A; 1OA4 A; 3AZX A; 2MPF A; 5CFD C; 2R07 2; 1CR7 A; 1RUE 2; 3D0J A; 2EAK A; 1M06 G; 3ILN A; 5SV5 A; 3WW3 A; 1ND3 B; 4CZS A; 2RS5 1; 1HV0 A; 2HWC 2; 4YZ4 A; 4FBF A; 2A6X A; 4C0B A; 3KLS A; 1W6P A; 1WUV A; 1NCR B; 3SCG A; 3WP3 A; 5BXX A; 5DUV A; 2OYZ A; 2HYK A; 1G8W A; 3FFZ A; 1G9B A; 1KXG A; 2IXK A; 1DQ1 A; 3WX5 A; 5K9Y A; 4Q4V 3; 1CAU A; 3ZXK A; 5IK5 A; 1C1L A; 1COV 1; 4R9A A; 3VL9 A; 1W6M B; 4F83 A; 4WBB A; 2CTV A; 3SLI A; 1DDL A; 3NAP B; 2BYV E; 5FRE A; 4ATE A; 1UZZ A; 3ZDS A; 2YXS A; 3VBF B; 2WFF 1; 3GHM A; 3HT2 A; 5BNQ A; 4QM9 A; 1C8M 3; 2RR1 1; 3AYC A; 4Q4X 2; 4NVP A; 1ND3 C; 1HRV 3; 3WDU A; 4MXW B; 5GZD A; 1IKP A; 4EN0 A; 1LGN A; 1RC6 A; 1QJZ A; 3E6C C; 2WLQ A; 3SXW A; 4AED A; 2GN7 A; 1NXM A; 2UY8 A; 5DZF A; 3WR0 A; 4LT5 A; 4AVT A; 3PME A; 4HPK B; 5ABJ A; 2K9Z A; 4HFS A; 1XND A; 5C33 A; 1RJ8 A; 3N7J A; 2C0Z A; 4LBM A; 1AXK A; 1VB2 A; 1VR3 A; 4A7Z A; 1U5X A; 3VBF C; 4BHP A; 1R08 3; 4JGZ A; 1UKR A; 2R06 1; 3VBO A; 2GMM A; 2QVS B; 2P3I A; 3TAY A; 1CN1 A; 1ND2 B; 1TMF 1; 1BYH A; 1H2N A; 2A3X A; 3VGI A; 1SLC A; 3WDY A; 1QXJ A; 5DG2 A; 1OA3 A; 1D4M 2; 1GKB A; 1DHK B; 2H6C A; 1AL2 1; 1V6N A; 3MGU A; 5CB7 A; 1POV 1; 1H0B A; 4CEY C; 3R05 A; 3OD0 A; 4RQP A; 4P9W A; 2WQZ C; 1WW5 A; 4IEV A; 1UU4 A; 2Z79 A; 3KQR A; 3M3O A; 4PDW B; 3M3I A; 1PZ7 A; 2O1Q A; 4Z1V A; 4NPR A; 5A33 B; 2EA7 A; 5JIA A; 4E27 A; 2B8M A; 1PIV 1; 2IC1 A; 4WVM B; 1EPW A; 4AFB A; 5AC9 3; 3ZN6 B; 1W6N B; 2HWB 3; 2VG9 A; 3VL8 A; 5FLJ A; 1SBF A; 1ND2 C; 4GAL A; 4AVB A; 1REF A; 3AK0 A; 5H9P A; 3W58 A; 3GBF A; 3VZM A; 5BY5 A; 1S2K A; 3R4D B; 4FPK A; 2ET1 A; 1I5Z A; 1UU5 A; 4FQZ A; 1YJK A; 3FUO A; 2JKB A; 2XVK A; 1U5Y A; 4DXZ A; 4E2Q A; 4V1P A; 3TNQ A; 1AXY A; 1VBD 2; 2LTN A; 1XRU A; 4WVV A; 4PDW C; 4YZ5 A; 4O9E A; 3H7J A; 