The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
104
|
sequence length |
341
|
structure length |
341
|
Chain Sequence |
MIVLFVDFDYFYAQVEEVLNPSLKGKPVVVCVFSGRFEDSGAVATANYEARKFGVKAGIPIVEAKKILPNAVYLPMRKEVYQQVSSRIMNLLREYSEKIEIASIDEAYLDISDKVRDYREAYNLGLEIKNKILEKEKITVTVGISKNKVFAKIAADMAKPNGIKVIDDEEVKRLIRELDIADVPGIGNITAEKLKKLGINKLVDTLSIEFDKLKGMIGEAKAKYLISLARDEYNEPIRTRVRKSIGRIVTMKRNSRNLEEIKPYLFRAIEESYYKLDKRIPKAIHVVAVTEDLDIVSRGRTFPHGISKETAYSESVKLLQKILEEDERKIRRIGVRFSKFI
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
Structure of Dpo4 DNA polymerase replicating across the genotoxic N7-methylguanine lesion
rcsb |
| molecule keywords |
DNA polymerase IV
|
| molecule tags |
Transferase/dna
|
| source organism |
Sulfolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
|
| total genus |
104
|
| structure length |
341
|
| sequence length |
341
|
| ec nomenclature |
ec
2.7.7.7: DNA-directed DNA polymerase. |
| pdb deposition date | 2015-10-22 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00817 | IMS | impB/mucB/samB family |
| A | PF11798 | IMS_HHH | IMS family HHH motif |
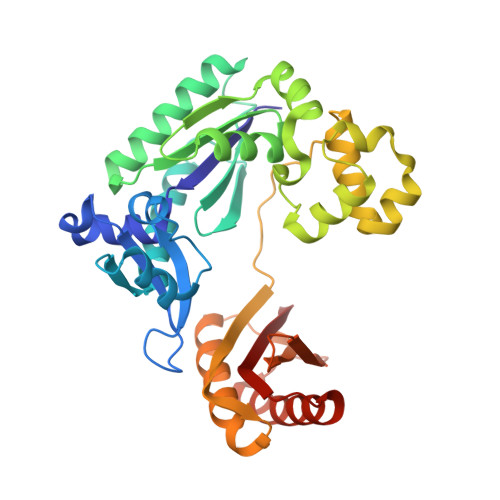
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Beta | Roll | Urease, subunit C; domain 1 | Urease, subunit C; domain 1 | ||
| Alpha Beta | 2-Layer Sandwich | Alpha-Beta Plaits | Alpha-Beta Plaits | ||
| Alpha Beta | 2-Layer Sandwich | Dna Ligase; domain 1 | DNA polymerase, Y-family, little finger domain |
#chains in the Genus database with same CATH superfamily 5KFZ A; 3IN5 A; 5K14 A; 3FFI A; 3HVA A; 1D0E A; 1HNV A; 4RU9 A; 1IKV B; 2IC3 A; 4TYB A; 4WFZ A; 2J6T A; 3M9N B; 4O3R A; 5DQG A; 4O3S A; 2BQU A; 3DYA A; 4NLS A; 4F4W A; 4ECY A; 4RW4 B; 4ZP7 A; 4HKQ A; 2W9B A; 4Q8E A; 2R8K A; 1LWF B; 3DOK B; 5KG4 A; 1LW2 A; 1REV A; 4EBD A; 5HRO B; 2FVQ A; 1HMV A; 1HQE B; 3UPF A; 1JLC B; 3OLA A; 2R2S A; 1TVR B; 5D3G B; 4KSE B; 3QZ7 A; 5CYM B; 3V6H A; 4G3I A; 1LW0 A; 3RB3 A; 2B6A B; 2AX0 A; 1RYS A; 4EO8 A; 2HWH A; 4RW9 B; 1T03 B; 3KK1 B; 2J6U A; 2FLL A; 1JX4 A; 3KLI A; 3PW5 A; 1S9F A; 2YKM A; 2JEF A; 5F8H A; 4WE1 A; 4RW7 A; 4PUO A; 1HNI B; 3DRP A; 4MIA A; 4ZMM A; 4NCG A; 1TKZ B; 3KJV B; 5KG3 A; 4NRU A; 4ZVG A; 4PQU A; 4F50 A; 4NLX A; 4RW6 B; 3OL8 A; 3KNA A; 5KFG A; 1NND A; 3BSC A; 5KFR A; 4KBI A; 4H54 A; 3GYN A; 1RTD B; 3KK2 A; 5KT6 A; 1IKY B; 2HNY A; 3LP2 B; 3QLH A; 5C24 B; 1TV6 A; 2OPR A; 2I5J B; 3M8P A; 4IG3 A; 1RT3 B; 2IM2 A; 5DG7 A; 5I42 B; 4IG0 B; 2RF2 A; 3PZP A; 4IRK A; 4DL5 A; 1W25 A; 3MED B; 2FVR A; 4Q43 A; 2BQ3 A; 5J2Q A; 4J9P A; 1FK9 A; 4NZ0 A; 1JXL A; 4MK9 A; 1K1Q A; 3QZ8 A; 4EBE A; 3CJ4 A; 2IAJ A; 1XR6 A; 3PR4 A; 1RT6 A; 4O4G A; 2FLN A; 1KLM B; 3H4B A; 1R0A B; 2AQ4 A; 2OH2 A; 3IS9 B; 4ECV A; 3GII A; 4IRC A; 5JXS A; 2D7S A; 2WOM A; 1DTT A; 5FDL A; 4TY8 B; 4K4Y A; 1TL1 A; 3UR0 A; 1JLB A; 4K4S A; 4ED8 A; 3DOL B; 5J2M A; 3H40 A; 3V6D A; 5J2N A; 1Z4U A; 3BR9 A; 1N5Y A; 1BQN A; 1HQU B; 4EYI B; 4JV0 A; 1FKO B; 4J9M A; 3V81 B; 5I3U A; 2W9C A; 3IRX A; 4F4X A; 1S6P B; 1XR7 A; 5HP1 B; 3KLH A; 2AGP A; 3JYT B; 4ZPB A; 4M95 A; 3UDL A; 2IMW P; 1HPZ A; 1S9E B; 4KV8 B; 3E01 A; 2YNH A; 4URS A; 4H4O B; 2WHO A; 1S97 A; 1K1S A; 4IFV A; 2V0N A; 1IM4 A; 2HND A; 5K14 B; 4O3N A; 2I1R A; 3N6M A; 3FFI B; 5F8L A; 4ICL A; 4IH7 A; 3FRZ A; 2XWY A; 3G86 A; 1ZTT A; 3KMQ A; 2IC3 B; 4ECU A; 4O3O A; 2C2R A; 2JC1 A; 3DLG A; 4IR9 A; 1EP4 B; 1KHW A; 4WZM A; 5KFK A; 4HYK A; 3MF5 A; 3RB6 A; 2W9B B; 3EZU A; 5KG7 A; 3KLE A; 4NLW A; 3MFI A; 2E9Z A; 2R8I A; 2QE5 A; 4PWD B; 3H5S A; 1LW2 B; 1REV B; 2HMI B; 1HYS A; 1HMV B; 4WYW A; 3MFH A; 3KHR A; 1RDR A; 4NLU A; 2R2R A; 3LP0 A; 5C42 A; 3OSP A; 1LW0 B; 1TKX A; 2V4Q A; 5KFE A; 3FQL A; 2OPP A; 3H59 A; 2W7O A; 4YR2 A; 4O44 A; 1EET A; 1JLG A; 2YKM B; 3MEC A; 3GV7 B; 3PW4 A; 4WE1 B; 1S1W B; 4MH8 A; 4LQ3 A; 1JLQ A; 1JLE A; 2CJQ A; 4QWD A; 4KAI A; 1BQM A; 1ZET A; 4YR3 A; 4IFY A; 4IH6 A; 4PQU B; 4K4Z A; 3DRS A; 5J1E A; 1IKW A; 2JLE A; 4GC6 A; 1RTI B; 2VG6 B; 2ALZ A; 1S1T B; 4ZPA A; 4DL7 A; 4TQS A; 4ZP9 A; 3KK2 B; 1RTJ B; 3C6T A; 4LSN A; 2OPR B; 4EYH B; 2VA2 A; 4IKA A; 4Y2A A; 3Q8Q B; 4IG3 B; 4KO0 A; 4TXS A; 4WXW A; 2J6S A; 1S9G B; 2RF2 B; 4ED7 A; 4XO0 A; 4OL8 A; 3H98 A; 4J9O A; 3T5H A; 4URG A; 2XCP A; 4LQ9 A; 5DGA A; 4ECQ A; 3QO9 B; 3GQC A; 1ZTW A; 3V6J A; 1RT6 B; 1QE1 B; 2BR0 A; 4O4G B; 1T05 B; 3RAQ A; 3HVW A; 4IDK B; 1RT5 A; 4H4M B; 3BGR B; 3MEE A; 2AGO A; 4ECX A; 4ECT A; 1UWB A; 2ASL A; 5KFM A; 2WOM B; 5FDL B; 3OL9 A; 4X2B A; 1C1C A; 1TL1 B; 2BE2 B; 1TP7 A; 4NLV A; 4ZVF A; 3KHG A; 3I0R A; 1U09 A; 4ECW A; 3RB0 A; 5KFI A; 3MR6 A; 3TAM A; 3GV8 B; 2XC9 A; 2YKN A; 2CKW A; 3V6D B; 1NHV A; 3N6L A; 2IJN A; 1N5Y B; 3FQK A; 1RT2 B; 2W7P A; 4M94 A; 3MEG A; 5KT5 A; 4DEZ A; 2W9C B; 2ILZ A; 3DLE B; 3IRX B; 3NBP B; 1RYR A; 4DG1 A; 5I3U B; 4NLO A; 4TN2 A; 3KK3 B; 1RTD A; 5HBM A; 3KLH B; 2BQR A; 4FBU A; 5KFU A; 2XI2 A; 1IKX A; 1DTQ A; 2VA3 A; 1SUQ A; 4FS1 A; 4I2Q A; 4URQ U; 5KFL A; 1RA7 A; 1WNE A; 4NLP A; 5KFA A; 3E01 B; 2YNH B; 2EC0 A; 4MKB A; 4R5P A; 3QIP A; 2IM0 A; 4WFX A; 4QW9 A; 4KE5 A; 4Y2C A; 2WON A; 3RAX A; 4IFV B; 3PW2 A; 3BQ2 A; 3V4I A; 5DLF A; 4YQW A; 1C1B A; 1DLO B; 2HND B; 4ZHR B; 3UQS A; 3LAK A; 1KHV A; 3OL7 A; 1LWC A; 3DLG B; 4TLR A; 1D1U A; 4U7C A; 2XGP A; 4RNM A; 1RTH A; 3H4D A; 4EBC A; 2VG7 A; 4XPE A; 5KG2 A; 2QV6 A; 3KLE B; 1N6Q A; 5KG6 A; 2V9W A; 4K4T A; 3QYY A; 4KB7 A; 4J9L A; 2XCA A; 3NL0 A; 3LP0 B; 4YR0 A; 3CDU A; 3OL6 A; 3GIK A; 4R8U A; 1S1U B; 2ZD1 A; 5CYQ B; 3G6X A; 1TKX B; 2BRK A; 5F8J A; 2R2U A; 3T5L A; 1JKH A; 1JLA A; 2OPP B; 5KT3 A; 1EET B; 3MEC B; 1GX5 A; 4YP3 A; 2UUW A; 1JLQ B; 3PJX A; 2C2D A; 4IFY B; 2AGQ A; 3IG1 B; 4I7F A; 2WTF A; 4KHM A; 2RKI A; 5KFB A; 3DRS B; 3FDS A; 4ID5 B; 5J1E B; 1IKW B; 2WB4 A; 2ZE2 A; 2E9R X; 1RT7 A; 2R8H A; 2FVC A; 4NRT A; 3C6T B; 1JLF B; 2BAN B; 4LSN B; 1SH2 A; 2JEG A; 3PHE A; 4LSL A; 2ILY A; 5KFY A; 1S10 A; 2GC8 A; 4TQR A; 1VRU A; 4J9Q A; 1S0O A; 4KKO A; 2AX1 A; 5F8I A; 4J9K A; 1GX6 A; 2FJV A; 3OHA A; 2ZKU A; 3DI6 A; 2WRM A; 4ED2 A; 2JEI A; 2YNI B; 4B3O B; 2VG5 A; 4WZQ A; 3DLK A; 3MR5 A; 3LAM A; 4MFB A; 5J2P A; 1QAI A; 3HVT A; 5KT2 A; 3TQ1 A; 1HNV B; 3H2L A; 5C25 A; 4ED3 A; 3PJW A; 1RT5 B; 1HQE A; 2GIR A; 1TVR A; 3MEE B; 5KFO A; 4RUA A; 2C2E A; 3CJ3 A; 3DYA B; 5KFJ A; 1UWB B; 3DRR A; 2OPS A; 4RW8 A; 5CYM A; 4EEY A; 4ECS A; 1C1C B; 3D28 A; 3KK1 A; 3HKW A; 3I0R B; 3I5B A; 3TAM B; 4KFB A; 4NLD A; 2YKN B; 4FS2 A; 3BJY A; 1HNI A; 4Q0B A; 3MEG B; 4NLQ A; 3BSO A; 2HAI A; 1SV5 A; 3ICL A; 1S1V A; 2C28 A; 1CSJ A; 1TKZ A; 4DG1 B; 3KJV A; 3SFU A; 4NLR A; 5HBM B; 4J9R A; 3Q8S B; 3VQS A; 3KLI B; 5HLF A; 3OLB A; 4PUO B; 1DTQ B; 1SUQ B; 5L1I A; 4IOB A; 4RW7 B; 3M8Q A; 5EDW A; 3DRP B; 2FJX A; 2UVU A; 1IKY A; 3LP2 A; 4NCG B; 4Y34 A; 5C24 A; 4YME A; 1S1X A; 3V4I B; 3GV5 B; 3LP1 A; 1C1B B; 1RT4 A; 2WK4 A; 5KG0 A; 5JUM A; 4JV2 A; 1LWC B; 4ZP8 A; 2HNY B; 4MIB A; 3QLH B; 1TL3 A; 3OHB A; 4ZPC A; 1RTH B; 1TV6 B; 2VG7 B; 3M8P B; 1N6Q B; 3CDE A; 3H5X A; 5KFD A; 2F8E X; 1C2P A; 2V4R A; 4Q44 A; 4R0E A; 2IBK A; 1R0A A; 4Q8F A; 3GIM A; 5J2Q B; 4RNN A; 4R8U B; 2DPJ A; 1FK9 B; 2BRL A; 2XGQ A; 1I6J A; 2HNZ A; 2B43 A; 4TY8 A; 1JKH B; 1JLA B; 5KFX A; 2IAJ B; 3CO9 A; 3GIL A; 1N48 A; 4QWA A; 1LWE A; 5KFQ A; 4MK7 A; 5C5J A; 2R8J A; 5KT7 A; 5DGB A; 3M9M B; 2D3Z A; 1DTT B; 3IGV A; 4I7F B; 2FJW A; 4K4U A; 4DL4 A; 2RKI B; 4Q45 A; 5F9N A; 1JLB B; 4MKA A; 5J2M B; 2OPQ A; 3JYT A; 2ZE2 B; 5J2N B; 4JUZ A; 4QWC A; 1MU2 A; 1BQN B; 3ITH A; 3T1A A; 3LAL A; 2B5J A; 4QW8 A; 4KV8 A; 4MK8 A; 1MML A; 4H4O A; 3KLV A; 3JSM A; 1S4F A; 2IJF A; 4LSL B; 4XPC A; 1C0U A; 3M9O B; 3CJ0 A; 3QID A; 5KG1 A; 2DPI A; 1JIH A; 1VRU B; 4KKO B; 3I0S A; 5DLG A; 1VRT A; 3KLG A; 1C0T A; 1HPZ B; 1RT1 A; 3MR3 A; 5KFS A; 3DI6 B; 4O3Q A; 3G6V A; 2D3U A; 3DDK A; 3BSN A; 3NAH A; 1EP4 A; 1S0M A; 2VG5 B; 1FKP A; 3DLK B; 4IRD A; 4MFB B; 5J2P B; 1UNN C; 3DMJ A; 3HVT B; 3NKY A; 3T19 A; 1YVF A; 3CJ5 A; 3EPI A; 4ICL B; 4PWD A; 3Q8P B; 2HMI A; 5C25 B; 4K4V A; 3T5J A; 4F4Z A; 3RBE A; 2R8G A; 2R2T A; 3DRR B; 2OPS B; 4RW8 B; 4NLY A; 3T5K A; 2IA6 A; 3OSN A; 1OS5 A; 3IGN A; 4KFB B; 4ED1 A; 1HYS B; 1S1W A; 3G6Y A; 4Q0B B; 5C42 B; 3H5U A; 4KHR A; 3BQ0 A; 1SV5 B; 1S1V B; 2YNF A; 1N4L A; 4RNO A; 3RBD A; 4G1Q A; 1S6Q A; 1J5O A; 5DQH A; 3I5C A; 3EPG A; 1XR5 A; 2UUT A; 4O44 B; 5HLF B; 5KFH A; 3SFG A; 5EWG A; 5L1J A; 1JLG B; 1RTI A; 1HAR A; 1S49 A; 4FBT A; 3Q8R B; 4O3P A; 2AU0 A; 1S1T A; 1TKT A; 2RDI A; 1JLE B; 2VG6 A; 1S0N A; 3C6U A; 3M8Q B; 1RTJ A; 1BQM B; 1S1X B; 3BQ1 A; 4O4R A; 4ZPD A; 3KLF A; 2YNG A; 3LP1 B; 1RT4 B; 3QGG A; 4TY9 A; 4GC7 A; 5KFP A; 4EO6 A; 1S9G A; 3KMS A; 4NLG A; 3V6K A; 3CJ2 A; 4QPX A; 3H5Y A; 1TL3 B; 5EUH A; 5KG5 A; 3NGD A; 4J9N A; 3CWJ A; 3DM2 A; 4ZP6 A; 3N6N A; 4I2P A; 4KO0 B; 2HWI A; 3PW7 A; 2W8L A; 3QO9 A; 2IM1 A; 1T05 A; 1QE1 A; 2W8K A; 1IKV A; 2WCX A; 4IDK A; 4H4M A; 2XI3 A; 3BGR A; 4K4X A; 2FLP A; 3E51 A; 2HNZ B; 4RW4 A; 1SH0 A; 3HHK A; 5KFV A; 4IR1 A; 4JV1 A; 3LAN A; 5F9L A; 2BE2 A; 4RNF A; 3RB4 A; 1LWF A; 4QWB A; 1RA6 A; 1LWE B; 2IM3 A; 3DOK A; 5HRO A; 3BRE A; 1JLC A; 2GIQ A; 2ASJ A; 5D3G A; 1RT2 A; 3DLE A; 3SI8 A; 3NBP A; 5L1K A; 2UVW A; 2B6A A; 1T94 A; 3KK3 A; 4RW9 A; 1T03 A; 2E9T A; 2OPQ B; 3PR5 