4MSV A; 4AFA A; 3IDC B; 1AR6 1; 1ULE A; 1YIP A; 2XH6 A; 4ZU5 A; 2Z5W A; 2A3W A; 5J37 A; 1AR7 3; 4LO5 B; 1NYW A; 2ARB A; 5I8T A; 1LOF A; 1TEI A; 5L36 A; 1G9C A; 5F6K A; 2ZX2 A; 4Q4Y 1; 1RUD 1; 5KBF A; 4E4D X; 4LBQ A; 1VBC 3; 3MW2 A; 4JDO A; 3J8F 3; 4FSJ A; 4NHL A; 1PM7 A; 2GAU A; 3FBM A; 3NV3 A; 3OTF A; 3WV6 A; 2QE3 A; 5E16 A; 4B7E A; 5K0U C; 2PAM A; 5I8Y A; 4GH4 B; 2OPK A; 2PHW A; 1IZ3 A; 5AC9 2; 2HWB 2; 4AHX A; 4XOG A; 4AHY A; 3UJQ A; 1RHI 3; 5J36 A; 2PLV 1; 4X6Q B; 5JLV A; 4SLI A; 1R1A 3; 4F3J A; 2FZ1 A; 1O4Y A; 5CFD A; 4MGK E; 4X8N A; 2PHF A; 4XFB A; 2GN3 A; 2DUQ A; 4ERO A; 4BWO A; 4MH0 E; 4KU7 A; 1VAK A; 2WNU B; 4YFZ A; 3HMY A; 2DCZ A; 3NVC A; 4BOW A; 5CAZ A; 1W6Q A; 2W68 A; 5FCC A; 3VKN A; 3VUO A; 3ZFG B; 4IEP A; 1SLA A; 4HSJ A; 4I3P A; 5AFE A; 1D0G A; 1DZR A; 2P3J A; 4GH4 C; 4EWA A; 1EPZ A; 1WLC A; 4GQ7 A; 2HWD 1; 2IFW A; 3WDW A; 1EV1 1; 5DZE A; 4DS0 A; 1LGC A; 2ATF A; 1SBD A; 1DQ5 A; 2JD4 A; 2RM2 3; 2GZW A; 3AKT A; 4AVS A; 1RUJ 1; 5FPZ A; 2AYH A; 3JD7 3; 3ENR A; 2WC2 A; 1IKQ A; 1SLI A; 2XL9 A; 1RUH 3; 3AMP A; 2MEV 2; 1ND3 A; 3DV8 A; 2DWR A; 4JQR A; 1VBB 1; 5EWS A; 5T55 A; 1Y43 B; 1BEV 3; 1ZZ6 A; 2ZTN A; 1BQP A; 1CX4 A; 2WNU C; 3U10 A; 4UW6 A; 1VBC 2; 3HN1 A; 1NCR A; 1YYN A; 3OCP A; 5IK8 A; 2PFW A; 3AMH A; 3IMN A; 3ZFG C; 2ZGR A; 4IES A; 1PO2 1; 3WP6 A; 4BQ1 A; 2RMU 1; 1W6M A; 5FPX A; 3FLR A; 1NE4 A; 3NAP A; 1HLC A; 3B01 A; 3SCH A; 3E6B A; 3VBF A; 1HQN A; 1PO1 1; 4FTS A; 1RHI 2; 3ZX7 A; 2E9Q A; 3UKV A; 3RQ0 A; 1H8T A; 2BNO A; 5I0U A; 1QDO A; 4NHM A; 2R0H A; 4Q0V A; 4I0A A; 4YGC A; 4MXW A; 3NKT A; 5EV7 A; 4LLF A; 5ACA 1; 3RYR A; 4KQ7 A; 3T1L A; 2WII B; 3DPR B; 3VZJ A; 3RYK A; 3V0B B; 4P9G A; 4XFG A; 4XF9 A; 4HPK A; 1V6L A; 1J1L A; 4PCR A; 2EXK A; 4FPG A; 2WS9 1; 3ALQ A; 3Q9O A; 