B; 3BSA A; 1MU2 B; 3CDW A; 5KFT A; 3KHL A; 3ITH B; 4WYL A; 2B5J B; 5DG9 A; 3TVK A; 4K50 A; 5KFC A; 3KOA A; 5L1L A; 3JSM B; 3PW0 A; 5F8N A; 2UVR A; 2JEJ A; 5F8G A; 1C0U B; 5L9X A; 4RW6 A; 4RUC A; 1DLO A; 3I0S B; 4ZHR A; 1IKX B; 1VRT B; 3KLG B; 4I2Q B; 1QAJ A; 1C0T B; 1RT1 B; 5KT4 A; 3MR2 A; 3NAI A; 4ECZ A; 3QGH A; 4R5P B; 3QIP B; 4ZVE A; 3CVK A; 2WON B; 2O5D A; 1FKP B; 2I5J A; 3I5A A; 4NLT A; 4RNH A; 3DMJ B; 1RT3 A; 5I42 A; 4IG0 A; 4WXO A; 4DL6 A; 4DL3 A; 3MED A; 5F8M A; 2C22 A; 2D41 A; 3ISN D; 5DG8 A; 1RAJ A; 3MTK A; 5KFF A; 1S1U A; 4Y3C A; 5KFN A; 5CYQ A; 3PVX A; 2FVS A; 4ECR A; 4RZR A; 1N56 A; 3ISN C; 1NHU A; 4J9S A; 2ATL A; 1SH3 A; 1KLM A; 4U6P A; 5EWF A; 2RDJ A; 3NMA A; 3FSI A; 3QGI A; 3IS9 A; 2JC0 A; 2ZD1 B; 3D5M A; 1T3N A; 2YNF B; 3IG1 A; 4WFY A; 1TQL A; 4G1Q B; 1S6Q B; 1J5O B; 4IQX A; 4ZVH A; 4DL2 A; 1S48 A; 3DOL A; 4ID5 A; 4B3O A; 4NYZ A; 4IH5 A; 4TYA A; 1TKT B; 4K4W A; 5KFW A; 1HQU A; 3C6U B; 1FKO A; 3V81 A; 1JLF A; 3GIJ A; 1S6P A; 2BAN A; 5HP1 A; 2W9A A; 1QUV A; 3KLF B; 2YNG B; 4ED6 A; 2UVV A; 2FVP A; 4ED0 A; 4QWE A; 3CIZ A; 1RT7 B; 4F4Y A; 2AWZ A; 5T14 A; 5EWE A; 1S9E A; 3KHH A; 5DQI A; 3DM2 B; 2YNI A; 2ASD A; 4I2P B; #chains in the Genus database with same CATH topology 1RWW B; 3EGZ A; 1Q7L B; 2VPS A; 1ACM B; 1SEI A; 4ECY A; 2N4C A; 3GB0 A; 3CPW R; 2R8K A; 5KG4 A; 1REV A; 3UPF A; 2PVP A; 2FHK A; 4KSE B; 1PSD A; 2Y0S A; 1YDL A; 2VC0 A; 3MAA A; 3M8Z A; 2HK3 A; 3T9N A; 3KNA A; 1XBW A; 1NND A; 1RP5 A; 3NWG A; 3SSR A; 1RHJ B; 2EK1 A; 1L3K A; 1OY8 A; 5ETV A; 4ELV A; 4IG0 B; 2CPD A; 2YY3 A; 3MED B; 2ICS A; 2DH9 A; 2UXB H; 5HBR B; 4KQN A; 2FB9 A; 2DJW A; 1XMO F; 2ZUG A; 2QLX A; 2EWH A; 1R0A B; 1FKA H; 1VC0 A; 2AQ4 A; 2WB1 E; 1G1X A; 1BXN A; 4K4S A; 1R8H A; 1ICE B; 3V6D A; 4PDB A; 3O3L A; 3MCO A; 2FMA A; 3P5T L; 4LR7 A; 4A48 A; 2OJ3 A; 3TNV A; 2G1H A; 1XEZ A; 3IBW A; 1YBA A; 2W9B B; 1AUS L; 3MFI A; 3HHD A; 2V8P A; 3D1Q A; 5B0B A; 5A0Y A; 1YKW A; 2HVR A; 1JLE A; 4A3F G; 2L08 A; 2FPG B; 3DRS A; 5FLM G; 4OYX A; 5D6G A; 4CLZ A; 2Y0B B; 1SC4 B; 4YY3 F; 3Q8Q B; 2R86 A; 2VOD A; 2YWW A; 1D9F A; 4CTA B; 1V5V A; 1HI1 A; 1QLN A; 3NGS A; 2B51 A; 3ZZP A; 4IDK B; 1R9Z A; 4OV1 A; 3BGR B; 3I9V 9; 5K20 B; 4AQY H; 3CCM S; 1N5Y B; 1CC7 A; 4FYY B; 2XI2 A; 4GLX A; 4F3M A; 3M8Y A; 2FE3 A; 1WAC A; 3DRY A; 1U0H B; 1TK5 A; 1UVK A; 3FZ2 A; 3UQS A; 7FDR A; 3ETM A; 2XGP A; 2OTL S; 1Q79 A; 4WKY A; 2K06 A; 5D1P A; 5BJR A; 3BV8 A; 4KB7 A; 4HLB A; 4FDF A; 1GX5 A; 3HF5 A; 1WI6 A; 3KJN B; 3UQ8 A; 4J90 A; 1FWI C; 4BXX G; 2YRX A; 1IBC B; 4DR5 H; 2ILY A; 3QGK C; 5LXX A; 1YZ6 A; 1NXI A; 2Y1R I; 4KKO A; 2DT9 A; 2ZKU A; 2D3O 1; 1TL5 A; 5IYD G; 2CAJ A; 4A8K A; 3DGP A; 2CW8 A; 3D9B A; 3L9Z A; 4ZVS B; 4DV2 F; 2QA4 S; 4C8O A; 4N7T A; 4UOG A; 3L4J A; 3TA1 A; 5GMK a; 1TK0 A; 1C30 A; 3BB5 A; 2VFS A; 3MEG B; 3EZO A; 3AOC A; 2C3U A; 3M8Q A; 1G2E A; 2MQ8 A; 4NCG B; 2BB0 A; 3LWL A; 2VUN A; 3GV5 B; 3N01 A; 4UZB A; 4ZP8 A; 1URR A; 3OHB A; 2QN6 B; 4L72 B; 1R0A A; 1DV2 A; 4ZLP A; 4BBR G; 2VQE J; 3PYE A; 5JVG Q; 4QWA A; 2AJ1 A; 3K57 A; 5DGB A; 1K0T A; 4DL4 A; 2MCT A; 1F9F A; 4MKA A; 3RU0 A; 2NS1 B; 1B0P A; 1RAH B; 1RAD B; 4DUZ F; 2JWN A; 4A8W A; 3D6N A; 1JIH A; 1KLN A; 4F3Q A; 3HHL A; 3L7P A; 2IVF B; 1N52 B; 3SCJ E; 2KL8 A; 2MB0 B; 1VQY A; 3LWB A; 2QA2 A; 4ICL B; 2UP1 A; 2V4J B; 3Q8P B; 4M9U A; 4Y2M A; 4DV4 H; 2E66 A; 2K1R B; 3EYZ A; 5DZ7 A; 3DRR B; 3KJF B; 5B09 A; 2YH1 A; 4OZJ A; 4C98 A; 1VK8 A; 2MQO A; 1WC4 A; 2BEF A; 3HD2 A; 3GR0 A; 2KTQ A; 1M1G A; 3D54 A; 2UU9 F; 3SE7 A; 5EWG A; 4V1A k; 4M5J A; 4Y28 C; 4LSX A; 2V94 A; 4DV0 F; 3QFW A; 2F4V H; 3TWZ A; 3RGI A; 4C5Z A; 4BJH A; 4OZ1 A; 3N6N A; 2JA5 A; 3A12 A; 4JI0 H; 3GG1 A; 3PIO Q; 2B4O A; 2EPI A; 4IDK A; 3BGR A; 1NJX A; 4O0N A; 2KG1 A; 2WBR A; 1FVI A; 5HRO A; 2EC2 A; 4M5M A; 3EGN A; 2YVS A; 2Q66 A; 2HHQ A; 1NM2 A; 3GTQ A; 3MTJ A; 5A76 A; 4F0H A; 2G0C A; 4CQB A; 1UVN A; 3MTW A; 4RW6 A; 3I0S B; 3IV8 A; 4TV9 F; 2E44 A; 4J91 A; 2UV8 G; 4ZLJ A; 2V2Z A; 1VQ9 S; 2NYH A; 4AYB E; 1TL7 B; 1WDD A; 4USV A; 3CJ8 A; 2U1A A; 1SH3 A; 2HFO A; 2K7K A; 5DDO C; 4DMZ A; 4QR0 A; 4DL2 A; 2VIC A; 3DOL A; 4NEZ A; 1EYZ A; 4C8N A; 9ATC B; 1I1G A; 5H80 A; 1RWX B; 4DR2 H; 3U7J A; 5T14 A; 1EKS A; 1KNZ A; 3OFG A; 2L9D A; 5DQI A; 2ZDQ A; 2N3O A; 1WC1 A; 1UJ5 A; 5KFZ A; 1D0E A; 2KT2 A; 4RL1 A; 4TYB A; 2H51 B; 3C14 A; 2R4F A; 2RRA A; 3R5J B; 2GHV C; 4H4K C; 3VZH A; 1N34 H; 4U7U K; 4RZQ A; 1Z2Z A; 1HNI B; 3G82 B; 2CW7 A; 4NCG A; 3GIP A; 5HJX A; 4NRU A; 2HHU A; 1A0I A; 3JQK A; 4RWW A; 1U2F A; 3HZG A; 2FC8 A; 2OPR A; 2IM2 A; 2VD3 A; 5B0C A; 4MKV A; 4IRK A; 5CD4 A; 5C8Y F; 4J9P A; 2DYA A; 3W5X A; 4ECV A; 5GAE I; 3GMG A; 4C65 A; 1MWQ A; 2QFJ A; 2IHR 1; 4CH0 S; 4DR1 H; 4X62 F; 5A2Q U; 4U7U D; 2GPW A; 1VJW A; 2EK6 A; 9RUB A; 4KYZ A; 4KV8 B; 4C8Z A; 3CQZ A; 4ZVT B; 4P2S A; 4KIA A; 2DXE A; 4FXV A; 1YSJ A; 3JSY A; 1WHV A; 4G1P A; 4ICL A; 1KQS R; 4ZIT A; 2DH7 A; 4O3O A; 3NGU A; 4C0B A; 4WZM A; 5KG7 A; 2H48 B; 1FJE B; 1EZ1 A; 4S0M A; 3OSP A; 4NA3 A; 2W70 A; 2BM1 A; 3PW4 A; 2CJQ A; 2WYO A; 3FCH A; 2OAU A; 5IT9 U; 2RS2 A; 1IKW A; 3VGS A; 1S1T B; 1RTJ B; 2GX8 A; 1HBO B; 3K5L A; 2MY8 A; 3KXD A; 4BCA A; 2VA2 A; 3GRI A; 2YPZ A; 4GKK J; 4IO1 A; 4QYZ J; 2R6F A; 4RBT A; 3R0D A; 2UXD J; 4ECQ A; 1NSP A; 2ZC3 A; 2I38 A; 3N4W A; 3FMW A; 4WK3 A; 1PKU A; 2VOO A; 3KHG A; 1FE0 A; 1DAR A; 2LA4 A; 5THW A; 1H3D A; 2Q2U A; 3DLE B; 1YMY A; 1UKU A; 4PG6 A; 1YLX A; 3B4D A; 4DPT A; 5H9E K; 4MKB A; 3BQ2 A; 3N8D A; 2YW2 A; 4OX6 A; 3PM9 A; 1NK0 A; 2BJ3 A; 1LWC A; 4TU8 A; 4C0H A; 4TLR A; 1ARO P; 4CNU A; 4AE5 A; 2NSC A; 1TL7 A; 1FER A; 4C5C A; 1YJT A; 1GYF A; 5KG2 A; 2PN1 A; 2VC1 A; 2U2F A; 4YR0 A; 1PCA A; 5CYQ B; 2DRA A; 2BRK A; 2HZC A; 2BW2 A; 2R7U A; 2ERR A; 5CQG A; 2WTF A; 1M0S A; 1VS0 A; 4B02 A; 2OSR A; 1Q82 T; 1UZD A; 4HI2 A; 2KFN A; 4O4L F; 1S0O A; 4WKR A; 4NA2 A; 4CLK A; 1APS A; 2R7S A; 3NP5 A; 3HVT A; 3T41 A; 1SL0 A; 4A3L G; 2H5J B; 3S7R A; 3W63 A; 1J5E F; 4DQQ A; 2YKN B; 1CJK A; 2R84 A; 1HNI A; 5FPR A; 2ZBP A; 4RDV A; 3BYP A; 4ME6 A; 5HBM B; 5HLF A; 3FMB A; 4JBC A; 3DFE A; 4F04 B; 4PAB A; 3OTC A; 4ZJQ A; 1HR0 J; 3QLH B; 2NPI A; 1U49 A; 2FHO B; 4L75 A; 3G4S S; 1Q1K A; 4LF7 F; 2XGQ A; 2YWK A; 4LRF A; 4A2A A; 2DVI A; 1O75 A; 3DLB A; 4M5D A; 2E7W A; 2D3Z A; 3BKF A; 5BPH A; 2RKI B; 3LA4 A; 4K2J A; 1R0C B; 2V8D A; 1PAU B; 3CCJ S; 4KV8 A; 2DK2 A; 3JSM A; 2CDR B; 3CTR A; 3QID A; 2ZOF A; 4ZPV R; 1O0P A; 5D0G A; 2D3U A; 3MM5 A; 4JI0 F; 3OVA A; 1UVI A; 2PD1 A; 3ZC4 A; 5KTQ A; 4HNT A; 2ZOM A; 4KGZ B; 1E9Y B; 4FVC A; 1YJ9 S; 3MM8 B; 4NZ6 A; 2DY1 A; 2YNF A; 1S1V B; 4DR4 J; 1B0V A; 1S49 A; 3Q8R B; 2AU0 A; 1S1T A; 2EFO A; 3C6U A; 1RTJ A; 1EX8 A; 1B7Y B; 3FVY A; 2P2C B; 1XER A; 1K6W A; 4Q2M A; 3LA0 A; 2AT1 B; 4HNU A; 5KG5 A; 5EN1 A; 1Q95 G; 4ZN1 A; 3B8H A; 4DBQ A; 2WVD A; 2HNZ B; 2RHQ B; 4JI2 J; 2GIQ A; 4A3J G; 3DLE A; 4PXC A; 5L1K A; 3BGF A; 2FK3 A; 2PII A; 1MU2 B; 3ITH B; 4KGK A; 3TRG A; 5E7O B; 4DR2 F; 5LP6 F; 3PNI A; 1ZS6 A; 4DYK A; 3ZZO A; 3KLG B; 1XWL A; 4R5P B; 1TMM A; 2B0G A; 4NLT A; 1XDN A; 3Q1K A; 4GOA A; 5CYQ A; 5F0U A; 2MQP A; 1Y1V G; 2ZD1 B; 4ZVH A; 5FD1 A; 4IH5 A; 2LEA A; 1JKJ B; 3DCA A; 2PT0 A; 4WBN F; 3GIJ A; 2YNG B; 2D69 A; 1N33 F; 2VR1 A; 2HFS A; 2IRU A; 3KHH A; 1FWL A; 4F0K A; 3SYZ A; 3SQG B; 3FBC A; 2KJW A; 3ISZ A; 2BQU A; 4F4W A; 2ROE A; 3QAU A; 1JNR B; 3PKM A; 3A4T A; 1KRP A; 4CO5 A; 4OQJ A; 4E0D A; 4OJ3 A; 5FD5 A; 1ZM2 A; 5F8H A; 2FTR A; 4MIA A; 4LF8 J; 4ZMM A; 3R2C J; 4CC6 A; 2AZ1 A; 1I7N A; 3N42 B; 3QLH A; 5T7L B; 2VFU A; 4DPW A; 5A8K B; 4OYZ A; 4P10 A; 5LMN H; 5ETO A; 2GAH C; 1K1Q A; 1GTH A; 4U95 A; 1M7J A; 2EBE A; 4W90 B; 2IDL A; 2WOM A; 5FLX U; 1MMN A; 2DNM A; 3FMA A; 1WG1 A; 1FKO B; 2H5I B; 1IBL H; 4HR2 A; 1WCM A; 4F26 A; 3HOW G; 2BJ8 A; 4OX9 H; 3OXF A; 2LXI A; 2V6A A; 3IWX A; 4IH7 A; 3FRZ A; 2KH9 A; 4DV6 J; 4IR9 A; 1KHW A; 1UBP C; 3LP0 A; 5D1O B; 3CC2 S; 2GMJ A; 1NEE A; 2I0X A; 4A3C G; 2IVY A; 1YJR A; 3HJV A; 4YB5 A; 3UD5 A; 1ZET A; 3Q83 A; 2R87 A; 5IM0 B; 1CJU B; 4ZA1 A; 3MKV A; 4DFK A; 4PPZ A; 2DNG A; 2JVR A; 4LSN A; 4P2X A; 2X3D A; 2MLY A; 1NZA A; 2IIH A; 5A8R A; 3I4M G; 4I6Y A; 1O8B A; 1XRF A; 5GAG I; 3I4P A; 1Y11 A; 2MKJ A; 4DPY A; 2OML A; 4KC5 A; 4ATN A; 4WTY A; 4A3B G; 3FCW A; 2W7P A; 4M94 A; 3QRQ A; 2W9C B; 3I12 A; 3H0G A; 3KG4 A; 3CJK B; 1YJN S; 2MGZ B; 2XZT B; 4F4K A; 1BHN A; 2HS0 A; 4URQ U; 2AD9 A; 4P71 A; 4B9U A; 2YNH B; 4R5P A; 2R7M A; 1TWF A; 1SBR A; 1KQF B; 4IZO A; 3GTY X; 1RGV A; 5K8P A; 3I9Z A; 1R9X A; 2YTC A; 3GLK A; 1U8W A; 2PI5 A; 3H4D A; 3I6P A; 1LOU A; 4P2M A; 4XPE A; 2BAC A; 1N6Q A; 5LOV F; 4OYO A; 1RAB B; 3QYY A; 2ZD1 A; 3CCL S; 1JKH A; 4L1K A; 4CLP A; 4AQL A; 1WEZ A; 5IS2 A; 1YRU A; 4X64 H; 4R88 A; 4KHM A; 4UST A; 4DV3 J; 4OYP A; 4RZB A; 3T3F A; 5GON F; 4X4O A; 4F2R A; 1IM6 A; 4B9T A; 3UN2 A; 3BG5 A; 3T1H H; 3P5B P; 4F7V A; 2VBW A; 1HWJ A; 2BZ2 A; 1Z2L A; 5HQM A; 3C0F B; 4L74 A; 2DGW A; 2H54 B; 1EJU C; 3A8K A; 2V8V A; 3D28 A; 3K5O A; 3J7A 1; 1TAU A; 4MTE A; 1YI2 S; 5HQL A; 4DN0 A; 3MM6 A; 4N8N A; 3RJM B; 2OHD A; 2UXC J; 3DRP B; 4PZV A; 4LFB H; 3VTI A; 5T7L A; 1FFG B; 1SKU B; 2EVZ A; 4N47 A; 2WK4 A; 2XHC A; 1GXT A; 5B0F A; 1XMQ F; 3G8S A; 2LQ5 A; 2AEM A; 5A8W B; 3VQT A; 3H5X A; 3M8R A; 4R0E A; 1S6U A; 1SKS A; 2C35 B; 