2JE7 A; 2VVD A; 4C90 A; 1HIX A; 1I6X A; 4Q4W 1; 1ZZC A; 2DCT A; 5KXD A; 1MVE A; 1JTZ X; 1ND2 A; 1XNK A; 1AL0 G; 1MAC A; 3WDT A; 3JD7 2; 3ZVX A; 3TN9 3; 2NZ9 A; 4LHL A; 4AK3 A; 1RUH 2; 4FOY A; 4HT1 T; 1HQL A; 2NYY A; 1BEV 2; 3DPR C; 3KGZ A; 2EXH A; 5H9R A; 1GNZ A; 1I3H A; 4CEW C; 2ZMK A; 5JMC A; 4XEZ A; 1EY2 A; 1GNH A; 4XW3 A; 1ZRC A; 1UPS A; 1PGW 1; 5A33 A; 5CNA A; 2P17 A; 2D7F A; 3B9C A; 4WVM A; 3EHK A; 3ZO0 B; 4YM1 A; 3PNA A; 3ZN6 A; 1W6N A; 2X53 S; 3B9I A; 1QQP 3; 4FOQ A; 1SQ4 A; 4MXV A; 2PA7 A; 3GZT B; 3DN7 A; 3ST7 A; 1Q8O A; 2Q8O A; 4XXP A; 4FTE A; 4Y1Z A; 3O0W A; 1W39 A; 2FMD A; 2BUK A; 2RHP A; 4H7L A; 4MX3 A; 5FLI A; 4AFD A; 2WZR 3; 4WM7 A; 1VJ2 A; 1HRI 1; 2VNG A; 2R1B A; 5FDB A; 1ZBA 2; 4L2N A; 3PVO A; 1R1Z A; 2DFC A; 1WA4 A; 4Y22 B; 4XMA A; 2XJQ A; 1S0I A; 4LA3 A; 2OA2 A; 3KCC A; 1H1A A; 5BNN B; 1O3Q A; 4N90 A; 2HWE 3; 5JSP A; 5IK7 A; 2CV6 A; 2ZGM A; 1ZZB A; 2D43 A; 4I01 A; 1UMZ A; 2GU9 A; 5KXB A; 4XFC A; 3M1H A; 1QJX 3; 1UN1 A; 2BW8 A; 3U4X A; 3N7K A; 3V0A B; 5DDJ 1; 1AYM 1; 2WFP A; 3L2Y A; 2VMI A; 1FMD 2; 1AYN 1; 2ZGL A; 1H1I A; 1S2B A; 4S2F A; 5DUW A; 4GB3 3; 3TN9 2; 2OII A; 4GWM A; 4A7K A; 5KHJ A; 4OHV A; 2X5I C; 1GZC A; 1V9U 2; 2RS1 1; 2A70 A; 3ILF A; 1T6G C; 4P5H A; 4DFS A; 2ZBJ A; 1QNY A; 4R52 A; 5BNN C; 3AFG A; 1RUG 1; 3CL1 A; 5JSO A; 3ZPY A; 1ALY A; 1F5J A; 1QQP 1; 1UIJ A; 1AR8 3; 4WM8 C; 4G3Y C; 3RI9 A; 2BWA A; 4K8F A; 4COV A; 2OUH A; 4M0H A; 3AXE A; 4M3C F; 4P9I A; 5HXV A; 5EYX A; 3MLL A; 1LU2 A; 3GYD A; 5A3M A; 1PNF A; 2R1D A; 2XJP A; 2XWJ B; 2VPV A; 4YGD A; 5T52 A; 1PVC 1; 2XJU A; 1HDK A; 3LCP A; 3WHU A; 1OMI A; 1RUC 2; 2PAK A; 4O9G A; 3ATG A; 4EV0 A; 2WNU A; 3MPB A; 4AED B; 2Y0O A; 1OA2 A; 1R09 3; 1YFY A; 5FRA A; 1LOD A; 4R9C A; 4COU A; 1RED A; 2G4I A; 2QJV