3KAL A; 3BZQ A; 4R8U B; 2DPJ A; 1HD0 A; 2BRL A; 2KVI A; 3D6M B; 2FHJ A; 2J0Q D; 1OSC A; 3GIL A; 3RRH A; 2P9C A; 5KFQ A; 3AD9 C; 3BDP A; 3IWL A; 3VAJ A; 2PC6 A; 3CT9 A; 1UEV A; 1B64 A; 1TA8 A; 3M6V A; 2W6Q A; 2AJ4 A; 2VHL A; 4A8S A; 1BGX T; 2Z30 B; 1T9V A; 3CJ0 A; 1FWD C; 3M4I A; 2XMW A; 4RKU C; 4LFA J; 4ES2 A; 3GJQ B; 5G0R B; 1IE7 C; 1X4B A; 4PXE A; 4G1Q A; 3HOZ G; 5IM0 A; 5KFH A; 3SFG A; 2W2O P; 4NZ7 A; 2NUU G; 4LF9 J; 1ITP A; 1QSS A; 4JI4 F; 1WG4 A; 1U6Z A; 4B9S A; 3GWC A; 3L4B C; 4ZHQ F; 3DM2 A; 4DS5 A; 1SKW A; 3PIP Q; 2KM8 C; 1NFG A; 2DHA A; 1VGY A; 4PWQ A; 3W62 A; 1P6T A; 4YBZ B; 4RNF A; 3W2W B; 2Z57 B; 3DOK A; 1LWE B; 4Y65 A; 4ZVO B; 4JYA H; 3QGP A; 1T03 A; 3CJK A; 2MGZ A; 1NZ8 A; 5KFT A; 1RK5 A; 2CQI A; 2IRY A; 1C0U B; 2DNO A; 1FWE C; 4AD6 A; 2N4D A; 1YQH A; 2OEK A; 1TUV A; 3Q86 A; 4X4Q A; 2JA5 G; 1JWW A; 2C22 A; 3SEO A; 4Y3C A; 2XLI A; 2EKN A; 5L46 A; 3GTG A; 1H6K X; 4NXM H; 1KLM A; 4LBI A; 2E7V A; 2A1B A; 1HNX F; 4FZV A; 3CCQ S; 3W8G A; 2XMT A; 4LFC J; 4P7V A; 3RV3 A; 5HP1 A; 2W9A A; 4HL9 A; 3TA2 A; 2J7U A; 4OI2 A; 4DX7 A; 1IOW A; 3PO2 G; 3LUY A; 2I9S A; 3IN5 A; 1Y00 A; 2CQY A; 2J0X A; 1B99 A; 2HVZ A; 2CQP A; 2CQD A; 2HHV A; 1YRR A; 2QJ3 A; 2ZVS A; 2A2D A; 3THP A; 1ZOT A; 1JLC B; 3RB3 A; 2AX0 A; 4F3O A; 4RW9 B; 1SCJ B; 5BYS A; 1S9F A; 3DRP A; 4LMZ A; 4PQU A; 3OL8 A; 3BSC A; 2EBG A; 5KFR A; 1H7X A; 2Y3Y A; 5A8W A; 2WAQ A; 2JFD A; 5I42 B; 4WCE Q; 3RW6 A; 3PZP A; 3UDC A; 4KY3 A; 1IR2 A; 3WIV A; 1XR6 A; 2M8D B; 5JXS A; 3QQC D; 3N41 B; 4JOU A; 5FII A; 1HQU B; 4OI3 A; 3HM9 A; 4I55 F; 3R97 A; 2DIS A; 3JYT B; 1PYS B; 3OJS A; 2GJF A; 3E01 A; 1Q78 A; 2HFN A; 3M1V A; 1MRO B; 1YJ7 A; 2BAB A; 4IFV A; 3OBI A; 3M2R A; 3G86 A; 2JC1 A; 1DBD A; 4PWW A; 3UR3 C; 4DV7 F; 4WF9 Q; 4ILR A; 4UCT A; 1TUW A; 2R8I A; 4PWD B; 1HMV B; 3DXS X; 2BW7 A; 1VC6 A; 5D15 A; 1ZA1 B; 1I51 B; 1VR6 A; 1Y5L B; 1EET A; 4LF5 F; 1S1W B; 2HHW A; 2B8P A; 1WKK A; 4DU8 A; 2RQK A; 2WSC C; 3KK2 B; 3C6T A; 1B04 A; 2WB1 D; 4EYH B; 4XO0 A; 2AIR B; 1C3O A; 2N7Y A; 2OO4 A; 2YKQ A; 3A1Y G; 1XIQ A; 2KFZ A; 1POK A; 1TL1 B; 3E0L A; 1WI8 A; 2D9O A; 3ZZT A; 1RTD A; 2XMJ A; 3QWP A; 4P52 A; 1EIY B; 5KFU A; 2QK4 A; 4UCO A; 3AHP A; 1SUQ A; 2DBB A; 4DV1 F; 4NLP A; 3I56 S; 4LRC A; 3GLJ A; 1M5H A; 2PI4 A; 2RIL A; 2YX4 A; 3DLG B; 3W8P A; 4JI5 F; 4J9L A; 1HD1 A; 3ADA C; 3NL0 A; 1HQ2 A; 5K5M A; 4I7F A; 2XQC A; 2YZM A; 4LFB F; 3D7S B; 4QBU A; 2DEI A; 4YUD A; 4DUY J; 2Q09 A; 4TU9 A; 2AX1 A; 1WH2 A; 3J81 W; 1FA0 A; 2WRM A; 5M2P A; 1QSY A; 5J2P A; 4FYX B; 1V3S A; 4JI3 J; 1U7L A; 1RT5 B; 2GIR A; 3MEE B; 2L9N A; 5LM7 E; 2Q6C A; 4U67 Q; 1ZPW X; 4DS4 A; 4DLE A; 4UZC A; 1M5P C; 1UFK A; 3NGT A; 2VQE H; 3K89 A; 2FJX A; 5JJB A; 2DNR A; 1EJW C; 1MN9 A; 2LU2 A; 4YME A; 3NOG A; 2X7I A; 3NNA A; 1H72 C; 3L2P A; 2HQF A; 2Z00 A; 4Q44 A; 2VAS A; 5IWA F; 1U45 A; 4QR2 A; 1XNR J; 1R6Y A; 2GHP A; 1VBX A; 1FK9 B; 2E1A A; 1CJT A; 1D9A A; 2QSW A; 2FVK A; 3M9M B; 3K8V A; 2B56 A; 3IGV A; 3OVB A; 1HKA A; 3NEK A; 4X4N A; 4E2F B; 3JYT A; 5J2N B; 1QY7 A; 5JVH Q; 2PO4 A; 4K0K H; 1MRO A; 2W6M A; 3GFZ A; 2DO4 A; 3DI6 B; 2ATC B; 4BY1 G; 2VG5 B; 4PWD A; 1VC5 A; 2GQQ A; 2R2T A; 1NJY A; 1V51 A; 1KCX A; 4RW8 B; 5LMU F; 4DR7 J; 2MQM A; 2VKR A; 1HYS B; 2H5E A; 2HRU A; 1S1W A; 3G6Y A; 5EX0 A; 1KVI A; 1WG5 A; 1J5O A; 1S7I A; 1X60 A; 2UUT A; 5IYB G; 1HAR A; 3UNG C; 4ANE A; 1BQM B; 2HHX A; 1GQE A; 1I94 J; 1KVJ A; 3PFO A; 1TL3 B; 3OR1 A; 1KJ9 A; 1E4F T; 5DS7 A; 2R7Z A; 3OPK A; 1NH8 A; 4HHU A; 1EUD B; 2NU8 B; 2GJ2 A; 4B9L A; 3A8J A; 1GSO A; 4JV1 A; 1NK8 A; 4QWB A; 2GGP B; 4C5B A; 4O23 A; 2UVW A; 1KN6 A; 3PR5 B; 2UXB J; 1IR1 A; 2M3D A; 1RAK A; 2ZH1 A; 4KH1 B; 4HDG A; 5C4X G; 1VRT B; 2HHT A; 4P2F A; 1KQK A; 4JB8 B; 1N5V A; 2ZDO A; 5ETR A; 1RT3 A; 2ZBC A; 5M2U A; 4M5I A; 4CT9 A; 2ZOG A; 4WFN Q; 1VIS A; 2XB2 D; 5HI7 A; 1TQL A; 4F0L A; 4IQX A; 4DZD A; 1HW8 A; 1IXK A; 1FD8 A; 1CQJ B; 4USK A; 3Q22 A; 1SJ1 A; 2HQG A; 4B17 A; 3S6E A; 3TIG A; 2FVP A; 4DFJ A; 1VJQ A; 5C1P A; 2ASD A; 5L8R C; 4A3M G; 5F3T A; 4ZIW A; 2FUG 9; 1VBK A; 1CQN A; 4M51 A; 4ED5 A; 2ZZN A; 1LEO A; 2W4P A; 3R6G B; 1BWV A; 1RIS A; 1FRI A; 3OLA A; 5D3G B; 2CPZ A; 2XSJ B; 3R7N B; 2DHX A; 3QRR A; 2LU1 A; 2XYG B; 3CMA S; 3K59 A; 2HWT A; 3N2C A; 2C3O A; 3NCP A; 2APO A; 1FXL A; 4AQY J; 1X31 C; 4LBP A; 3HB9 A; 3KJQ B; 4TV8 F; 2G2Z A; 4WHB A; 2QFI A; 2HHH F; 3C16 A; 1FK9 A; 4DQI A; 1H2V Z; 3KNG A; 2QLB B; 4EBE A; 3D3B J; 1RT6 A; 1KRC C; 2QT3 A; 1ZM9 A; 3UI3 A; 4ZCI A; 2R92 A; 2YPY A; 3SCK E; 5J2N A; 3MPG A; 4FN5 A; 3MPW A; 4UOH A; 4F02 A; 1V5I B; 1KEE A; 3CJQ A; 3LPY A; 1UA0 A; 2IC3 B; 2ACM A; 2XUL A; 2HG4 A; 1AFJ A; 1WF2 A; 2CPX A; 1SL2 A; 2VV5 A; 4PCQ A; 3GFX A; 4GKK H; 2HMI B; 4NX9 A; 1HYS A; 4DR5 J; 2ZKT A; 1FRX A; 2RML A; 2V4Q A; 2UXD H; 3H59 A; 4ZPT R; 2YKM B; 1RCX B; 2WSF C; 4MH8 A; 1BQM A; 4IFY A; 4K4Z A; 1WCM G; 4DL7 A; 2AJF E; 2ZNZ A; 1U25 A; 4DJB A; 3QJJ A; 1NH7 A; 2J6S A; 5F41 A; 2J76 E; 3C8Y A; 4K7Q A; 2V58 A; 2HLT A; 3DU5 A; 3H7H B; 3ENQ A; 1G4C A; 5H9E J; 2M5Y A; 2XZD B; 2KU7 A; 2Q43 A; 1UWB A; 2ASL A; 4LBH A; 2PGC A; 2UUA H; 3KLH B; 1FTR A; 4DPU A; 1DTQ A; 1PYO B; 1RK6 A; 5HO4 A; 3WO1 A; 1F6T A; 5LSR A; 1C1B A; 3HPO A; 2H4Y B; 2BBE A; 4OI4 A; 4OHZ A; 4CIO A; 3N5B A; 3KLE B; 1F2G A; 1IR0 A; 1KTV A; 3CDU A; 3GIK A; 1MMD A; 3CW2 C; 3T5L A; 5CB4 F; 3EE3 A; 5IYZ F; 1FEE A; 3R27 A; 1OY6 A; 1TEL A; 3N43 B; 4YDX A; 3H0G G; 1F3V A; 3JTN A; 3UCG A; 5LYJ F; 4DV4 J; 3MWE A; 4QIW E; 3CXC R; 5KFY A; 3MZI A; 1GO3 E; 2KVQ E; 1R89 A; 1SJ3 P; 2F8H A; 5LL6 b; 3L4K A; 1HNV B; 3GJS B; 4J0M A; 4G7E B; 2GFF A; 2XSJ A; 2F4V J; 3TAM B; 4KFB A; 3KDN A; 2E1R A; 1UC9 A; 1SIW B; 4NLQ A; 3OOQ A; 2B9X A; 4C11 B; 4O4H F; 2OFH X; 4Y34 A; 2NYI A; 1T9Y A; 3KUH A; 1FDB A; 4FF4 A; 4RBU A; 1BC6 A; 2MEW A; 2AZ3 A; 1RTH B; 1TV6 B; 5IZH A; 1VBZ A; 2IBK A; 3G8T A; 2Y9H A; 1J27 A; 2D3O R; 1FWP A; 2VUM A; 3AQP A; 4NXN F; 1FJ7 A; 2BJD A; 3T1A A; 3W65 A; 3R2D J; 3KTQ A; 4B3S J; 3VAM A; 1BGW A; 2QL9 B; 1VRU B; 4J7C A; 1RT1 A; 2M7S A; 1ZBT A; 4ZK7 M; 3CCV S; 1UNN C; 2DNN A; 4PG7 A; 2HMI A; 2XNQ A; 4B08 A; 4B3M J; 2OP5 A; 1IWG A; 4JI8 J; 2HKE A; 2CKZ B; 4MV3 A; 4HT7 A; 5DDP C; 5E0O A; 2RH3 A; 4USH A; 1K9M T; 4USU A; 5L1J A; 1T8E A; 3LS9 A; 4ZCL A; 4WD3 A; 4Q0Y A; 5KFP A; 3EZ5 A; 2R7N A; 1P1L A; 2MJU A; 4J9N A; 1JJ2 R; 3M32 B; 1RBL A; 4DX5 A; 2QYG A; 3B4M A; 5HVH A; 3C15 A; 4LRD A; 1LWF A; 1RA6 A; 1CE8 A; 1R6V A; 5IV3 A; 2XLJ A; 2B6A A; 2ZJR Q; 2C2O B; 2EG2 A; 4CLW A; 3B78 A; 3OFF A; 2FLO A; 5K8N A; 4KGM A; 3UCU P; 4OIX A; 3KOA A; 3PCO B; 2P5V A; 1X5P A; 3VAF A; 5IYC G; 3V2U C; 3QAT A; 3UFC X; 3ICJ A; 4OYA A; 4DR1 J; 2W4D A; 1X7V A; 3BR8 A; 2HW3 A; 2RRB A; 2C2Z B; 5EWF A; 3IAS 9; 3D5M A; 4LH6 A; 4H4K A; 3RZA A; 2X1A A; 4QQB A; 4P73 A; 2C3M A; 2DHS A; 3QFF A; 1KOH A; 3R5F A; 3BAA A; 1OQ6 A; 4ED6 A; 2UVV A; 3WHI A; 3IRW P; 1P9Y A; 3CIZ A; 4AT1 B; 2AWZ A; 3BH6 B; 5EWE A; 1S9E A; 1K8A T; 4LF8 H; 4ILL A; 5CZV A; 1IQT A; 4MYT A; 1HNV A; 3HXL A; 1IKV B; 3OTE A; 3D0H E; 4O3R A; 1KEK A; 4G7E A; 4HKQ A; 2WAQ E; 1QAX A; 4EGL A; 4OYB A; 4KTQ A; 3PRD A; 4USI A; 5GMD A; 4CO4 A; 3W3S A; 1LW0 A; 1A9X A; 2HWH A; 4WP8 A; 4AC7 C; 3JAM W; 3L09 A; 1ONW A; 5CA0 F; 2HS3 A; 5E3R A; 2DKO B; 3ZNJ 1; 1VQS A; 3BBF A; 5KOW A; 5KFG A; 4CH7 A; 4KRW A; 2KYX A; 3B82 A; 3HD1 A; 3M2U A; 1TV6 A; 4C3M A; 4TNO A; 1IUJ A; 5DG7 A; 3GE9 A; 3TA0 A; 3NOC A; 1ZM3 A; 1SB6 A; 1KR4 A; 3U1M A; 4GKK F; 4WFB Q; 1UPM B; 1KOO A; 1NJW A; 2FLN A; 3D2W A; 5IT9 W; 3IS9 B; 2UXD F; 4ZIV A; 5HMW A; 2G0J A; 3P6Y C; 1X4E A; 3HKZ A; 4CEX C; 2OOD A; 4J9M A; 3TZW A; 4ZPB A; 1TL4 A; 2FPI B; 4B3T J; 1N8R T; 2Y8W A; 2EW9 A; 2DIK A; 4NML A; 1P9Q C; 4XYM B; 2C2R A; 4TU7 A; 5KFK A; 4C3L A; 4B9N A; 1PBA A; 1RDR A; 4DCP B; 3TAP A; 2K3I A; 2LDI A; 3ONQ A; 1SPB P; 2W7O A; 3P8B B; 2KB2 A; 2ZH9 A; 3MEC A; 4WE1 B; 5K8O A; 3D1O A; 1JLQ A; 3J7A N; 1B70 B; 5CD4 H; 2KDO A; 1HBU A; 2JLE A; 1RBA A; 1U1R A; 1EAR A; 2FGC A; 4Y2A A; 1E4E B; 2KT3 A; 3RGZ A; 5D4N A; 4I69 A; 1X4C A; 3Q0A A; 5IP7 G; 1Y9O A; 5DGA A; 3QO9 B; 2GSE A; 2XZW A; 4H4M B; 1C1C A; 3MR6 A; 1AZS A; 1RAE B; 4I67 A; 3FSX A; 4HI1 A; 3K8W A; 4IAJ A; 1U6B A; 4K59 A; 4OMF G; 4F2S A; 4A3I G; 3M6W A; 1B33 N; 4FXW A; 4KE5 A; 1KS2 A; 4YQW A; 3OL7 A; 3I4H X; 4LF4 H; 4F8R A; 4RNM A; 2EKY A; 4K5S A; 1VQ6 S; 1HR0 F; 2HGM A; 3TAR A; 3P3D A; 2RT3 A; 1RWN B; 2VZ9 A; 2W7A A; 3IG0 A; 1CJV A; 3PO3 G; 2GS8 A; 4LR8 A; 1GTE A; 2AGQ A; 3HT3 A; 5D4L A; 5J1E B; 2ZE2 A; 4DN9 A; 1M5V C; 3OW2 R; 2UUC F; 4OHV A; 1H74 A; 3AA9 A; 4TQR A; 3QSI A; 3HJF A; 4B3R H; 2F4F A; 2XTJ P; 2CQ0 A; 3GG0 A; 1FE4 A; 1HNZ H; 3SDE A; 2QC3 A; 3OD2 A; 4ZN3 A; 2DGU A; 4BDP A; 1SVD A; 4ED3 A; 2OPS A; 4ECS A; 3W64 A; 1T9T A; 3I0R B; 3AFG A; 4NLD A; 4ITS A; 1AW0 A; 3Q2T C; 4Q0B A; 1IVZ A; 3J80 Y; 3CD7 A; 4JI1 J; 3JSN A; 3LNP A; 1F5A A; 4DR4 F; 3Q8Y A; 3LP1 A; 1ZPV A; 4WIQ A; 1RT4 A; 3AB2 B; 1SCU B; 4CRJ A; 1N32 J; 1V9O A; 3M8P B; 1XMO J; 4A3E G; 3R6L B; 1C2P A; 3TEH B; 2CPH A; 1F3F A; 1U26 A; 1XNQ H; 3ILO A; 4EWT A; 1F9E B; 3ZZR A; 3W5Y A; 4MK7 A; 4YJ2 F; 4LFA H; 5F9N A; 1KDN A; 1JLB B; 1OO0 B; 5J2M B; 4JI2 F; 4QWC A; 3ZTP A; 3NNH A; 3SV3 A; 2B5J A; 3K2Y A; 4LSL B; 3W9H A; 2DPI A; 4RB1 A; 3MMB A; 3DDK A; 2E5J A; 3DLK B; 3EPI A; 2LCW A; 2G13 A; 3CC4 S; 2P6S A; 2BVZ A; 4EI2 A; 2IA6 A; 2WDT A; 3UFX B; 4C60 A; 3MPV A; 2AHO B; 3VAG A; 4A47 A; 5MAO A; 3R8Y A; 1TKT A; 2V59 A; 3HHL D; 4KT0 C; 4J5O A; 1E4E A; 4YY3 J; 5E3P A; 4GO5 X; 2HFU A; 4CYU B; 1S79 A; 2CJB A; 4DQP A; 2Z4P A; 1PO9 A; 3CWJ A; 3QO9 A; 1L3S A; 4H4M A; 2VQF F; 4KJI A; 4I6A A; 3WNZ A; 1RK8 A; 3BRE A; 2YKP A; 1FO1 A; 3LOU A; 3CC7 S; 3PX0 A; 2N7C A; 4YHH J; 4N8O A; 2WW4 A; 2W2Q P; 2OOF A; 5LMN J; 4GVL A; 5F8N A; 4IYQ A; 2LXF A; 4LF8 F; 4NA1 A; 4QPT A; 3MR2 A; 4FF2 A; 3L3C A; 4ETS A; 1R9Y A; 4NL5 A; 2CPE A; 4LFC H; 3MHY A; 1GSH A; 3NG8 A; 2MKS A; 1MWY A; 3TUP A; 2R7Z G; 3K58 A; 5HK4 A; 1RQ0 A; 1EUC B; 4DV5 F; 3BPD A; 3ISN C; 4EEQ A; 1IQZ A; 3PN1 A; 2UZA A; 3IXQ A; 4A3G G; 4OX9 J; 1YRX A; 2R7V A; 3B54 A; 4KHP F; 2YX1 A; 2DGT A; 1X5O A; 5DDQ C; 2VFV A; 3ETJ A; 7FD1 A; 3KLF B; 5B0A A; 5BYQ A; 2AEF A; 4OHY A; 5LSO A; 3SBM A; 1S99 A; 4DZ6 A; 3FFI A; 1GMV A; 5GAG U; 5FLX W; 2BYC A; 4K5R A; 4B9M A; 2F8M A; 1U1Q A; 3CUL A; 4DV6 F; 1UEU A; 5CYM B; 1RYS A; 2PMH A; 4EZ9 A; 2FD2 A; 4U8Y A; 4RLE A; 4BY7 G; 4UFZ A; 3GJT B; 3KJV B; 4P9S A; 2J5A A; 4DF4 A; 2AYE A; 1H7W A; 1IWA A; 3J7A K; 2HBY B; 1FHT A; 3P45 B; 2I5J B; 1UFW A; 3M8P A; 1AA1 B; 2CQH A; 1JJH A; 1OWX A; 2HAF A; 2BQ3 A; 4NZ0 A; 4JI7 H; 3HM7 A; 3QZ8 A; 5LA6 F; 2V5A A; 2R85 A; 2FK2 A; 2A3J A; 5JQW A; 2OI2 A; 1JLB A; 5J2M A; 4M5K A; 1FWF C; 4DUZ J; 3V81 B; 5F3Z A; 5I3U A; 3IRX A; 1S6P B; 1SC3 B; 5E3A A; 1YBQ B; 1JDB B; 2C3P A; 3MPU B; 4ES3 A; 2V0N A; 2ZWO A; 2HND A; 3U1N A; 2NR0 A; 2RB7 A; 2Z2Z A; 2G4B A; 4X4V A; 4X64 J; 4MV9 A; 1REV B; 4WYW A; 3T1H J; 4XYL B; 2OPP A; 2R92 G; 3FK4 A; 3JB9 k; 4NMX A; 3BKU A; 3D68 A; 3J7Z T; 3MAA B; 1HWK A; 1TK8 A; 2CWX A; 3P4U B; 3TI0 A; 4KAI A; 3PYF A; 2DO0 A; 2ZZM A; 1SIZ A; 1PJ7 A; 4CYU A; 4YDD B; 2CG8 A; 3DN9 A; 4DV3 F; 3GGM A; 3ZZU A; 3NMR A; 4WXW A; 1KJ8 A; 3T5H A; 5FSE C; 1VQL S; 4MAK A; 3J81 U; 3ORR A; 3MCN A; 2CJK A; 3VGU A; 1NK6 A; 4NCA A; 1X9N A; 4ECT A; 5KFM A; 2LEP A; 3V6D B; 1QSL A; 2GO9 A; 2BV3 A; 1FWH C; 3BBC A; 2CPJ A; 5D0R A; 1LQ9 A; 2I6W A; 2BQR A; 3IP0 A; 1Q8K A; 3ZFV A; 2C42 A; 4Y2C A; 1FWA C; 1TWG A; 1FWB C; 2UXC F; 2LDY A; 1FJC A; 4MKY A; 2VG7 A; 4FYD A; 1VDH A; 2V9W A; 2B8Q A; 4DUY H; 2HYI B; 4MV4 A; 1P27 B; 1BQX A; 1VFJ A; 5A0Y B; 4AND A; 3PJX A; 1OPZ A; 4JI3 H; 2FPH X; 4HH1 A; 3DRS B; 5D0H A; 2HQC A; 3FBB A; 2FVC A; 1Q5V A; 1EJV C; 3FC9 A; 1NPK A; 3HT0 A; 5JVL A; 2A18 A; 1AFI A; 4LF1 A; 4ED2 A; 2JEI A; 1VIX A; 1QAI A; 2RQM A; 1WUU A; 4C6A A; 2MDA A; 5C25 A; 4XK8 C; 2C2E A; 5CYM A; 1UFL A; 1EKR A; 2Q1L A; 4LFA F; 1XNR H; 2Y8Y A; 2AMC B; 4PG4 A; 3KJV A; 3Q8S B; 1AN7 A; 4RW7 B; 3KXY T; 2ZHO A; 4LF9 F; 3OXL A; 2VJU A; 4QIG A; 2IYG A; 1V6H A; 4JYA J; 3AXM A; 4DSL A; 3AD7 C; 2PFF B; 2HJW A; 1L3U A; 2C1E B; 4FPI A; 3TJ8 A; 3F73 A; 5KFX A; 4X62 J; 4C9D A; 2OPQ A; 5IQQ A; 1YBQ A; 4RDW A; 5IT4 A; 4HR7 A; 2BJ9 A; 3IAM 9; 1NK9 A; 1UL3 A; 3M9O B; 3PKY A; 1KSP A; 4KKO B; 4NXM J; 1C0T A; 3RTX A; 5KFS A; 3LW5 C; 1TWC A; 2CJY B; 4CH1 A; 1KQG B; 4LJM A; 4RPF A; 5U47 A; 4IRD A; 3DMJ A; 1I94 H; 2DNQ A; 3DGP B; 2QX0 A; 3K0J A; 3NGR A; 4K4V A; 1YK9 A; 2DHH A; 4IQM A; 1X0P A; 2XO7 A; 3E2E A; 4EFA C; 4OZ3 A; 1RLD A; 2AEJ A; 2R7L A; 1Y7P A; 2VUM G; 2OLW A; 3EIC A; 3E5N A; 1PYT A; 3RBX A; 3M8Q B; 2RDI A; 3S8S A; 3OFH A; 2PA8 D; 3AY0 A; 2LYV A; 4EO6 A; 2CZ9 A; 4LFC F; 4X4U A; 1ZAW A; 4A3D G; 5EUH A; 4X66 H; 3NGD A; 3H2V E; 1N5S A; 2CZ4 A; 1HL6 A; 1PG5 B; 5AE2 A; 4K4X A; 4HEA 9; 3GIR A; 1Q8I A; 3H3V H; 2MMY A; 1E6V A; 2JA6 A; 5GAH U; 1VRQ C; 2E9T A; 2YWH A; 5KX5 F; 3MWD A; 1AXQ A; 3MUM P; 1RHK B; 3HHL B; 4I2Q B; 1QAJ A; 3QGH A; 2JSX A; 2O5D A; 2DGQ A; 1W7W A; 2QIF A; 4FKY A; 1EJR C; 5KFF A; 4A8Q A; 2G2O A; 1FX4 A; 5HAO A; 4U6P A; 3QGI A; 2JC0 A; 4WFY A; 4R25 A; 1Y3J A; 2DR5 A; 2FJD B; 2MZJ A; 1WOO A; 5BMV F; 1QUV A; 2CG4 A; 3EXC X; 3VWS A; 3HFV A; 3ETH A; 2M52 A; 2PAJ A; 2CNN B; 2E5I A; 3EMT A; 5E3Q A; 2J6T A; 1L5U A; 1WJ9 A; 4O3S A; 5E5N A; 3ZTS A; 5HRO B; 5KKN B; 3QZ7 A; 2XMV A; 2VJV A; 4G3I A; 2W01 A; 2FLL A; 1GAO A; 4RW7 A; 2HBQ B; 2ZDG A; 2BKO A; 4C5Y A; 3CF5 Q; 3R5D A; 4RW6 B; 1AB8 A; 4H54 A; 2CVI A; 2RJZ A; 4M1K A; 3U02 A; 4JI6 F; 2QLW A; 1WR2 A; 2GQ2 A; 2RF2 A; 1TTH B; 3DRX A; 1SFO A; 2RF4 A; 1Q86 T; 4YIW A; 3EXY A; 3DOM A; 2KXH A; 1FDD A; 2OH2 A; 3MUT P; 3UCZ P; 3G87 A; 4RAY A; 2DIT A; 2QG9 B; 3DOL B; 3C2P A; 3BR9 A; 2ADC A; 2QS8 A; 2E1C A; 2CJX B; 2V2Q A; 4URS A; 1KRA C; 3BDE A; 1IBK H; 1VMB A; 2R93 A; 4PRF A; 2B63 A; 2HGL A; 3JSR A; 2FZK B; 3C14 B; 1S3T C; 3EJM A; 1SJQ A; 2QE5 A; 3OTO J; 1H38 A; 1QUP A; 3WDO A; 3M2V A; 1LXN A; 5B0D A; 3MM9 B; 4YR2 A; 1WF1 A; 4HHH A; 3LGN A; 1VQ4 S; 3HSZ A; 4EGQ A; 4P7T A; 2NXU A; 2ZBQ A; 2HVS A; 2NU6 B; 3QNW B; 3LGH A; 2P6T A; 2FZC B; 2OPR B; 1FI4 A; 1HNW F; 1LK7 A; 1LFP A; 2ZH8 A; 2ADB A; 1NK5 A; 3OVS A; 4DUY F; 2IBO A; 2M2B A; 3GQC A; 1ZTW A; 1U0S A; 4K8R A; 3R7S B; 1KFS A; 2AP6 A; 3W5Z A; 1U02 A; 4ECX A; 1NHK L; 4JI3 F; 4ECW A; 3RB0 A; 2FJZ A; 4LUB A; 2CKW A; 5C3E G; 1UEK A; 4G6V B; 4NLO A; 4RB0 A; 4DR3 F; 2Z2L A; 3I4N G; 4LSA A; 3R5X A; 4I2Q A; 1M0W A; 4FF1 A; 3K3P A; 4WFX A; 2HLU A; 1FCA A; 2E2H A; 2VPW B; 3RUP A; 3VF0 B; 4XZ3 B; 1HI0 P; 2UUB J; 5A2Q Y; 1YIT S; 2BE7 D; 4K0K J; 1XNR F; 1YJU A; 2JOQ A; 1CQI B; 5LT5 A; 1IKW B; 2C3Y A; 2VPY B; 4CLU A; 1TFY A; 1A5N C; 1VLO A; 2LLZ A; 3D4J A; 2RUG A; 1TKD A; 2NXE A; 3LAM A; 4MFB A; 5LJ5 Y; 1Y0H A; 1KVK A; 4AXI A; 3IA0 A; 3PJW A; 1D09 B; 5ETQ A; 3DYA B; 3OR2 A; 4EEY A; 4OZ0 A; 3CJT A; 1YBT A; 2OEJ A; 2E5H A; 2QL5 B; 3AAW B; 4EFE A; 1SV5 A; 2X51 A; 4M5H A; 2C28 A; 1CSJ A; 4EU0 A; 4DR7 F; 3K5I A; 3VQS A; 3OLB A; 4PXD A; 4LRA A; 1S1X A; 1FNX H; 4LF2 A; 4DL0 C; 2GMH A; 4Y2K A; 1M1H A; 1YNY A; 1F5B A; 1GMW A; 4JV2 A; 1LWC B; 2C00 A; 1F9H A; 4MIB A; 1I94 F; 2B7C B; 2HPU A; 5CA5 A; 4B3S H; 3H25 A; 5J2Q B; 3BA8 A; 4PG5 A; 3V4Z A; 4QIW D; 3ZTQ A; 3RN6 A; 1A9N B; 3A3P B; 1KFD A; 1NSQ A; 2QT7 A; 1QTM A; 2HBR B; 4DSE A; 4B3M H; 4JI8 H; 5HMD A; 5M2R A; 4F67 A; 2B4U A; 4QW8 A; 4DQS A; 3WC0 A; 4Q7A A; 4FKX A; 2CFX A; 1FRJ A; 1T02 A; 4MT1 A; 2UXB F; 3UDV A; 3BSN A; 1L3T A; 1S0M A; 1FKP A; 2FHJ D; 1FKA F; 3HVT B; 1YVF A; 3SZ2 A; 2K3K A; 2NUA B; 2FQQ B; 2MQL A; 1IBM H; 3MM9 A; 4DF8 A; 1CJK B; 4QIE A; 2RU3 A; 3WC2 A; 1S6Q A; 4LF6 H; 2DLN A; 5HLF B; 3O2I A; 2JX2 A; 2FC9 A; 2MSS A; 2HGS A; 2YH0 A; 4X65 J; 3R5G A; 2IP4 A; 5IT0 A; 1K73 T; 4ZP6 A; 2B4V A; 4I2P A; 4OHX A; 2HHP A; 2IM1 A; 1XRT A; 4KPY A; 2JLF A; 2FLP A; 3E51 A; 1YTY A; 2R9L A; 4RW4 A; 2HFV A; 5G4H C; 3EX7 B; 4M5D B; 1OQ3 A; 5AXK A; 1OYD A; 2W57 A; 4I6W A; 1YGY A; 1L3V A; 3JSM B; 5L9X A; 3JAM U; 4WGX A; 2MKC A; 2NPF A; 4UY8 5; 5KT4 A; 1HW9 A; 2I8C A; 3MM5 B; 1RBO B; 5L39 A; 4AQY F; 3G9C A; 2ROG A; 3DGV A; 4RZR A; 6FD1 A; 3M6U A; 2XY5 A; 1KJQ A; 2RDJ A; 2VB6 A; 2YNF B; 1T95 A; 3GP9 A; 1Q7Y T; 4ZVU B; 3DF7 A; 3C6U B; 1M5K C; 4Z37 A; 3G96 A; 4FPR A; 3CJS A; 4F4Y A; 1U2C A; 1I7L A; 1WD6 A; 2V2V A; 3MI7 X; 2KHD A; 1UP1 A; 1S0V A; 3DYA A; 5EX3 A; 2DRB A; 4Q8E A; 4GKJ J; 2R7T A; 2F40 A; 2WBM A; 1LW2 A; 3CED A; 1H0H B; 3CD0 A; 3M30 A; 3FZB A; 3I96 A; 2QMU B; 4DR5 F; 1TKZ B; 1BWE A; 4CZC A; 1GSA A; 5KG3 A; 5BR8 H; 1VIO A; 2Z8Q A; 3NY0 A; 4NI2 B; 2XBP A; 4IG3 A; 2L41 A; 3OQ2 A; 2N3L A; 3OUY A; 3UN5 A; 2J7W A; 5J2Q A; 1JXL A; 2UUA J; 4B3T H; 3FEQ A; 2AV5 A; 4O4G A; 3RIZ A; 3VPB A; 2OEL A; 5BPF A; 1NDC A; 4OV6 A; 3DLL Q; 4DXL A; 2BA9 A; 1RWV B; 3LF0 A; 3BGO P; 2F65 A; 2E5L J; 2NU9 B; 3CVZ A; 4PAA A; 3W9I A; 2WAQ D; 3PRB A; 3HOU G; 1QJH A; 1RZM A; 1S97 A; 3WBZ A; 1RAA B; 4A8O A; 3A13 A; 3N6M A; 1S5Z A; 2VON A; 3MDF A; 2MTL A; 4MA0 A; 1BLU A; 4KQZ A; 2MZ1 A; 1EP4 B; 3EZU A; 1O6X A; 2QA4 G; 1RA0 A; 2DY9 A; 3MFH A; 4BWM A; 3RW7 A; 2VF3 A; 1TKX A; 4D3G A; 3FQL A; 1R8P A; 4HAC B; 1Q5Y A; 1JMT A; 2IVM A; 5MLC V; 3HO1 A; 4QWD A; 1U1N A; 1ZH5 A; 4PR6 A; 1RTI B; 2MLX A; 1HLW A; 4JI4 J; 2ALZ A; 4ZP9 A; 4MV6 A; 1VLY A; 3GTM A; 4HVZ A; 4IKA A; 3FKI A; 4DV4 F; 2W6N A; 4F25 A; 2W24 A; 2XS2 A; 4AYB A; 2MY7 A; 5IQ6 A; 3DC2 A; 1WC6 A; 1FRL A; 4FNH A; 3BYR A; 4WTO B; 2FYX A; 2CJ9 A; 5ETN A; 3RAQ A; 4PQA A; 3WX4 A; 1KBL A; 2WOM B; 3OL9 A; 1R31 A; 4UCS A; 1TP7 A; 4TUY F; 3GV8 B; 3VAH A; 2DR6 A; 2XYH B; 4DEZ A; 4DG1 A; 4U96 A; 2F4V F; 4YOE A; 3KK3 B; 3D6T B; 3QJP A; 2LMI A; 2MQQ A; 3T1Y J; 4CQD A; 2FKL A; 2XMA A; 4HDH A; 3FEY B; 1GFW B; 3V4I A; 1FNM A; 4CEU C; 5KG6 A; 4K4T A; 2YYA A; 5LXT F; 3LP0 B; 2DIV A; 2FJA B; 5CCL A; 2YU9 A; 5F8J A; 4X4P A; 4A46 A; 1HNX J; 5KT3 A; 3PO5 A; 4JV5 J; 2YZN A; 2P8I A; 2Y1L B; 3FGV A; 3VHQ A; 4CP5 A; 3IM9 A; 4ID5 B; 4X84 A; 3WZ0 A; 3F44 A; 4LSN B; 1SH2 A; 2WW8 A; 3MXH P; 2ZH6 A; 4JI1 H; 4PYA A; 3MUV P; 4ZI7 F; 4L6V 3; 3BEG B; 5A8R B; 5KT2 A; 3H42 A; 2CA9 A; 1Y10 A; 3LGM A; 1NK7 A; 1RAG B; 1RWO B; 1N32 H; 1RAF B; 3TZY A; 1XMO H; 2AJQ A; 4C4W A; 5HJY A; 4B3M F; 3BJY A; 4JI8 F; 3EVO A; 2HAI A; 5AE1 A; 3SQO P; 1TKZ A; 3BH7 B; 3SFU A; 1X9M A; 5LXY A; 2CQ3 A; 4PUO B; 2G1D A; 4NI2 A; 2VKZ G; 2FJB B; 4WP9 A; 3K5P A; 3I31 A; 3NWR A; 2O01 C; 3P96 A; 4YJ3 F; 2V68 A; 5LOQ A; 1M0T A; 1N6Q B; 1N34 F; 1IOV A; 3GIM A; 4DPO A; 2YWF A; 1I6J A; 1JKH B; 3MD1 A; 2IAJ B; 5HL7 Q; 3GPA A; 1UVL A; 1JNZ B; 2W2M P; 4C97 A; 1S6O A; 2Y0S E; 4GBD A; 2JNJ A; 4JUZ A; 3D3C J; 1BQN B; 2RVJ A; 1WOR A; 3L7O