A; 1BMV 2; 2C5D A; 3ZFG A; 2HWE 2; 4LA2 A; 1G9D A; 3UKN A; 4YYO A; 3OJB A; 2UYA A; 1F5F A; 3KT7 A; 4NWV A; 5IUQ A; 4ISR A; 2ZZJ A; 1RUF 3; 3VBO B; 1HV1 A; 1A8M A; 4EN9 B; 3OYW B; 4XQ4 A; 1QJU 3; 4PIX A; 2UU8 A; 2R16 A; 4Y5Z 0; 2XJT A; 1RVF 3; 4LE4 A; 3SCF A; 1QJY 2; 3K48 A; 3B93 A; 5A3L A; 2KXL A; 4AFC A; 2P2K A; 3LA7 A; 1U0A A; 4QT6 A; 4AED C; 5J96 B; 5CBL A; 3OGG A; 3VLB B; 4OHY A; 4NN0 A; 4RQP B; 1MS8 A; 4K6B A; 1ZA4 A; 1MQT B; 1OSG A; 3ZMO A; 2XHK A; 1TME 3; 1AR8 2; 1H2M A; 4IEO A; 1YXW A; 4IEY A; 3JQW A; 2ZX4 A; 3NKG A; 2ZAH A; 4M0N A; 3RZZ A; 1ASJ 3; 1HQW A; 2VOK A; 1AXZ A; 4M01 A; 3VBO C; 1N3O A; 4SBV A; 3JB4 C; 4S2H A; 4FAG A; 2VNR A; 1HX6 A; 1YIF A; 3EQE A; 1LU1 A; 1FOD 2; 3PMS A; 3K51 A; 4P9Y A; 1GV9 A; 4KWK A; 3LOB A; 3AYA A; 2ZHN A; 5T5O A; 4RQP C; 3DPR A; 3V0B A; 1X36 A; 2WA3 A; 1NOV A; 1HXS 3; 4FOV A; 3AP7 A; 1MQT C; 3N36 A; 2A6W A; 3V64 A; 2PHR A; 2MNG A; 4XQD A; 1KJR A; 1VAL A; 3WDV A; 4AVC A; 4Q2F A; 2GC3 A; 2VQA A; 2WA4 A; 1WMD A; 2PTM A; 2RJK A; 2ZAB A; 3CO2 A; 3WW2 A; 1QJU 2; 2HJI A; 3LA3 A; 2R04 1; 3WNX A; 2WL1 A; 4DY5 A; 2R07 3; 1MZ5 A; 1RVF 2; 1PHM A; 2VVF A; 2GNM A; 1CGP A; 1WW7 A; 2HWC 3; 4BPZ A; 1YLL A; 1FPN 1; 2ES3 A; 4IEQ A; 3ZN4 A; 2I74 A; 1J1T A; 1ENX A; 1VRH 2; 1ULG A; 3AP4 A; 5E89 A; 5GZG A; 1MEC 2; 4QXK A; 4XF1 A; 4HSL A; 4XIO A; 4Z82 A; 3ELN A; 2V5Y A; 1WLD A; 3J4U H; 5JPN B; 4K40 A; 2GME A; 2DVF A; 1VP6 A; 3FHI B; 4N9I A; 5FBZ A; 1S0G A; 1JHN A; 5KC6 A; 1QHD A; 1LOF C; 2JE9 A; 4Q26 A; 2A71 A; 3BIW E; 4OI4 A; 3VZO A; 5J48 A; 3MEP A; 4LE3 A; 4NR1 A; 4PIZ A; 4Q4X 3; 4YZ3 A; 2ZGN A; 3MZH A; 5JAX A; 3QH6 A; 4L9B A; 1PGL 2; 4XHB A; 1DF0 A; 1VBE 1; 2QDR A; 3AMM A; 1DYP A; 4BIO A; 5BNP B; 1GIC A; 4Y22 A; 4EWD A; 3D6E A; 3H3Z A; 4B29 A; 3LAG A; 5LEK A; 