A; 3AXK A; 1U2R A; 2JJ4 D; 1A0O B; 4C48 A; 1NO8 A; 1EP4 A; 1WC5 A; 2PLM A; 1T11 A; 1S4E A; 4YY3 H; 1RSC A; 2R8G A; 4NLY A; 4UW2 A; 3TYT A; 1OS5 A; 4DR1 F; 4HAC A; 4LF5 J; 3MM6 B; 3KFW X; 1KON A; 2FK1 A; 1RTI A; 1PJ6 A; 3JS9 A; 3M7V A; 4ENO A; 1RWK B; 4O4R A; 8ATC B; 3PO4 A; 3RTV A; 2WVF A; 4CDI A; 2A10 A; 1DQ9 A; 4YHH H; 3H5Y A; 2CQ4 A; 4GY7 A; 3MMJ A; 3GIQ A; 1U8S A; 1UVM A; 1WOP A; 3UO0 A; 1DV1 A; 3HFZ B; 1GXU A; 3TUZ C; 1FNO A; 2GOK A; 3KK3 A; 4FVM A; 4J9U E; 3N6X A; 1Q53 A; 2DD8 S; 5L1L A; 3PX6 A; 3GCW P; 4MV7 A; 1V3R A; 3QIP B; 1B3T A; 2RGV A; 4JI5 J; 4WXO A; 2LP4 A; 2D41 A; 5DG8 A; 1N56 A; 1NK4 A; 1NME B; 3D0I E; 1RAI B; 2MJN A; 2JA6 G; 4G1Q B; 4DV2 H; 4PYD A; 4EPE C; 4ID5 A; 2CQ1 A; 3QT7 A; 4OX8 A; 5DDR C; 3IGH X; 3SSQ A; 2YL2 A; 1UC8 A; 3JRX A; 3BN4 A; 1KRB C; 3BO2 A; 2E1M C; 4QJF A; 2H65 B; 3HMJ G; 3G71 S; 5ETT A; 3DM2 B; 3H90 A; 1H98 A; 5K14 A; 4EK2 A; 3ENV A; 2NLW A; 4RU9 A; 1FVS A; 4MMO A; 3VGV A; 4QJV A; 5LJ3 Y; 3JSL A; 4ZP7 A; 3K5N A; 5IC4 B; 3PTW A; 3PV8 A; 3DOK B; 4EBD A; 1X8D A; 1Q8B A; 3TVY A; 3LML A; 3QPI A; 4Z42 C; 2UZ9 A; 2M9K A; 2P0A A; 1T03 B; 2PMZ A; 2YQ3 A; 2VIH A; 1A7G E; 3PW5 A; 4PUO A; 3R7B B; 2RAQ A; 2GZO A; 3GYN A; 4DSK A; 3DDI A; 5C24 B; 4DUZ H; 1RLC L; 2M4M A; 5IWA J; 3B6B A; 1W25 A; 4DL5 A; 2CIM A; 4BC9 A; 2HHS A; 1FT8 A; 2HVQ A; 2IAJ A; 1ZAX A; 1KLM B; 3H4B A; 1M6V A; 1VBH A; 1WC0 A; 3V7P A; 4ANC A; 3ZZZ A; 2ZC4 A; 5FDL A; 4TY8 B; 2J9D A; 4UQT A; 4LF4 J; 1DGS A; 1BQN A; 3PFE A; 5D4O A; 1VCT A; 3ZO7 A; 5HP1 B; 3S1T A; 1WF0 A; 1A5M C; 4M95 A; 2IMW P; 2UU9 H; 1HBN A; 2F3J A; 5HMZ A; 2EI5 A; 2V5H G; 2R3V A; 3ET6 A; 2WHO A; 1K1S A; 2LEB A; 4UCU A; 3KMQ A; 1RWP B; 3BGL A; 4I56 A; 1RA5 A; 2J9E A; 4M63 A; 2KT5 A; 4M5N A; 2CNL B; 2AF6 A; 4NLW A; 3G77 A; 3QMQ A; 5CAA A; 1U48 A; 3KHR A; 1R8A A; 5AJ3 F; 3MMA A; 2RU9 A; 2VFT A; 3CDB A; 4B3R J; 2W1B A; 1HNZ J; 3ZZY A; 4IH6 A; 4PQU B; 5C2G A; 4BLG A; 3NS6 A; 1AUV A; 5FSD C; 2GNK A; 2OUG A; 4KGV B; 2R93 G; 3HBL A; 2B63 G; 2WVB A; 2J9C A; 1KWM A; 1YHQ S; 1WTB A; 2RGR A; 4DU7 A; 3D6F B; 1T05 B; 1K0V A; 5B0E A; 1AUX A; 2MQN A; 4PKD B; 1I3Q A; 4DSF A; 2DXD A; 1JXE A; 1RT2 B; 4XDC A; 3HSJ A; 1P1M A; 2EC0 A; 3E01 B; 1XNQ J; 3QIP A; 2IM0 A; 5G4D A; 5J1P A; 3M1V B; 4IFV B; 4JI1 F; 1DLO B; 3TOQ A; 3O7U A; 3M2R B; 1KHV A; 1X4D A; 1W2B R; 4LIW A; 4O0I A; 2VPZ B; 1PJ5 A; 1N32 F; 1JLA A; 1S77 D; 4PWU A; 3DNC A; 4ETZ A; 3DHW C; 4Z7C A; 1EET B; 3OV7 A; 1RB0 A; 2MHE A; 4M8L A; 5AXL A; 2MZQ A; 3EUN A; 2JZ5 A; 4FF3 A; 4C8K A; 4IHJ F; 3C6T B; 3PHE A; 3MWB A; 4YB1 P; 1U6F A; 1GX6 A; 2KG0 A; 1QVG R; 4UF4 A; 3TQ1 A; 3NS5 A; 3BO3 A; 2M0G B; 1FES A; 1PX2 A; 5C44 G; 1Y5N B; 1RTD B; 2IYI A; 3Q23 A; 2VFR A; 4RU2 A; 1J6P A; 1GK8 A; 1SUQ B; 5HN0 A; 5C24 A; 4X62 H; 4CQC A; 1G3O A; 3Q2S C; 3BS9 A; 2HHH J; 2Q6B A; 1LFW A; 3ILL A; 2JDJ A; 2FTY A; 4RNN A; 1V3Z A; 3CJR A; 2JLG A; 4FYW B; 3MJF A; 2A2C A; 1XK8 A; 3CCW A; 1DAX A; 2R7K A; 3LPE A; 4TY8 A; 4MAM A; 2ZH7 A; 2HQD A; 3K17 A; 1DTT B; 2PDA A; 4I7F B; 4NYO A; 2ZH2 A; 4K4U A; 5CYJ A; 2Z2M A; 1HHQ A; 3J7Y I; 1X5S A; 2OTJ S; 1U1P A; 1S4F A; 2FP4 B; 4YHH F; 4FYV B; 2ZNY A; 5AXM A; 5LMN F; 3R5A A; 2N82 B; 5T41 A; 3G6V A; 4CO3 A; 5J2P B; 3VPC A; 1BD6 A; 4WPA A; 4ES1 A; 3POT A; 5IP9 G; 2IMR A; 3BKT A; 1YX2 A; 2GJI A; 4NOH A; 4OJH A; 3IGN A; 3CCE S; 1Q8L A; 4YAK B; 1U1L A; 1IBL F; 4ZVP B; 4CQ1 A; 1ZAV A; 4DFM A; 3NNC A; 3R5C A; 4OX9 F; 4ZPD A; 3BQ1 A; 3KMS A; 2BKP A; 1RHQ B; 3FRY A; 1RI7 A; 5E3C A; 2HWI A; 2W8L A; 4R0J A; 1T05 A; 4DLK A; 1Y1W A; 4F0M A; 3MAQ A; 3BO4 A; 4DR6 J; 1SH0 A; 3HHK A; 5F9L A; 1CJT B; 1RT2 A; 1N0U A; 5GAH I; 1JQG A; 2KKH A; 1LV5 A; 4WYL A; 3TVK A; 4JI7 J; 4I1T A; 2GW8 A; 3DUG A; 1DLO A; 5DO2 A; 3CGI A; 3CVK A; 1U1K A; 4IG0 A; 1SQG A; 2CZV C; 3MED A; 1TWA A; 1T9X A; 1RAJ A; 1GMU A; 3Q2O A; 3F41 A; 1WWH A; 3NMA A; 2KRR A; 1A5O C; 1J5O B; 2OFG X; 4ZDN A; 4NYZ A; 4IWY A; 1NKB A; 4K4W A; 1J5E H; 4OJ1 A; 3N9U C; 4UCV A; 1X0F A; 4BWS C; 2MKH A; 4QWE A; 4RUH A; 4RB3 C; 2D74 B; 3OR1 B; 3AYH B; 3HPA A; 1N36 J; 4AL7 A; 3HVA A; 1SC6 A; 2CJA A; 4LH8 A; 4WFZ A; 4NLS A; 1WHX A; 2W9B A; 2QA1 A; 1RTZ A; 2NVX A; 4X64 F; 3MR7 A; 2ZVI A; 4IOC Q; 1OJ4 A; 4LF7 H; 2CQ2 A; 2IMO A; 3KKF A; 2HGN A; 3QWU A; 3T1H F; 2V8G A; 4YB7 A; 1JLL B; 4F50 A; 5FJ4 A; 1CUL B; 5KT6 A; 4EZ6 A; 1RT3 B; 3PXG a; 2KZZ A; 2VDH A; 3QFH A; 3KAJ A; 4TLQ A; 2NZ4 A; 4CTA A; 3R23 A; 5K8M A; 2F63 A; 1MW7 A; 4CWB A; 4IRC A; 1DTT A; 2J0S D; 4K4Y A; 2P53 A; 4NXN J; 3H40 A; 1N5Y A; 3OTO H; 4EYI B; 5AKQ A; 2Z58 B; 5I3P A; 1XC9 A; 2ZIT A; 1FRK A; 4X4S A; 3IR5 B; 4CC5 A; 1U0H A; 3ELH A; 4O3N A; 3L95 X; 4ZQI A; 2XIG A; 2FIU A; 3D6H B; 3PEH A; 1UW9 A; 3HKZ D; 4Y0F A; 4NLU A; 4WP3 A; 3QAE A; 1UTA A; 1O51 A; 3W60 A; 1M5S A; 3UW1 A; 2HIQ A; 3JQM A; 3O1L A; 1X4G A; 4L76 A; 2BMB A; 3IO1 A; 2BXJ A; 1PAE X; 2F7V A; 1RJQ A; 2ZFH A; 3DLJ A; 1I3O B; 3C16 B; 4M5G A; 1RT6 B; 4UTG A; 4NLV A; 3HSG A; 2K7J A; 1U09 A; 1WKL A; 1D9D A; 2UUB H; 1EJX C; 3MEG A; 3NBP B; 5DM6 Q; 1RCO B; 5FPO A; 3UBP C; 2CNO B; 3TII A; 2ZM6 J; 3GYG A; 2SXL A; 3HLU A; 4MV1 A; 2CZG A; 1R8B A; 2DNH A; 3FKI G; 1Y5I B; 1N33 H; 4I4B A; 3LP8 A; 5CH7 B; 2NSB A; 1NBE B; 4JYA F; 2VQD A; 3GTJ A; 1S1U B; 4CNY A; 4QR1 A; 3T9Z A; 3KTM A; 2C2D A; 4IFY B; 2J7K A; 4R7R A; 1K83 A; 3FDS A; 3RRG A; 3VPD A; 5FD6 A; 2XSF A; 1R7I A; 3HVR A; 2JL9 A; 4I64 A; 4L3N A; 4OZ2 A; 4NXM F; 1HBM B; 1R8C A; 4NWB A; 4PXB A; 2V4J A; 2B9Y A; 1R0B G; 4MIW A; 2K1R A; 4M7E A; 1XTZ A; 3OJU A; 1FXR A; 1UWB B; 3DRR A; 3VVU A; 2VHG A; 3PYG A; 4X66 J; 3O44 A; 1RJR A; 1DTQ B; 5L1I A; 1CUL A; 4IOB A; 2MKI A; 1LK5 A; 2LVW A; 2ZHA A; 1FJG J; 2G0I A; 3LWM A; 1C1B B; 2MZT A; 1FFS B; 3FKB A; 4UOF A; 3RUB L; 3MMC B; 4UUX A; 2QSF A; 4BKN A; 2AYM A; 1T0T V; 3W9J A; 1WEX A; 3CD6 S; 2B43 A; 2W25 A; 2E5G A; 3C3L A; 2RS8 A; 2MZR A; 1BY9 A; 1I50 A; 3R1H A; 1S12 A; 4MK8 A; 4N76 A; 2V67 A; 2PN6 A; 3I0S A; 3ENW A; 2VU5 A; 4RO0 A; 3OTB A; 1HPZ B; 2Z56 B; 5CZX A; 3UVJ B; 2EXU A; 1UA1 A; 1H2U X; 4X65 H; 2D9P A; 2GAG C; 4KFB B; 3NGK A; 1A6L A; 2X1G A; 4RJV A; 2VBX A; 3BQ0 A; 4RNO A; 3R3T A; 4Z7Y A; 5CAE B; 3KDO A; 2KN4 A; 4LMY A; 2R42 A; 4OZ5 A; 3RFC A; 4U9R A; 2CPI A; 3G6E S; 4F36 A; 4EG0 A; 1R27 B; 2W6O A; 2IDE A; 5H1S V; 2XHW A; 3PW7 A; 5GAD I; 3PDN A; 1ZM4 A; 2Z0G A; 2I80 A; 3GFY A; 2DR9 A; 5KFV A; 1N54 B; 5A6W A; 2YXL A; 5C7P A; 4HT5 A; 2CYY A; 3NBP A; 3HHN B; 4YAJ B; 1T94 A; 2ABY A; 4F0S A; 4WWS A; 3MM7 B; 1P8L A; 3G82 A; 1DFD A; 4RUC A; 1IKX B; 4LVO P; 1RT1 B; 2V9K A; 1F60 B; 3QT5 A; 1EAY C; 2KSW A; 4ZLN A; 3MPD A; 1S1U A; 2B9E A; 4KR0 B; 3TX0 A; 4NYP A; 1VOM A; 3VVT A; 3F8N A; 3OT9 A; 3OXG A; 3FSI A; 1IBK J; 1B6R A; 4DDF A; 3EYY A; 1RHM B; 5JH7 F; 1S48 A; 2GQC A; 4GKJ H; 4EMD A; 3D67 A; 1SJF A; 2UUC J; 3MGJ A; 2PMZ E; 3LO3 A; 1RT7 B; 1QD1 A; 4Y8V B; 3E8O A; 1HBM A; 5CAB A; 1VHF A; 2DHG A; 3HZE A; 4UB9 A; 1XMQ H; 4XTT A; 3C15 B; 1LWF B; 4QZV B; 1VBG A; 1YBU A; 1HQE B; 4G65 A; 1TVR B; 2O1P A; 3V6H A; 2B6A B; 4EO8 A; 4I7H A; 3OTO F; 1GEH A; 4OYM A; 2AQV A; 2CWK A; 4LR9 A; 1YIJ S; 2DH8 A; 3QBC A; 4IO9 Q; 2J9D E; 4BWJ A; 1DHM A; 3LP2 B; 2E5L H; 1HBO A; 3MMC A; 5C2C A; 3ORQ A; 3TZX A; 2AJ2 A; 5DM7 Q; 3HP6 A; 3MDW A; 3IR6 B; 2VQE F; 2M70 A; 2GVZ B; 4ED8 A; 1Z4U A; 1F2H A; 1DUR A; 1RUS A; 4JV0 A; 5ITH A; 1SXL A; 1HPZ A; 1S9E B; 2KXN B; 5L38 A; 2XS5 A; 4AL5 A; 3TQT A; 1T3O A; 4Z2X A; 3PAC A; 3UVJ A; 1IM4 A; 2JA7 A; 2I1R A; 5F8L A; 1TR0 A; 2GO8 A; 1ZTT A; 4JI4 H; 3NCQ A; 2I9U A; 3JTP A; 1SL1 A; 2DRD A; 1A5K C; 3A3N B; 5B7W A; 3THV A; 4UOC A; 1LW0 B; 2UUB F; 5CHC B; 2DCU B; 4K0K F; 1NU4 A; 4ID8 A; 4WBF A; 4GC6 A; 4J9V A; 2VG6 B; 1WHY A; 1BXR A; 2FZG B; 3M2U B; 1B6S A; 4XAK A; 1HWI A; 1S9G B; 3T1Y H; 2XCP A; 2E29 A; 4EP8 C; 2VQF J; 1S72 S; 2OLS A; 5IFM A; 1QE1 B; 4B3Z A; 3AJD A; 1AT1 B; 4OYI A; 1SI9 A; 3ETO A; 5KAK A; 2DNP A; 2XC9 A; 2YKN A; 2KZM A; 3MM7 A; 2IJN A; 4OI0 A; 2BOA A; 2W6Z A; 3RR7 A; 3R5B A; 3ZZ0 A; 5HBM A; 2KO1 A; 1W96 A; 1IKX A; 1HNX H; 2CAD A; 1WNE A; 4JV5 H; 1RAC B; 2BUN A; 2MLZ A; 3HOV G; 1XMB A; 3AOA A; 4BC7 A; 3RAX A; 3TUJ C; 4ELU A; 2BJ1 A; 3LAK A; 1NSK L; 5HQ8 A; 2KFY A; 1PIE A; 4DV5 J; 4EPD C; 2GLT A; 1WSR A; 1WKJ A; 3OL6 A; 3MEC B; 4KHP J; 1JLQ B; 2XS7 A; 4CNZ A; 2E1P A; 1DZ5 A; 4PPD A; 2AQO A; 1RXO B; 2MST A; 3SFW A; 2RKI A; 1HBU B; 4KPC A; 5C1O A; 1RT7 A; 3QGA C; 2R7Q A; 1GH8 A; 2E7X A; 2BAN B; 2RNE A; 2XMK A; 1IBM J; 2GC8 A; 3OHA A; 4KH0 B; 4GRS A; 4C8Y A; 1CEZ A; 5AT1 B; 1UJ6 A; 5HV6 A; 5IXU A; 4LF6 J; 3S01 A; 1HHS A; 3MR5 A; 2B4P A; 3HSD A; 2HSE B; 2HIY A; 1HQE A; 2QTW A; 1TAE A; 1TVR A; 5KFO A; 3LTO A; 5KFJ A; 1C1C B; 4HQ6 A; 5D77 A; 3MM8 A; 1AZS B; 3GFH A; 3TX8 A; 1S1V A; 4LSZ B; 5BYR A; 3HOX G; 3CSK A; 4J9R A; 3KLI B; 3RBZ A; 3OFE A; 2IRX A; 2H4W B; 3LP2 A; 1HWU A; 2JHE A; 3M05 A; 3DLH A; 1T36 A; 2B7B B; 4BHQ A; 2BJ7 A; 4N5S A; 3QRP A; 2KYZ A; 1FFW B; 4CLF A; 5KFD A; 2AKW B; 4DCO B; 4CLY A; 4X65 F; 3RR8 A; 4O4I F; 3V5R A; 1S59 A; 3RAM A; 3OUZ A; 2HNZ A; 4CO0 A; 1CJV B; 2QMX A; 2GVZ A; 4DV7 H; 5I47 A; 1UCN A; 3ZTO A; 1QDU B; 3GID A; 3N44 B; 4A2B A; 2ZE2 B; 3TAN A; 1MU2 A; 3ITH A; 4PEO A; 3W7B A; 1VQM S; 3Q89 A; 2FVM A; 3J7Z 5; 4LF5 H; 2DXF A; 5DET A; 2MXY A; 4ER8 A; 2XLK A; 5D4P A; 4K2X A; 3KLG A; 3KG1 A; 3CIM A; 3WIU A; 1TU0 B; 4RWX A; 3SDE B; 3TP2 A; 4L73 A; 1RAO A; 4FM9 A; 5LSL A; 5A6T C; 2OPS B; 3CCT A; 4FU0 A; 3OSN A; 1P1T A; 4Q0B B; 4OZN A; 1REG X; 3ULH A; 5DQH A; 2VOB A; 4O44 B; 5IOL A; 1EF2 A; 4CNT A; 2VG6 A; 1GKQ A; 4O3P A; 1GT8 A; 1YJW S; 1S0N A; 4W98 A; 4UY8 T; 2YNG A; 3LP1 B; 5LL6 d; 1RT4 B; 2RDD A; 1S9G A; 2DIU A; 2LQB A; 3V6K A; 3T6B A; 4DV1 H; 3GR1 A; 4KO0 B; 3CE8 A; 1G7C B; 2NRE A; 3SQG A; 5GME A; 1QE1 A; 2RQC A; 2Y0S D; 2WCX A; 4MYU A; 2MKK A; 5BR8 J; 1ONX A; 3NRB A; 2HS4 A; 1JJ4 A; 3K7R A; 5B08 A; 4JI5 H; 4RBV A; 1GNK A; 3LSC A; 2HRT A; 1EJT C; 2VM8 A; 3SI8 A; 4GKJ F; 3NCR A; 1Z1Z A; 4A4J A; 3BSA A; 4ERD A; 2B5J B; 5DG9 A; 4QIW A; 4ILM A; 5ETL A; 2OWO A; 4BBY A; 3MMB B; 5JMN A; 3NAI A; 4V1Y A; 1JK9 B; 3RRK A; 1QAY A; 1HFE L; 4OYW A; 3GTP A; 4DLG A; 1ZA2 B; 3AJ1 A; 3HXM A; 1OYE A; 5A8K A; 4Y6I A; 2HBZ B; 4KIR A; 1Y1W G; 2DNK A; 4N77 A; 3IBF B; 4CCH A; 2O03 A; 2CQB A; 2UUA F; 3N40 P; 4IOA Q; 3FEZ A; 3MCM A; 1TKT B; 5KFW A; 2ZEJ A; 1FKO A; 2BAN A; 4DFP A; 1D8Z A; 4ED0 A; 2E5L F; 2EFQ A; 4TVA B; 1F1B B; 3HFK A; 4H9M A; 3M9N B; 1CS0 A; 2P9G A; 3JZI A; 2NUH A; 5IWA H; 5D1O A; 5AXN A; 3VAL A; 2YWG A; 2AYB A; 1PK8 A; 1L2Z A; 3KLI A; 4EPB C; 2NVY A; 1S7H A; 4ZVG A; 1CJU A; 2ASY A; 5IV4 A; 3G8C A; 1IKY B; 2NXN A; 5HAT A; 3EVM A; 1VYA A; 5HMY A; 4LH7 A; 4MK9 A; 3DU6 A; 2YQ0 A; 2D7S A; 3P49 B; 2HFZ A; 3UDE A; 5LMU H; 1IN0 A; 2XMM A; 1OSD A; 2W9C A; 3TJ9 A; 4C3K A; 4HTJ A; 3VGT A; 2E2I A; 3UDL A; 1T7P A; 2YNH A; 2J8S A; 1LKZ A; 3T1Y F; 1BE4 A; 4JP8 A; 3FFI B; 3H8Y A; 2VPQ A; 2XWY A; 5B0G A; 4V19 X; 2AW0 A; 2R7O A; 3T6J A; 2R2R A; 2CPF A; 1OMS A; 4O44 A; 5A6P A; 2CQC A; 3GV7 B; 1B7F A; 2CQG A; 4F0R A; 1S57 A; 2YBV A; 4Q5V A; 4JV5 F; 2FDN A; 2YCH A; 3LH2 S; 4KO0 A; 2UBP C; 2QLJ B; 2I2Y A; 1RE1 B; 4DI6 A; 2D1C A; 2GA7 A; 1KJI A; 3Q24 A; 1KC8 T; 1FWC C; 2AGO A; 2FY1 A; 3KAK A; 1NHV A; 3N6L A; 3FQK A; 4EGJ A; 3BM7 A; 5I3U B; 3IRX B; 3NXZ A; 4DZH A; 4FBU A; 1DY3 A; 2BDP A; 2NU7 B; 2HVH A; 3PW2 A; 5DLF A; 2HND B; 1FWJ C; 3GON A; 3M8W A; 1TUG B; 3QT6 A; 3BF4 A; 1HIY A; 1EQM A; 2XCA A; 3MEK A; 3U9T A; 4R8U A; 5ETK A; 3G6X A; 3L7U A; 1OIA A; 2W2P P; 2VH7 A; 2HVI A; 1FWG C; 2OPP B; 3AD8 C; 3CD5 A; 4YP3 A; 4MTP A; 4CLL A; 2KRB A; 3NQB A; 4MTD A; 3MOZ A; 4NRT A; 2K2P A; 1NPR A; 2JEG A; 3UNY A; 4DX6 A; 2XM3 A; 2ZDH A; 2GCF A; 2ZH5 A; 5F8I A; 3J80 W; 3K3V A; 2ZJQ Q; 2FJV A; 2VL1 A; 5I4M A; 1D8Y A; 3V4M A; 4CNS A; 4APM A; 1EHW A; 1PIL A; 4UQG A; 2SCU B; 4P6Q A; 2CUY A; 3FCV A; 2HHH H; 2X1F A; 5G0R A; 4FS2 A; 1ROF A; 1MLA A; 4W4M A; 3O5T B; 5J2T F; 4HHJ A; 2ZDP A; 1ZZN A; 3J81 Y; 3RG6 A; 1TWH A; 5EDW A; 4ZCM A; 1SC1 B; 1IKY A; 3RJ0 A; 2M88 A; 4NCB A; 2KAU C; 3UE2 A; 1BNC A; 1GKP A; 4JI6 J; 5KG0 A; 2GPS A; 2HNY B; 5I3Q A; 4ZPC A; 5HVG A; 2ZJP Q; 2VG7 B; 3O6Q A; 2YS7 A; 3TJA A; 2V4R A; 3KYL A; 3MOJ B; 2XO6 A; 5H9F K; 1NKC A; 1TFW A; 3BPS P; 1FRH A; 2CRL A; 3W2W A; 1LWE A; 2F9J A; 4ZJL A; 5C5J A; 1WOS A; 2V63 A; 3US5 A; 1GMW D; 2LQ9 A; 4ZCK A; 1EQ0 A; 2GWN A; 4XPC A; 1C0U A; 4ELT A; 3H20 A; 3I82 A; 2IPO B; 4DOZ A; 1V9P A; 4O3Q A; 4DV2 J; 1EJS C; 3MCS A; 2Z2Y B; 3T19 A; 3GCX P; 5C25 B; 1MWZ A; 3GJR B; 1W2I A; 4F4Z A; 3LQV A; 2QRR A; 4HH0 A; 3W66 A; 1ELO A; 4ED1 A; 1ZHS A; 1B4S A; 3H5U A; 3PJ9 A; 3RGX A; 4DR6 H; 4WU3 A; 4E98 A; 3X3U A; 4CO2 A; 4FBT A; 1Z2O X; 1SJ4 P; 4A8X A; 2COL A; 2EEY A; 3HHL C; 4TY9 A; 2VDI A; 1HNW J; 1U1O A; 2QZ8 A; 6AT1 B; 3MDU A; 2O66 A; 1URN A; 3UD3 P; 1UZH A; 3BN7 A; 2EFP A; 4WJB A; 4IWX A; 2M8H A; 1JLC A; 2ASJ A; 1MSW D; 4ZKA A; 2JFK A; 4RW9 A; 4DR3 J; 2OPQ B; 3CDW A; 4ZOS A; 4W92 B; 2XCL A; 2F5G A; 1KJJ A; 1FF2 A; 5KFC A; 2A6O A; 1E9Z B; 4C8M A; 3FNN A; 3GSI A; 1C0T B; 1NRK A; 2CNK B; 1C4C A; 4LRE A; 2WON B; 1ZQ7 A; 3DMJ B; 5I42 A; 2KM8 B; 5F8M A; 1G6B A; 3RV4 A; 2M05 A; 2CDQ A; 2FVS A; 4DWI A; 4ECR A; 1M5O C; 4A8M P; 3OTD A; 2PBZ A; 1N36 H; 2UU9 J; 4GVB B; 1ZHQ A; 4DV0 J; 1JJC B; 4TYA A; 4NE9 C; 1W93 A; 1HQU A; 3GTO A; 4LF4 F; 1VK3 A; 4OZL A; 2HEK A; 1DRZ A; 4B9V A; 3PRV A; 1KD1 T; 4YEA A; 2DNL A; 5DQG A; 3CUN A; 3CME S; 2KXF A; 2FVQ A; 1HMV A; 1E6V B; 3KK1 B; 3I87 A; 4C0O C; 4OFZ A; 1NUE A; 1NB7 A; 2W4C A; 3M8S A; 4B3R F; 4NXN H; 1HNZ F; 3KK2 A; 1ZZF A; 2HNY A; 5LXR A; 2IA0 A; 1AUD A; 3MWM A; 2WZ1 A; 2W71 A; 4UT4 A; 2GVD B; 3CJ4 A; 5EOY A; 3PR4 A; 1FWK A; 2ZHB A; 5K9F A; 1IJF B; 1E6Y B; 2XEX A; 3CB4 A; 3TR5 A; 2FY8 A; 1K1D A; 4UUW A; 4OHW A; 1TL1 A; 2HK2 A; 2G2Y A; 4BBS G; 3TW0 A; 1JDE A; 3I32 A; 7AT1 B; 4Y00 A; 3HHE A; 3EDR B; 5CCM A; 4H4O B; 3SSS A; 1HA1 A; 1XNQ F; 3EGV A; 4WFA Q; 4RB2 C; 2QYZ A; 3DLG A; 3MF5 A; 3TUI C; 4DX8 H; 5KFE A; 5CA1 F; 4CO1 A; 1M90 T; 2P9E A; 3ENA A; 3JSB A; 2HZV A; 2OIH A; 4YR3 A; 5IC6 B; 3JQJ A; 2PE8 A; 3U1L A; 4ZPA A; 1FD2 A; 4TQS A; 3HUL A; 4HTI A; 4O2B F; 2RF2 B; 2OKQ A; 2JA7 G; 2ZM6 H; 5HAN A; 4URG A; 1CPZ A; 5J2U F; 2EFN A; 1FDA A; 3DOM B; 4GO7 X; 3V6J A; 1RT5 A; 3MEE A; 1QTN B; 3J7Y U; 2FJW A; 5KFI A; 1YJV A; 3X1L B; 4OX7 A; 4X4R A; 2PEB A; 4FS1 A; 1RA7 A; 2YWE A; 4AYB D; 3UWT A; 2WON A; 2RHS B; 3AA8 A; 1H73 A; 1KP6 A; 1WEY A; 2YX7 A; 1D1U A; 3EGW B; 4QIF A; 3HX9 A; 4UCR A; 4BS2 A; 2Y1I A; 4GVB A; 3M2V B; 1PO6 A; 4DHV A; 2R2U A; 2DNY A; 6FDR A; 2R82 A; 5DRK A; 4OJG A; 2DGX A; 1ZYQ A; 4LVN P; 3IG1 B; 1LWX A; 5KFB A; 2C2N A; 4U8V A; 1N88 A; 3WO0 A; 4O2A F; 2O3D A; 2IJR A; 4OI6 A; 2V50 A; 4J9Q A; 3ZXW A; 1I5O B; 4X4T A; 3DI6 A; 3VCD A; 2YNI B; 4YN3 B; 2VG5 A; 1C4A A; 1FJG H; 2OSQ A; 1RJP A; 4P72 A; 3TZZ A; 4ZPW R; 4RW8 A; 2FJE B; 3PEJ A; 3BBB A; 2KHC A; 3KK1 A; 4OI1 A; 3GZ7 A; 2A6M A; 1R9T A; 3PG9 A; 2ROP A; 1VKZ A; 1YWX A; 2HVD A; 3TIN A; 3D66 A; 2W6P A; 2MYF A; 3K5M A; 4DSI A; 2H9Z A; 1VHI A; 1EHI A; 4LT6 A; 1TL3 A; 1PGZ A; 2QLF B; 2F8E X; 2GVD A; 1EB0 A; 4CM0 A; 1E6Y A; 2C2K B; 3DXB A; 3CO9 A; 2GGP A; 1VQO S; 3T8L A; 4I68 A; 4DR6 F; 5BXI A; 4Q45 A; 5DZ6 A; 4V19 J; 4YB6 A; 3LAL A; 4H4O A; 2G3F A; 1JB0 C; 2YZG A; 5DLG A; 1VRT A; 3QT8 A; 4JI7 F; 3MR3 A; 2KI2 A; 4RUB A; 3JRW A; 3NAH A; 3PYD A; 3AJ2 A; 4MFB B; 1J5E J; 3CCZ A; 3RBE A; 4PG8 A; 3OR2 B; 1E4G T; 3T5K A; 2ZH4 A; 3FBE A; 2V2K A; 3R5H A; 1V4Y A; 3FEX B; 4KHR A; 1SV5 B; 2ALY B; 3MD3 A; 3RBD A; 3I5C A; 1XN9 A; 3HKZ E; 1JLG B; 2PEH A; 3BAB A; 3Q8V A; 3C19 A; 1K70 A; 1S1X B; 3AOB A; 5EGY A; 2YRW A; 1CLF A; 2HZA A; 4QPX A; 2JVO A; 4FP9 A; 4LF7 J; 2QEX S; 4C5A A; 2QGF B; 1NDP A; 2W8K A; 2Q2T A; 1VQ8 S; 3SV4 A; 3MPY A; 1NAQ A; 1N36 F; 1FRM A; 3RB4 A; 1A5L C; 3JTO A; 5D3G A; 1HR0 H; 2NXJ A; 1WEL A; 3RF2 A; 4NPO A; 3KHL A; 2G9O A; 5D8D A; 2DR7 A; 1UAW A; 3PW0 A; 5F8G A; 2L9W A; 4JI0 J; 1RWU A; 3IMQ J; 3W61 A; 4C7Q A; 3VR1 A; 2OGU A; 2DGO A; 1FKP B; 3I5A A; 4HNV A; 4RNH A; 2UUC H; 2GIF A; 1EZZ B; 5KFN A; 3PVX A; 1NHU A; 2XMU A; 2ACY A; 2I8E A; 3IG1 A; 3D1H A; 1S6Q B; 1YRT A; 1VQN S; 1O12 A; 2F1F A; 2B3T B; 2ZBR A; 2NCK L; 4CM2 A; 2RRN A; 4MA5 A; 5GVQ A; 2YNI A; 4I2P B; 4USW A; 4AUK A; 3EDQ B; 4DR4 H; 4QCL A; 2YKA A; 2IC3 A; 3SCI E; 1TZ0 A; 1B0T A; 1MNE A; 4RW4 B; 2EFV A; 1KKH A; 3E74 A; 2R7X A; 3AOD A; 3GSD A; 4DR2 J; 1RHR B; 1JX4 A; 2YKM A; 2JEF A; 2GML A; 3JXO A; 4XIU A; 1N5T A; 3LSB A; 1VI7 A; 3R1L A; 4NLX A; 1XJW B; 4CI0 B; 4JI2 H; 1X4F A; 5L9N A; 2PSZ A; 3UD4 P; 1ULZ A; 1GKR A; 4QU6 A; 1BUX A; 3UN3 A; 1N34 J; 4Q43 A; 2LXU A; 2PMW A; 2PMZ D; 2Q3P A; 3JAM Y; 4L9X A; 3ZTR A; 3GII A; 5GAE U; 2RIJ A; 3PY8 A; 2QO3 A; 5HMX A; 3HDS A; 1N33 J; 4QU7 A; 3ILI A; 4WK1 A; 5EZY F; 1XR7 A; 2ZZT A; 5HVF A; 3KLH A; 4FNI A; 2XY6 A; 3NS7 B; 2DCL A; 1DQA A; 1DIK A; 1PC5 A; 2ZM6 F; 1RKI A; 4IUF A; 3WX8 A; 1POJ A; 5IT9 Y; 1Y4Z B; 4ECU A; 4ED4 A; 2WB1 A; 3KLE A; 1RWM B; 4ZLL A; 1LW2 B; 3ZNU A; 4LIR A; 1VBY A; 3M30 B; 5EHH A; 3J81 j; 2VQF H; 1JLG A; 1Z2N X; 4LQ3 A; 3THT A; 1EJ7 L; 2BJE A; 4P74 A; 2WVE A; 4IG3 B; 4FXD A; 2YS6 A; 2R7R A; 2QYC A; 3BGU A; 5ETS A; 3OUY B; 3H98 A; 4J9O A; 1BJT A; 1FX2 A; 2JRS A; 4LQ9 A; 3A3O B; 4O4G B; 3EVW A; 3HVW A; 2CXE A; 1ULR A; 2DEJ A; 4ZVF A; 4DV5 H; 3F56 A; 3I55 S; 4QYZ K; 3TAM A; 5A2Q W; 4UBP C; 1UMJ A; 3GTK A; 5KT5 A; 4A8F A; 4DUT A; 4TN2 A; 2X1B A; 4C11 A; 1FTC A; 2V34 A; 2ZWP A; 2HQ3 A; 4X66 F; 2H1Y A; 8AT1 B; 5KFL A; 4R85 A; 4KHP H; 4QW9 A; 1R3N A; 1BMQ B; 1D3W A; 4U7C A; 1FJG F; 2BOP A; 1RTH A; 1X5T A; 1F0X A; 1SQF A; 3N0V A; 1TKX B; 3M4X A; 1HI8 A; 1Y14 B; 2YQ1 A; 3SCL E; 4QYR A; 4DV6 H; 4A8Y A; 3NLC A; 1CS4 B; 4ZOL F; 1JLF B; 4KGX B; 4AFF A; 1Q81 T; 3O8W A; 1S10 A; 1VRU A; 1UET A; 5GAD U; 1FXD A; 1VS3 A; 4C8L A; 4B3O B; 2HRY A; 2W2N P; 4WII A; 4XX0 B; 2DR8 A; 3CJ3 A; 5HME A; 2FB0 A; 2AJ0 A; 2C2M B; 2P9B A; 4DPX A; 3BSO A; 4DG1 B; 4P75 A; 5HSP A; 1K44 A; 4A93 G; 1FVQ A; 3V4I B; 3HK2 A; 1S03 G; 3CDE A; 3M32 A; 2Q79 A; 3IR7 B; 1I6U A; 1CG2 A; 4V1X A; 4Q8F A; 4AXJ A; 3W2V B; 4G1I A; 1N48 A; 3MUR P; 1IJE B; 5KT7 A; 4RR5 A; 1MN7 A; 2NXC A; 4N56 A; 3TAQ A; 5D78 A; 2F9D A; 1FDN A; 1MML A; 4GMK A; 3KLV A; 1RHU B; 2LQJ A; 3IWN C; 4ALZ A; 1CQM A; 2BE9 B; 3KG0 A; 4A3K G; 3IM8 A; 2FPP B; 3NKY A; 5F0W A; 3CJ5 A; 4DV3 H; 3SIP B; 4USM A; 5K2M A; 3T5J A; 3WQP A; 4L3K A; 2FGO A; 3U9S A; 1MZB A; 1U47 A; 5AJ3 J; 5D0E A; 2CVE A; 3DHX A; 3T81 A; 4RAZ A; 5C42 B; 5E33 A; 3AQO A; 2V8H A; 3EPG A; 2ZJT A; 1XR5 A; 4LRB A; 2MZS A; 1IBK F; 3C46 A; 1HWL A; 4MV8 A; 3H0P A; 1XMQ J; 1RJJ A; 3VV2 B; 2VBY A; 5DJB A; 3PX4 A; 2ZUA