3A4U A; 1A78 A; 1W6Q B; 4I02 A; 1Q8Q A; 4W6X A; 5BNN A; 5T5J A; 1APN A; 2XVG A; 2DU1 A; 1ZNP A; 3FX3 A; 1ZBE 1; 3VBU B; 4Y1U A; 4AEJ A; 3SH5 A; 4YVW B; 1NY7 2; 2Z2S B; 3WQ1 A; 2R07 1; 4HBN A; 1RUE 1; 3DGT A; 3SNM A; 2JXA A; 2I07 B; 3R4U A; 4JVA A; 3AB0 A; 4UBG A; 2W08 A; 1SDW A; 3V0A A; 5F1R A; 1W6O A; 4XJ8 A; 3LA2 A; 3HIF A; 2E7T A; 4ZU4 A; 1SAC A; 1RMU 2; 2GH8 A; 1D4M 3; 4XTM A; 1VBA 2; 5BNP C; 1GR3 A; 1LTE A; 1C8N A; 3O0X A; 3WZ1 A; 2C1F A; 2DCJ A; 3SIS A; 2JDZ A; 2VUJ A; 1EAH 2; 2JEC A; 4BZ4 A; 2VOV A; 3WG3 A; 2BPA 2; 5C4W B; 3H9A A; 4W6Y A; 2DUP A; 3VBU C; 3M2M A; 2UUR A; 1QOO A; 4YVW C; 2PEL A; 3NJZ A; 2ZZQ A; 4MV2 A; 3IM0 A; 2AR6 A; 4CP2 A; 1AZD A; 1PZ8 A; 2Y38 A; 3B3Q E; 1NQJ A; 1H8T B; 4AEK A; 2XLA A; 3N7M A; 4BH9 A; 2RS5 2; 3J26 N; 1S55 A; 4H55 A; 2VOX A; 3ZSL A; 4BYY A; 4K1Y A; 5A32 B; 3FFQ A; 1FXZ A; 1NCR C; 5GLT A; 1FT9 A; 4ISQ A; 2IZW A; 4D69 A; 2UWA A; 1TQ5 A; 5IT6 A; 5C4W C; 1JOJ A; 2D6P A; 1MS9 A; 2C9A A; 4LNV A; 1X33 A; 4GMP 3; 1VBD 3; 4RD7 A; 1KJL A; 1YWK A; 3AYE A; 3AJY A; 3J9S A; 4FPH A; 5AVA A; 1AR9 3; 3AM2 A; 1KIT A; 4QQH A; 2D5F A; 3NAP C; 1ENR A; 2D3R A; 2X8K A; 3PJY A; 1SLL A; 3RMX A; 1LED A; 4GMP 0; 2KA3 A; 2Q0A A; 2RR1 2; 1I1E A; 3A0N A; 3OYW A; 5C6C A; 3SH4 A; 1H8T C; 4UW5 A; 2QFE A; 2CWS A; 3DL3 A; 1MS5 A; 2R49 A; 2SBA A; 2ZA9 A; 3NST A; 1JUI A; 2NN8 A; 1Q8P A; 4LBL A; 4Q4V 1; 1QOS A; 4R9B A; 5CG9 A; 2XVL A; 2ZGK A; 3N4M A; 1UPI A; 4FPL A; 3WOG A; 3OBR A; 1J59 A; 2YZ4 A; 3LKJ A; 4OI0 A; 4B4R A; 1MQT A; 5FMO L; 4FOW A; 1RUI 3; 1X8M A; 1C8M 1; 2AZ5 A; 4BLJ A; 3H1M A; 1SBE A; 4YW2 A; 4S2D A; 2HKX A; 2ZX3 A; 3H8U A; 4EN7 B; 2DVD A; 2YDE A; 1HRV 1; 1LOB A; 1HW5 A; 3AZZ A; 3S7E A; 2EF6 A; 4XFF A; 4AQS A; 1UW8 A; 2XKP A; 2VNO A; 1XQN A; 1V6K A; 1G86 A; 2G9F A; 3L2H A; 3AKR A; 1PK6 B; 5JR7 B; 3UV9 A; 3ASI A; 4QGN A; 4WM7 B; 2QQP A; 3WLU A; 3MIH A; 2WT1 A; 2RS3 2; 4JTO A; 1CAW B; 5C9A B; 3RDI A; 3HD8 B; 4LO6 B; 4Z2W A; 2UWB A; 3G7D A; 3RYP A; 5FQ0 A; 4C1W A; 4JV4 A; 4Q1P A; 1R08 1; 1YJL A; 1AR9 2; 5J09 A; 2D6O X; 4FP3 A; 1N1T A; 2AEN A; 4QQ2 A; 4S2G A; 2EXJ A; 1AVB A; 1U12 A; 1X82 A; 4Z8B A; 1TE1 B; 2AH2 A; 4K1Z A; 4RS5 A; 3O12 A; 4XF4 A; 2IWG B; 3MFC A; 1FAT A; 1LOC A; 4Y1V A; 2LAL A; 1LOE A; 1PK6 C; 3JBG 3; 4Q0S A; 2ETE A; 4HVR A; 4WM7 C; 4I2O A; 1TNR A; 4XPV A; 2MEV 3; 2MHF A; 3IIA A; 2PN5 A; 5C9A C; 4FO9 A; 4OHX A; 4UW4 A; 2VUA A; 2GNT A; 3FUQ A; 3VV1 A; 1DIW A; 4B3N A; 3NV4 A; 5AC9 1; 4XJZ A; 3MW4 A; 2HWB 1; 4EJR A; 3IKW A; 1NA1 B; 1RUI 2; 1AR6 2; 5KHK A; 4L92 A; 2PYT A; 2BTV C; 1QMJ A; 1LHO A; 4PIY A; 1RIN A; 3RSJ A; 3CF6 E; 3LZZ A; 4CDX A; 4FT8 A; 2W52 A; 5FFO C; 2VEC A; 4QU2 A; 4COW A; 5H9S A; 4B8E A; 4DB5 A; 2XLG A; 3H3U A; 3KSC A; 1DGR A; 3GZA A; 1RUD 2; 1QGL A; 4BLI A; 4YZ2 A; 2XJS A; 3U0Z A; 1H1M A; 2Q1M A; 5BNP A; 4YW1 A; 4IQP A; 1GZ9 A; 2IXH A; 4CP0 A; 1AR7 1; 1I9R A; 3OGJ A; 1CQ9 A; 5FOJ B; 5HI0 A; 2HEY F; 3B02 A; 3BCW A; 4OUS A; 3VBU A; 4YVW A; 1C4R A; 4ZU7 A; 2GMP A; 4YG0 A; 2PLV 2; 2QUO A; 1KD7 A; 1XYN A; 4M3C E; 3SH3 A; 1WMF A; 3JBG 2; 1VBC 1; 1S0F A; 4QX5 A; 1YHF A; 3J8F 1; 2PQQ A; 1LKN A; 4XFI A; 2ZKN A; 2GDF A; 2E53 A; 1RL3 A; 1GFF 2; 2X29 A; 2QKD A; 4COZ A; 3NWK A; 4LEJ A; 4ORF A; 4AQT A; 4RGA A; 2DVG A; 2WNV A; 2D5H A; 5C4W A; 4D7S A; 1YRZ A; 2B7Y A; 3SMH A; 3TWI A; 2UYB A; 4HLT A; 2GC2 A; 1WLT A; 2HWD 2; 4QPG B; 4AEM A; 1U5Z A; 4CRQ A; 4WVW A; 1RHI 1; 4RFT 0; 1X50 A; 1R1A 1; 1H4H A; 1Z7S 3; 5LEJ A; 4Z92 B; 3OY8 B; 2V73 A; 3KS7 A; 2WLP A; 2GH2 A; 2NMN A; 1SFY A; 5J7M A; 4QM8 A; 1IS5 A; 4Y1X A; 5EYY A; 4B4P A; 1CVN