A; 4EFB A; 2UV8 A; 4ETX A; 4M5L A; 2XYP B; 1IKV A; 3CCR S; 3D0G E; 3HI9 A; 2UXC H; 3KWM A; 8RUC A; 4IR1 A; 2EFG A; 2MKY A; 2KJV A; 3CDA A; 1SYX B; 3IC1 A; 4XDD A; 3IFE A; 2JEJ A; 3KUG A; 4CLT A; 4ECZ A; 4ZVE A; 2OD4 A; 2J0W A; 3EWG A; 4DL6 A; 4ZS6 A; 4DL3 A; 1Q4R A; 3ISN D; 1F5C A; 3VMM A; 1UPP A; 3MTK A; 2ZH3 A; 3IS9 A; 3PHT A; 1NDK A; 2RQ4 A; 4USJ C; 1CS4 A; 4WU2 A; 3HOY G; 2X3G A; 1GXL A; 1JLF A; 2P50 A; 2AYG A; 1WSC A; 2QL7 B; 1UVJ A; 1NKE A; 3Q8U A; 4B3O A; 2HQI A; 2XNR A; 3VZI A; 3TO8 A; 4LF9 H; 3MAH A; 1CBK A; 2DWC A; 4YB8 B; 2R2S A; 2JA8 G; 3ILJ A; 1NB2 A; 4OLP A; 3TVZ A; 5KOX A; 2J6U A; 1AYE A; 4WE1 A; 3N79 A; 4KBI A; 1JXV A; 2DGS A; 5FLX Y; 4ZVR B; 5E2Q A; 1RU2 A; 3W1Y C; 3GYX B; 1GGO A; 1S68 A; 2NQP A; 5JVD F; 2FVR A; 3W2V A; 1NJI T; 2WE6 A; 2FHM A; 4Q2L A; 4YS2 A; 4I50 F; 4NAS A; 3OUU A; 5SUH A; 5SZT A; 4EUV A; 4R7W A; 4QIV A; 1PBU A; 2POK A; 3UR0 A; 3EGJ A; 4N41 A; 2VPX B; 3G2X A; 4F4X A; 2BKN A; 1CVJ A; 1T9U A; 2RUS A; 5HMF A; 2AGP A; 3CO0 P; 4B3S F; 1U4B A; 5K14 B; 3BA9 A; 2VOP A; 1LXJ A; 1X4H A; 2CPY A; 4RX6 A; 3C66 A; 1NPP A; 3AT1 B; 4HYK A; 1N0V C; 3RB6 A; 1HHT P; 1PC4 A; 2HVE A; 2E9Z A; 3QJL A; 3H5S A; 1RKJ A; 1NJZ A; 2XWX A; 4I4T F; 5C42 A; 1IBL J; 5D8H B; 4J37 A; 3QSY B; 4QOT A; 1SQE A; 1VAJ A; 2H3E B; 1U1M A; 1Q0N A; 5J1E A; 3CCS S; 2LEC A; 1VQK S; 1WTF A; 1OY9 A; 3AH6 A; 4MJK A; 2Y1J A; 2RD5 C; 2PUZ A; 1Q16 B; 4TXS A; 1CC8 A; 1SKR A; 4ED7 A; 3DDN A; 5RUB A; 3G8Q A; 1IBM F; 3J80 U; 2OGJ A; 5J1N A; 2BR0 A; 1P8G A; 2XD4 A; 4LF6 F; 1LNQ A; 4CLS A; 4X2B A; 1TMJ A; 5FDL B; 2BE2 B; 2OEM A; 3I0R A; 2LKZ A; 4ZVQ B; 4CT8 A; 2PA3 A; 5FJ8 G; 2ILZ A; 1RYR A; 2GV1 A; 3IBC B; 2VA3 A; 5KFA A; 1HBN B; 2RSQ A; 3ET6 B; 2B9W A; 4DV7 J; 4ZHR B; 4Y2I A; 1H2T Z; 1UWA A; 4EBC A; 2QV6 A; 3VAK A; 1YG0 A; 2MHN A; 1X9S A; 4C2M A; 2DGV A; 2R7W A; 2DTJ A; 3MMA B; 1X9W A; 3R9L A; 1WHW A; 3N5F A; 2UUW A; 3CNQ P; 2OMO A; 1VQ7 S; 2WB4 A; 2PFD A; 2E9R X; 2R8H A; 1M1K T; 1YZ7 A; 2L48 A; 3BE7 A; 4LSL A; 1N5Q A; 2L3M A; 2I87 A; 4J9K A; 4E8U A; 2VUG A; 2WSE C; 4WZQ A; 3N28 A; 3DLK A; 3HCX A; 2XY7 A; 4JI6 H; 3H2L A; 1KC7 A; 3FJ2 A; 3CMQ A; 2IY5 B; 3AB4 B; 2LFV A; 4RUA A; 1XQI A; 2HUR A; 1WDT A; 1NLK L; 1UW4 A; 3HKW A; 3I5B A; 2RE1 A; 4DV1 J; 3ICL A; 4NLR A; 3K5H A; 2DZD A; 4I0W A; 5H9F J; 1NCL A; 2UVU A; 1T9W A; 2K8G A; 4OLO A; 5JUM A; 2LQ8 A; 3DKD A; 1WSV A; 1Y3K A; 3EM1 A; 1MG7 A; 1VQ5 S; 1JLA B; 4LFB J; 1GLV A; 2R8J A; 3TQE A; 5BR8 F; 1NDL A; 3GTL A; 4ZJO A; 2LA6 A; 3FSY A; 2NZC A; 1S76 D; 1R43 A; 1UJ4 A; 2IJF A; 3KUE A; 5KG1 A; 2J8A A; 1K47 A; 3GU0 A; 5CXT A; 2MPU A; 2QMW A; 3VAI A; 4B3T F; 1W26 A; 3H2U B; 4TKT A; 1HNW H; 5AE3 A; 2O67 A; 4AL6 A; 1FEH A; 3U27 A; 2BM0 A; 2BX6 A; 1OWT A; 1N4L A; 4DQR A; 4OL8 A; 5E41 A; 1JLE B; 1DQ8 A; 4D3H A; 1Z2P X; 1DJ0 A; 3KLF A; 1OPI A; 3QGG A; 4GC7 A; 3F0N A; 4NLG A; 4DR3 H; 3CJ2 A; 1KBR A; 1O5J A; 2A0F B; 3A8I A; 1TAQ A; 3A8E A; 1X4A A; 3SXL A; 3SMZ A; 2XI3 A; 4NEY A; 3LAN A; 1Y1V A; 2BE2 A; 2V69 A; 2IM3 A; 3FBF A; 5ETM A; 1U24 A; 4K50 A; 4F4A A; 5ETP A; 2UVR A; 4ZHR A; 4DSJ A; 4DV0 H; 2EG1 A; 2GHW A; 2I5J A; 5LMU J; 3POT B; 3TEG A; 1RU1 A; 3CCU S; 4YFU A; 3V4S A; 1SJR A; 2X0S A; 2OD6 A; 4J9S A; 2ATL A; 2VBZ A; 4E4T A; 1T3N A; 1WC3 A; 5ADZ A; 2WVC A; 3RYL A; 4DCJ B; 1KTQ A; 3E3X A; 1J2V A; 1QM9 A; 2ZBI A; 2DB1 A; 2JH3 A; 2XG8 A; 3V81 A; 3M6X A; 2YKO A; 1S6P A; 2J9G A; 2DNZ A; 2IFX A; 4DR7 H; 1VQP S; 1QVF R; 1NSA A; 3FUY A; #chains in the Genus database with same CATH homology 5F3T A; 3EGZ A; 3HFK A; 1Q7L B; 1UP1 A; 1S0V A; 2FUG 9; 4ED5 A; 1CQN A; 3M9N B; 2ZZN A; 2W4P A; 3DYA A; 4ECY A; 5EX3 A; 2N4C A; 3GB0 A; 2DRB A; 2P9G A; 4Q8E A; 3CPW R; 2R7T A; 2F40 A; 2R8K A; 2WBM A; 2NUH A; 5KG4 A; 1LW2 A; 1REV A; 1RIS A; 3CED A; 1FRI A; 3UPF A; 3OLA A; 5D3G B; 2FHK A; 2CPZ A; 1H0H B; 3CD0 A; 4KSE B; 3M30 A; 5AXN A; 3VAL A; 2YWG A; 2AYB A; 1PSD A; 2XSJ B; 2DHX A; 3FZB A; 3I96 A; 3KLI A; 2VC0 A; 4DR5 F; 2LU1 A; 3CMA S; 1TKZ B; 1BWE A; 3T9N A; 4CZC A; 1S7H A; 5KG3 A; 3K59 A; 2HWT A; 4ZVG A; 3KNA A; 1VIO A; 1XBW A; 2Z8Q A; 1NND A; 1RP5 A; 2ASY A; 2C3O A; 3NWG A; 3NCP A; 2APO A; 2EK1 A; 3SSR A; 1IKY B; 1L3K A; 1FXL A; 2XBP A; 4IG3 A; 4ELV A; 2L41 A; 4IG0 B; 2CPD A; 2YY3 A; 3MED B; 5HMY A; 3OQ2 A; 2DH9 A; 2HHH F; 2N3L A; 3OUY A; 5J2Q A; 2DJW A; 2J7W A; 4DQI A; 1FK9 A; 1XMO F; 1JXL A; 4MK9 A; 1H2V Z; 3KNG A; 2ZUG A; 4EBE A; 2QLX A; 2EWH A; 1RT6 A; 2AV5 A; 4O4G A; 1R0A B; 3DU6 A; 1VC0 A; 2AQ4 A; 1G1X A; 1ZM9 A; 2D7S A; 3UI3 A; 4ZCI A; 3DLL Q; 3P49 B; 2HFZ A; 4K4S A; 1R8H A; 2BA9 A; 3LF0 A; 3V6D A; 3SCK E; 5J2N A; 4FN5 A; 1IN0 A; 2XMM A; 1OSD A; 2W9C A; 4C3K A; 3MPW A; 4HTJ A; 4F02 A; 2WAQ D; 3PRB A; 3UDL A; 1QJH A; 1T7P A; 2YNH A; 3O3L A; 1LKZ A; 1S97 A; 3WBZ A; 3P5T L; 3T1Y F; 4A8O A; 3N6M A; 3FFI B; 3LPY A; 3H8Y A; 2VON A; 2XWY A; 3MDF A; 4A48 A; 1UA0 A; 2IC3 B; 2OJ3 A; 5B0G A; 3TNV A; 4V19 X; 1BLU A; 1XEZ A; 4KQZ A; 2MZ1 A; 1EP4 B; 2XUL A; 3IBW A; 2HG4 A; 1YBA A; 1AFJ A; 1WF2 A; 2AW0 A; 2W9B B; 2CPX A; 2R7O A; 2VV5 A; 3EZU A; 3MFI A; 1SL2 A; 4PCQ A; 3GFX A; 2QA4 G; 2HMI B; 3HHD A; 4NX9 A; 3T6J A; 1HYS A; 3MFH A; 4BWM A; 2R2R A; 3RW7 A; 2CPF A; 3D1Q A; 2ZKT A; 1FRX A; 1TKX A; 2RML A; 2V4Q A; 4D3G A; 3FQL A; 3H59 A; 5B0B A; 4O44 A; 4ZPT R; 1R8P A; 5A6P A; 2YKM B; 2IVM A; 1JMT A; 2CQC A; 3GV7 B; 5A0Y A; 5MLC V; 1B7F A; 2WSF C; 2CQG A; 4MH8 A; 1JLE A; 3HO1 A; 4QWD A; 1U1N A; 2L08 A; 1ZH5 A; 1BQM A; 4IFY A; 4Q5V A; 4K4Z A; 3DRS A; 4JV5 F; 2FDN A; 4PR6 A; 1RTI B; 2ALZ A; 5D6G A; 4DL7 A; 4ZP9 A; 2AJF E; 1VLY A; 2ZNZ A; 3LH2 S; 1U25 A; 4HVZ A; 4IKA A; 4YY3 F; 4DJB A; 3Q8Q B; 4KO0 A; 3QJJ A; 1NH7 A; 4DV4 F; 2J6S A; 2I2Y A; 2VOD A; 4F25 A; 2W24 A; 2XS2 A; 1D9F A; 2MY7 A; 5IQ6 A; 5F41 A; 2D1C A; 2J76 E; 2GA7 A; 3C8Y A; 3DC2 A; 4CTA B; 1FRL A; 4FNH A; 2HLT A; 1HI1 A; 3DU5 A; 3ENQ A; 1QLN A; 2CJ9 A; 3Q24 A; 1KC8 T; 2B51 A; 3ZZP A; 5H9E J; 3RAQ A; 2M5Y A; 4IDK B; 2KU7 A; 4PQA A; 4OV1 A; 3BGR B; 3WX4 A; 3I9V 9; 2AGO A; 2Q43 A; 2WOM B; 1UWB A; 2ASL A; 2FY1 A; 3OL9 A; 1R31 A; 3CCM S; 1TP7 A; 3GV8 B; 1N5Y B; 1NHV A; 3N6L A; 3FQK A; 3VAH A; 3BM7 A; 1CC7 A; 4DEZ A; 2PGC A; 3IRX B; 4DG1 A; 5I3U B; 2F4V F; 4YOE A; 3KK3 B; 3KLH B; 4FBU A; 3QJP A; 1FTR A; 2LMI A; 2XI2 A; 2MQQ A; 1DTQ A; 2BDP A; 4F3M A; 1WAC A; 5HO4 A; 3DRY A; 2HVH A; 4HDH A; 3FEY B; 3PW2 A; 3V4I A; 1FNM A; 5DLF A; 2HND B; 1C1B A; 5LSR A; 3HPO A; 1TK5 A; 1UVK A; 3FZ2 A; 3UQS A; 7FDR A; 2BBE A; 2XGP A; 2OTL S; 4OI4 A; 1Q79 A; 4OHZ A; 4WKY A; 3N5B A; 4CIO A; 3KLE B; 5KG6 A; 3BF4 A; 4K4T A; 5BJR A; 4KB7 A; 2XCA A; 3MEK A; 1F2G A; 1IR0 A; 1KTV A; 3CDU A; 3LP0 B; 3GIK A; 4R8U A; 2DIV A; 2FJA B; 3G6X A; 1MMD A; 5CCL A; 1OIA A; 5F8J A; 2VH7 A; 4X4P A; 2HVI A; 4HLB A; 3T5L A; 4A46 A; 2OPP B; 5KT3 A; 1FEE A; 3R27 A; 3PO5 A; 3CD5 A; 1GX5 A; 4YP3 A; 4MTP A; 3HF5 A; 3FGV A; 1WI6 A; 2KRB A; 4YDX A; 4ID5 B; 3JTN A; 3MOZ A; 3UCG A; 4NRT A; 4X84 A; 3WZ0 A; 3F44 A; 2K2P A; 3CXC R; 4LSN B; 1SH2 A; 2JEG A; 2ILY A; 2WW8 A; 5KFY A; 3MZI A; 3MXH P; 2GCF A; 2Y1R I; 2ZH5 A; 4KKO A; 5F8I A; 2DT9 A; 2ZH6 A; 2ZJQ Q; 2FJV A; 2ZKU A; 2VL1 A; 3MUV P; 1R89 A; 5I4M A; 1D8Y A; 1SJ3 P; 3V4M A; 1TL5 A; 2F8H A; 4L6V 3; 3BEG B; 4A8K A; 5A8R B; 5KT2 A; 3DGP A; 1PIL A; 1HNV B; 4UQG A; 4DV2 F; 2QA4 S; 4C8O A; 3LGM A; 4P6Q A; 1NK7 A; 2GFF A; 3TA1 A; 2XSJ A; 5GMK a; 1TK0 A; 2X1F A; 2AJQ A; 4C4W A; 3TAM B; 4KFB A; 5G0R A; 4FS2 A; 2E1R A; 3BB5 A; 4B3M F; 2VFS A; 1ROF A; 3BJY A; 4JI8 F; 1SIW B; 3MEG B; 4NLQ A; 2HAI A; 5AE1 A; 1TKZ A; 3SFU A; 3O5T B; 2B9X A; 4C11 B; 1X9M A; 5LXY A; 4HHJ A; 2ZDP A; 2CQ3 A; 4PUO B; 1ZZN A; 2G1D A; 2C3U A; 3J81 Y; 3M8Q A; 5EDW A; 2OFH X; 4ZCM A; 1G2E A; 1IKY A; 2M88 A; 4NCG B; 4NCB A; 2VKZ G; 4Y34 A; 3UE2 A; 3LWL A; 2NYI A; 2FJB B; 3K5P A; 3GV5 B; 3I31 A; 1FDB A; 4FF4 A; 3N01 A; 4RBU A; 5KG0 A; 1BC6 A; 2O01 C; 4ZP8 A; 3P96 A; 1URR A; 3OHB A; 2HNY B; 5I3Q A; 4ZPC A; 2ZJP Q; 1RTH B; 1TV6 B; 4L72 B; 2VG7 B; 5IZH A; 1N6Q B; 3O6Q A; 2V4R A; 3KYL A; 1VBZ A; 3MOJ B; 2IBK A; 3G8T A; 1N34 F; 1R0A A; 1NKC A; 3GIM A; 1TFW A; 4DPO A; 2YWF A; 4ZLP A; 2D3O R; 1I6J A; 1JKH B; 3MD1 A; 2IAJ B; 1FRH A; 5HL7 Q; 5JVG Q; 2CRL A; 3W2W A; 1UVL A; 4QWA A; 1LWE A; 2AJ1 A; 2F9J A; 3K57 A; 5C5J A; 1JNZ B; 5DGB A; 4C97 A; 1K0T A; 4DL4 A; 3AQP A; 4NXN F; 1S6O A; 1F9F A; 4MKA A; 1FJ7 A; 3US5 A; 3RU0 A; 2NS1 B; 1B0P A; 4JUZ A; 2BJD A; 1BQN B; 3T1A A; 4DUZ F; 2RVJ A; 2JWN A; 2LQ9 A; 4ZCK A; 3KTQ A; 3L7O A; 3VAM A; 1U2R A; 2JJ4 D; 4XPC A; 1C0U A; 4ELT A; 3H20 A; 3I82 A; 4DOZ A; 1VRU B; 1JIH A; 4A8W A; 1KLN A; 4F3Q A; 3HHL A; 3L7P A; 1RT1 A; 2M7S A; 1ZBT A; 2IVF B; 4O3Q A; 1NO8 A; 1N52 B; 3SCJ E; 1EP4 A; 4ZK7 M; 3CCV S; 3MCS A; 2MB0 B; 1UNN C; 3T19 A; 1VQY A; 2QA2 A; 4ICL B; 2UP1 A; 2V4J B; 2DNN A; 3Q8P B; 4PG7 A; 5C25 B; 4Y2M A; 1MWZ A; 2HMI A; 1W2I A; 2E66 A; 4F4Z A; 2K1R B; 2XNQ A; 4B08 A; 3EYZ A; 3LQV A; 2R8G A; 2OP5 A; 3DRR B; 2QRR A; 4HH0 A; 5B09 A; 2YH1 A; 4OZJ A; 4C98 A; 4NLY A; 1VK8 A; 4UW2 A; 3TYT A; 2MQO A; 4HT7 A; 1ELO A; 1OS5 A; 5DDP C; 4DR1 F; 4ED1 A; 5E0O A; 2RH3 A; 3MM6 B; 3H5U A; 3GR0 A; 3KFW X; 1KON A; 2KTQ A; 4USH A; 1K9M T; 4WU3 A; 3D54 A; 2UU9 F; 4E98 A; 5EWG A; 5L1J A; 1RTI A; 3X3U A; 4CO2 A; 1T8E A; 4FBT A; 1SJ4 P; 4Y28 C; 4ZCL A; 4O4R A; 2V94 A; 4A8X A; 4DV0 