A; 1GYK A; 2CNA A; 4P9L A; 1CPN A; 4N7I A; 3C2U A; 1EP0 A; 1VBB 2; 4U2A A; 2P3K A; 5A32 A; 1ZBA 3; 4AI1 A; 2F6B A; 4MUV A; 4AI3 A; 2E7Q A; 2IXC A; 3I59 B; 3S6P A; 1CJD A; 3MIG A; 1H8V A; 2AUY A; 3M8L A; 4E2S A; 3ZSK A; 1QXP A; 5C8W A; 3N7L A; 5HBA A; 2DF7 A; 1BBT 2; 4RGG A; 1PVX A; 4YVS B; 1ZRF A; 4QPG C; 4N53 B; 3HR9 A; 1S0D A; 1E57 A; 4GIQ A; 2RM2 1; 4ES9 A; 5I93 A; 4D3W A; 1FYU A; 2XWB B; 3JD7 1; 3NV2 A; 3ROU A; 1KDM A; 1RUH 1; 2B46 X; 1LCL A; 1PO1 2; 1LOA A; 3LGR A; 1BEV 1; 1FMD 3; 2WZP P; 1XXN A; 1NLR A; 1DG6 A; 4YGB A; 1S0C A; 4RA0 A; 5JSR A; 1FI2 A; 4XJW A; 4COY A; 1V9U 3; 2NP0 A; 1ZZ8 A; 5F5Q A; 4FPF A; 4UW3 A; 2ZHK A; 3TRS B; 5CA6 A; 1QU0 A; 2DVA A; 4AFM A; 1MS4 A; 4YVS C; 2H6B A; 3EO6 A; 1V70 A; 2WS9 2; 1LOG A; 3TB0 A; 3WG1 A; 1HKD A; 4HK8 A; 4PBO A; 1MS0 A; 4IV1 B; 2A6Y A; 3AXD A; 4IXL A; 2DUR A; 1Z7S 2; 3FC0 A; 1IS4 A; 3N3H A; 4Y1Y A; 4F7Z A; 2CL2 A; 1YO8 A; 4AI2 A; 5JZG B; 5FO7 B; 4Q0U A; 4LTA A; 3ZXF A; 2ARE A; 3KT1 A; 3P3N A; 3FBY A; 1LJ7 A; 5HCF A; 1PK6 A; 1OLR A; 1BVV A; 3CJI B; 1RUC 3; 5DUX A; 1Q5O A; 4C91 A; 3ZXE A; 3H7Y A; 4IV3 A; 3ZXG A; 5C9A A; 1CAW A; 4KG0 A; 4ZKT B; 1H4G A; 2P37 A; 2I45 A; 3B00 A; 4AI0 A; 2H0B A; 4P14 A; 4L11 A; 5JOX A; 3ES4 A; 1PGW 2; 5I8S A; 1YI9 A; 4IV1 C; 4NO4 A; 3WVJ A; 3VBH A; 1DFQ A; 1YNA A; 2WJS A; 4Q8L A; 3FZ3 A; 4RSU A; 3TN9 1; 2FHR A; 4YAR A; 1PMI A; 3CU7 A; 1ONA A; 3KT4 A; 5JZG C; 1W6O B; 3R4S A; 5DXD A; 1RB8 G; 4MZU A; 1OPM A; 2OVU A; 2VUL A; 5JIU A; 1J3P A; 1QJY 3; 3CJI C; 3MDP A; 1NG0 A; 4ITC A; 3GBW A; 2A6V A; 4YFW A; 3ZYJ B; 4ASM B; 5KXE A; 3GZU C; 4YW4 A; 2QZ2 A; 1K5M B; 1DBN A; 1MVQ A; 2Z69 A; 4YNI A; 1IHM A; 1GBG A; 4GKX A; 1LEN A; 2JX9 A; 4C1X A; 1NA1 A; 5GJ6 A; 3QCW A; 3MIB A; 4W6W A; 1IOA A; 4B4Q A; 2JG8 A; 4BIF A; 4XMI A; 2ET7 A; 4HVO A; 1MZF A; 4OFG A; 5DDJ 2; 1AYM 2; 3APB A; 3BOP A; 4FXK B; 3RNS A; 3VN4 A; 1LRH A; 2B39 A; 3ES1 A; 2WZR 1; 2JG9 B; 5E1R A; 3U07 A; 3IDE A; 1YI7 A; 3WHT A; 4JCK A; 2D6M A; 4L8Q A; 3MI8 A; 1FOD 3; 3ACL A; 1C5H A; 4XBQ A; 2LM3 A; 4DOU A; 1ULF A; 5T5L A; 2PHD A; 3ZSE A; 2DUO A; 1N3P A; 4JRA A; 2HWE 1; 4YLZ A; 2YUE A; 1UKG A; 2XJV A; 4XHX A; 1O7F A; 4Y26 A; 1A3K A; 1PZ9 A; 1QJX 1; 4FQ4 A; 4HAN A; 2VU9 A; 1CJP A; 4L97 A; 4AHZ A; 4PBP A; 3WIN E; 3LSE A; 5EFU A; 1ZZ7 A; 4LHK A; 4GB3 1; 3WG4 A; 1W2U A; 4DNY A; 1QJY 1; 1KRI A; 2DU0 A; 2ZAC A; 3PVB B; 4XDQ A; 3MW3 A; 1FX5 A; 1H9W A; 3NV1 A; 1WBF A; 3G6J B; 4HKW A; 4ES8 A; 4FVV A; 4IEW A; 4RHV 3; 3TNP B; 2QNK A; 1QH7 A; 2XGX A; 1AR8 1; 2ZX1 A; 2BGC A; 2WSV A; 4IEZ A; 4RR3 B; 5GLV A; 3ZSM A; 2ARX A; 5L19 A; 2Y0I A; 1OFS A; 1VRH 3; 3BOW A; 1FV3 A; 4YGE A; 1J58 A; 5HCA A; 3C3V A; 3MIC A; 1MEC 3; 2TNF A; 3ZYP A; 4QPG A; 4YM0 A; 4CDQ B; 2XBO 3; 4K21 A; 4M00 A; 5JCT A; 1B35 B; 2XB6 C; 4XXA A; 3OY8 A; 4FXG B; 1QOT A; 4YW5 A; 3E5X A; 1VB4 A; 1GQH A; 2JJ6 A; 4FAH A; 1XE8 A; 3POY A; 3KD4 A; 1C5I A; 3IWZ A; 4NHK A; 5KHG A; 2W0X A; 1ZX5 A; 1R09 1; 4IND A; 3V65 A; 1H9P A; 5HCB A; 3RRD A; 4CDW A; 3IT8 A; 4FPC A; 4CDU B; 3JUU A; 3PVE A; 1BZW A; 3R6S A; 5BYN A; 2KM2 A; 3I59 A; 4TSV A; 1F8V A; 2HWF 2; 1G7Y A; 1Q3E A; 4JGY B; 4QHB A; 1GZW B; 5CAD A; 5FQ0 B; 2CGP A; 4MGY E; 1CIW A; 2VW2 A; 4XBL A; 4YVS A; 4IEX A; 4N53 A; 1RUF 1; 3I54 A; 1JUH A; 5JI9 A; 3B5L B; 2E63 A; 1QJU 1; 4H3W A; 4AHW A; 2R0F A; 4XJ9 A; 1B35 C; 1LR5 A; 2OZ6 A; 2GND A; 4RS5 B; 1RVF 1; 3WY6 A; 3KMH A; 5AVP A; 2B9U A; 3FJS A; 3IPV B; 4KL1 A; 1RIR A; 1ZRR A; 3BOD A; 4E9K A; 4MYE A; 1DZQ A; 2D93 A; 4Q1R A; 4KU8 A; 1UIK A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...