F; 2COL A; 3HHL C; 3PO4 A; 3RTV A; 4TY9 A; 5KFP A; 2A10 A; 1U1O A; 2QZ8 A; 1DQ9 A; 3EZ5 A; 3H5Y A; 2CQ4 A; 1P1L A; 2MJU A; 3MMJ A; 4OZ1 A; 4J9N A; 2O66 A; 1JJ2 R; 3N6N A; 3M32 B; 3GG1 A; 1URN A; 3PIO Q; 1U8S A; 2B4O A; 2EPI A; 4IDK A; 3UD3 P; 1UVM A; 3BN7 A; 3BGR A; 1NJX A; 2KG1 A; 3B4M A; 2WBR A; 2EFP A; 4WJB A; 1LWF A; 1RA6 A; 1GXU A; 5HRO A; 2M8H A; 1JLC A; 2ASJ A; 3TUZ C; 2XLJ A; 1MSW D; 1FNO A; 4ZKA A; 2B6A A; 3KK3 A; 2ZJR Q; 4FVM A; 2EG2 A; 2JFK A; 4RW9 A; 3B78 A; 2OPQ B; 3EGN A; 3OFF A; 2YVS A; 2FLO A; 3CDW A; 5K8N A; 4KGM A; 1Q53 A; 2DD8 S; 4ZOS A; 4W92 B; 3UCU P; 2Q66 A; 2HHQ A; 1FF2 A; 5KFC A; 3MTJ A; 5L1L A; 4OIX A; 3KOA A; 3PX6 A; 2G0C A; 1UVN A; 2P5V A; 4RW6 A; 4C8M A; 3FNN A; 3I0S B; 1X5P A; 1C0T B; 1NRK A; 3VAF A; 1V3R A; 1C4C A; 2E44 A; 3QIP B; 2UV8 G; 1VQ9 S; 2WON B; 3V2U C; 3DMJ B; 3UFC X; 5I42 A; 2KM8 B; 4WXO A; 5F8M A; 2D41 A; 1G6B A; 5DG8 A; 2W4D A; 1X7V A; 2CDQ A; 3BR8 A; 2FVS A; 4DWI A; 4ECR A; 2RRB A; 1M5O C; 1N56 A; 1NK4 A; 2HW3 A; 4A8M P; 3OTD A; 2U1A A; 1SH3 A; 2HFO A; 5EWF A; 3IAS 9; 3D0I E; 2K7K A; 2MJN A; 3D5M A; 5DDO C; 4H4K A; 4G1Q B; 3RZA A; 4DMZ A; 2X1A A; 4QR0 A; 4QQB A; 4DL2 A; 4ID5 A; 3DOL A; 2C3M A; 2CQ1 A; 4OX8 A; 4TYA A; 5DDR C; 1HQU A; 3SSQ A; 4C8N A; 2DHS A; 1I1G A; 1KOH A; 3BN4 A; 1OQ6 A; 3BO2 A; 4ED6 A; 2UVV A; 2E1M C; 1VK3 A; 4LF4 F; 3IRW P; 3U7J A; 3CIZ A; 2AWZ A; 5T14 A; 5EWE A; 3OFG A; 1S9E A; 3HMJ G; 4OZL A; 1KNZ A; 1K8A T; 2L9D A; 5DQI A; 3G71 S; 1DRZ A; 2N3O A; 3DM2 B; 4ILL A; 1H98 A; 5CZV A; 1UJ5 A; 4B9V A; 5KFZ A; 5K14 A; 1D0E A; 2KT2 A; 3ENV A; 1IQT A; 4MYT A; 1HNV A; 2NLW A; 4RU9 A; 1IKV B; 1FVS A; 3D0H E; 3OTE A; 4MMO A; 4TYB A; 4YEA A; 1KD1 T; 2DNL A; 4O3R A; 4QJV A; 5LJ3 Y; 5DQG A; 1KEK A; 4HKQ A; 4ZP7 A; 1QAX A; 4EGL A; 3K5N A; 2R4F A; 3PV8 A; 3CUN A; 3CME S; 2KXF A; 3DOK B; 2RRA A; 2FVQ A; 4EBD A; 1X8D A; 3PRD A; 1HMV A; 1E6V B; 1Q8B A; 4USI A; 3TVY A; 2GHV C; 4CO4 A; 3W3S A; 3QPI A; 4H4K C; 3VZH A; 1LW0 A; 2M9K A; 4U7U K; 2HWH A; 1T03 B; 3KK1 B; 2YQ3 A; 1A7G E; 3I87 A; 4C0O C; 3PW5 A; 4OFZ A; 1Z2Z A; 4PUO A; 3L09 A; 1NB7 A; 1HNI B; 2HS3 A; 5E3R A; 4NCG A; 1VQS A; 2W4C A; 4NRU A; 2HHU A; 5KOW A; 3M8S A; 4B3R F; 5KFG A; 3GYN A; 1HNZ F; 4CH7 A; 4DSK A; 3KK2 A; 1ZZF A; 2HNY A; 4KRW A; 5LXR A; 5C24 B; 2KYX A; 3B82 A; 1U2F A; 3HZG A; 3M2U A; 1TV6 A; 2FC8 A; 4RWW A; 2OPR A; 4C3M A; 2IA0 A; 1AUD A; 4TNO A; 1IUJ A; 2IM2 A; 2M4M A; 5DG7 A; 2VD3 A; 5B0C A; 3GE9 A; 4IRK A; 4DL5 A; 1W25 A; 3TA0 A; 1ZM3 A; 2CIM A; 1SB6 A; 4J9P A; 1KR4 A; 4BC9 A; 3U1M A; 4GKK F; 4WFB Q; 2HHS A; 1FT8 A; 1KOO A; 3CJ4 A; 2IAJ A; 3PR4 A; 1ZAX A; 1NJW A; 2FLN A; 1KLM B; 3H4B A; 3D2W A; 2ZHB A; 3IS9 B; 4ECV A; 1IJF B; 1E6Y B; 2XEX A; 5K9F A; 3CB4 A; 3TR5 A; 3ZZZ A; 5GAE I; 2ZC4 A; 5HMW A; 2UXD F; 3P6Y C; 3GMG A; 4UUW A; 4OHW A; 4TY8 B; 5FDL A; 1TL1 A; 2J9D A; 1X4E A; 4UQT A; 2QFJ A; 2IHR 1; 1BQN A; 3PFE A; 5D4O A; 4CH0 S; 4J9M A; 5HP1 B; 3TW0 A; 3S1T A; 3I32 A; 4X62 F; 4Y00 A; 3HHE A; 1VJW A; 2EK6 A; 4ZPB A; 5CCM A; 1WF0 A; 4M95 A; 2IMW P; 4KV8 B; 1HBN A; 2F3J A; 5HMZ A; 2EI5 A; 2V5H G; 4H4O B; 4C8Z A; 1TL4 A; 2WHO A; 1K1S A; 4P2S A; 4KIA A; 3SSS A; 1N8R T; 2EW9 A; 1YSJ A; 4FXV A; 2LEB A; 1HA1 A; 3JSY A; 1WHV A; 4G1P A; 4NML A; 4ICL A; 1KQS R; 1XNQ F; 2DH7 A; 4WFA Q; 3KMQ A; 3BGL A; 1P9Q C; 4O3O A; 2C2R A; 3DLG A; 4I56 A; 2J9E A; 4M63 A; 4C0B A; 4WZM A; 4TU7 A; 5KFK A; 2KT5 A; 3MF5 A; 4C3L A; 5KG7 A; 2AF6 A; 4NLW A; 1FJE B; 4B9N A; 3QMQ A; 1U48 A; 3KHR A; 1RDR A; 3TAP A; 2K3I A; 1R8A A; 3TUI C; 2LDI A; 3OSP A; 4NA3 A; 4DX8 H; 5AJ3 F; 3ONQ A; 5KFE A; 3MMA A; 2RU9 A; 2W7O A; 2ZH9 A; 2KB2 A; 2VFT A; 3CDB A; 3MEC A; 4CO1 A; 2BM1 A; 4WE1 B; 3PW4 A; 1M90 T; 2P9E A; 5K8O A; 3D1O A; 1JLQ A; 3JSB A; 2CJQ A; 2OIH A; 4YR3 A; 3ZZY A; 5CD4 H; 2KDO A; 3FCH A; 4IH6 A; 4PQU B; 1HBU A; 2PE8 A; 2OAU A; 3U1L A; 1IKW A; 2JLE A; 2RS2 A; 1S1T B; 4ZPA A; 1FD2 A; 4TQS A; 1RTJ B; 3NS6 A; 2GNK A; 2GX8 A; 1HBO B; 3K5L A; 1U1R A; 2MY8 A; 2FGC A; 4Y2A A; 2VA2 A; 4BCA A; 4HTI A; 2KT3 A; 4B9M A; 1W2B R; 2RF2 B; 4IO1 A; 4QYZ J; 2OKQ A; 5D4N A; 4RBT A; 4I69 A; 1X4C A; 2J9C A; 3Q0A A; 4URG A; 1CPZ A; 1Y9O A; 1YHQ S; 2EFN A; 5DGA A; 1FDA A; 4ECQ A; 1WTB A; 3QO9 B; 4GO7 X; 3V6J A; 1T05 B; 1K0V A; 5B0E A; 1RT5 A; 2XZW A; 4H4M B; 2ZC3 A; 2I38 A; 2MQN A; 3N4W A; 3FMW A; 3MEE A; 4WK3 A; 4PKD B; 1C1C A; 2VOO A; 3KHG A; 1FE0 A; 1DAR A; 3J7Y U; 2FJW A; 4DSF A; 5KFI A; 3MR6 A; 1YJV A; 2LA4 A; 1H3D A; 5THW A; 4I67 A; 1JXE A; 1RT2 B; 3DLE B; 3FSX A; 3X1L B; 4HI1 A; 1UKU A; 3K8W A; 4OX7 A; 4XDC A; 4X4R A; 1U6B A; 4PG6 A; 4K59 A; 3B4D A; 4OMF G; 4F2S A; 4FS1 A; 3M6W A; 1RA7 A; 2YWE A; 4AYB D; 3E01 B; 2EC0 A; 4MKB A; 3QIP A; 4FXW A; 2IM0 A; 3UWT A; 5G4D A; 4KE5 A; 5J1P A; 1KS2 A; 2WON A; 3M1V B; 4IFV B; 4JI1 F; 3BQ2 A; 3AA8 A; 4YQW A; 1DLO B; 4OX6 A; 3TOQ A; 3PM9 A; 1NK0 A; 3M2R B; 1KHV A; 1WEY A; 1LWC A; 1X4D A; 2YX7 A; 3I4H X; 4C0H A; 4TU8 A; 4TLR A; 3OL7 A; 4LIW A; 1ARO P; 1D1U A; 4AE5 A; 3EGW B; 2NSC A; 4QIF A; 4F8R A; 1FER A; 4RNM A; 4O0I A; 1YJT A; 2EKY A; 2VPZ B; 5KG2 A; 3HX9 A; 4K5S A; 2VC1 A; 1VQ6 S; 1HR0 F; 2HGM A; 4BS2 A; 2U2F A; 2Y1I A; 1N32 F; 3TAR A; 4YR0 A; 3M2V B; 3P3D A; 2RT3 A; 1PO6 A; 5CYQ B; 2DRA A; 2VZ9 A; 2BRK A; 4DHV A; 2W7A A; 2R2U A; 1JLA A; 2DNY A; 6FDR A; 1S77 D; 4PWU A; 3DNC A; 2HZC A; 4ETZ A; 3DHW C; 5DRK A; 1EET B; 3OV7 A; 2BW2 A; 2DGX A; 4OJG A; 2MHE A; 2ERR A; 4M8L A; 1ZYQ A; 2R7U A; 4LVN P; 5CQG A; 1GTE A; 2AGQ A; 3IG1 B; 5AXL A; 2WTF A; 3HT3 A; 1M0S A; 5D4L A; 5KFB A; 4B02 A; 5J1E B; 2MZQ A; 3EUN A; 2ZE2 A; 2JZ5 A; 4DN9 A; 2OSR A; 1N88 A; 1Q82 T; 1M5V C; 4FF3 A; 4C8K A; 3C6T B; 3OW2 R; 2UUC F; 2O3D A; 3PHE A; 4HI2 A; 2KFN A; 4OI6 A; 4OHV A; 3MWB A; 3AA9 A; 4TQR A; 4YB1 P; 4J9Q A; 1S0O A; 1U6F A; 1GX6 A; 4WKR A; 3HJF A; 4X4T A; 4NA2 A; 3DI6 A; 2KG0 A; 2CQ0 A; 1APS A; 3GG0 A; 3VCD A; 1FE4 A; 2YNI B; 2R7S A; 1QVG R; 2VG5 A; 3SDE A; 1C4A A; 3NP5 A; 3HVT A; 2DGU A; 4BDP A; 3TQ1 A; 3NS5 A; 2OSQ A; 4ED3 A; 1SL0 A; 3BO3 A; 3S7R A; 2M0G B; 4ZPW R; 2OPS A; 4RW8 A; 2FJE B; 3PEJ A; 4ECS A; 2KHC A; 1FES A; 3KK1 A; 1J5E F; 3I0R B; 4DQQ A; 4NLD A; 4OI1 A; 2YKN B; 3GZ7 A; 4ITS A; 1Y5N B; 1AW0 A; 1HNI A; 3Q2T C; 4Q0B A; 2ROP A; 1RTD B; 2IYI A; 3Q23 A; 5HBM B; 2VFR A; 3J80 Y; 3CD7 A; 5HLF A; 4RU2 A; 1SUQ B; 3FMB A; 1YWX A; 1F5A A; 4DR4 F; 4JBC A; 5HN0 A; 3DFE A; 2MYF A; 5C24 A; 3K5M A; 4DSI A; 3LP1 A; 1ZPV A; 2H9Z A; 1RT4 A; 3OTC A; 3AB2 B; 1G3O A; 3Q2S C; 4LT6 A; 3QLH B; 1TL3 A; 1PGZ A; 3BS9 A; 2Q6B A; 2NPI A; 1V9O A; 1LFW A; 3M8P B; 1U49 A; 2FHO B; 2F8E X; 1C2P A; 2CPH A; 2JDJ A; 3G4S S; 1U26 A; 4RNN A; 1E6Y A; 1V3Z A; 2JLG A; 1Q1K A; 4LF7 F; 2A2C A; 2XGQ A; 1XK8 A; 4EWT A; 2YWK A; 3CCW A; 1DAX A; 4TY8 A; 3DXB A; 3CO9 A; 3ZZR A; 2GGP A; 2DVI A; 1VQO S; 3DLB A; 4MK7 A; 4M5D A; 2E7W A; 4I68 A; 2ZH7 A; 4DR6 F; 2D3Z A; 1DTT B; 2PDA A; 4I7F B; 4NYO A; 2ZH2 A; 4K4U A; 2RKI B; 2Z2M A; 5CYJ A; 4Q45 A; 5F9N A; 1JLB B; 1OO0 B; 5J2M B; 3J7Y I; 4V19 J; 3RR7 A; 1X5S A; 2V8D A; 4JI2 F; 4QWC A; 4YB6 A; 2OTJ S; 3CCJ S; 3LAL A; 3NNH A; 3SV3 A; 2B5J A; 1U1P A; 4KV8 A; 3K2Y A; 2DK2 A; 4H4O A; 3JSM A; 1S4F A; 4LSL B; 3CTR A; 1JB0 C; 3QID A; 2DPI A; 4YHH F; 2ZOF A; 5DLG A; 1VRT A; 2ZNY A; 5AXM A; 5LMN F; 3MMB A; 4JI7 F; 3R5A A; 3MR3 A; 1O0P A; 4ZPV R; 2N82 B; 2KI2 A; 5T41 A; 3G6V A; 2D3U A; 3DDK A; 3MM5 A; 3NAH A; 4JI0 F; 2E5J A; 3OVA A; 3AJ2 A; 3DLK B; 4MFB B; 4CO3 A; 5J2P B; 1UVI A; 3EPI A; 1BD6 A; 2LCW A; 4ES1 A; 3POT A; 2PD1 A; 2G13 A; 2GJI A; 3CCZ A; 3CC4 S; 2P6S A; 3RBE A; 2BVZ A; 3ZC4 A; 4PG8 A; 5KTQ A; 3OR2 B; 2ZOM A; 3T5K A; 2ZH4 A; 4NOH A; 4FVC A; 2IA6 A; 2V2K A; 3IGN A; 3MM8 B; 4OJH A; 3CCE S; 1Q8L A; 1YJ9 S; 3MPV A; 1U1L A; 1IBL F; 4NZ6 A; 4CQ1 A; 1ZAV A; 3FEX B; 4DFM A; 4KHR A; 1SV5 B; 2YNF A; 1S1V B; 3MD3 A; 3RBD A; 1B0V A; 3I5C A; 3NNC A; 3VAG A; 3R5C A; 1XN9 A; 4A47 A; 5MAO A; 1JLG B; 1S49 A; 2PEH A; 4OX9 F; 1TKT A; 3Q8R B; 2AU0 A; 1S1T A; 3HHL D; 4KT0 C; 2EFO A; 3C6U A; 1RTJ A; 1S1X B; 3BQ1 A; 4ZPD A; 4J5O A; 3FVY A; 5EGY A; 1XER A; 5E3P A; 4GO5 X; 3KMS A; 1CLF A; 4QPX A; 4Q2M A; 1S79 A; 2JVO A; 2CJB A; 4DQP A; 5KG5 A; 5EN1 A; 3FRY A; 2Z4P A; 4FP9 A; 1RI7 A; 3CWJ A; 5E3C A; 2QEX S; 2HWI A; 2W8L A; 3B8H A; 3QO9 A; 4DBQ A; 4R0J A; 1T05 A; 2W8K A; 1VQ8 S; 3SV4 A; 3MPY A; 1L3S A; 4H4M A; 3MAQ A; 3BO4 A; 1NAQ A; 2HNZ B; 2VQF F; 1SH0 A; 1N36 F; 4I6A A; 1FRM A; 4KJI A; 3HHK A; 1RK8 A; 5F9L A; 3RB4 A; 3BRE A; 2YKP A; 3JTO A; 1FO1 A; 2GIQ A; 5D3G A; 1RT2 A; 1N0U A; 3DLE A; 4PXC A; 5L1K A; 3BGF A; 3LOU A; 5GAH I; 3CC7 S; 3PX0 A; 1WEL A; 2PII A; 2KKH A; 4NPO A; 2N7C A; 1LV5 A; 1MU2 B; 3ITH B; 4N8O A; 3KHL A; 2G9O A; 4WYL A; 4KGK A; 3TVK A; 3TRG A; 2DR7 A; 1UAW A; 3PW0 A; 5E7O B; 4DR2 F; 5F8N A; 4I1T A; 5F8G A; 2GW8 A; 2L9W A; 3PNI A; 4IYQ A; 1RWU A; 4C7Q A; 1DLO A; 3ZZO A; 2LXF A; 3VR1 A; 3KLG B; 4LF8 F; 4NA1 A; 4QPT A; 5DO2 A; 3MR2 A; 1XWL A; 3CGI A; 4FF2 A; 3L3C A; 4R5P B; 2DGO A; 3CVK A; 4NL5 A; 2B0G A; 2CPE A; 1FKP B; 3I5A A; 3MHY A; 4NLT A; 1U1K A; 4RNH A; 3NG8 A; 2MKS A; 4IG0 A; 1MWY A; 2CZV C; 3MED A; 3K58 A; 1RQ0 A; 1RAJ A; 4DV5 F; 5CYQ A; 5KFN A; 3PVX A; 3ISN C; 5F0U A; 3F41 A; 2MQP A; 1IQZ A; 1NHU A; 1WWH A; 2XMU A; 2UZA A; 3IXQ A; 2ACY A; 2I8E A; 3NMA A; 1YRX A; 2R7V A; 2KRR A; 2ZD1 B; 3IG1 A; 3D1H A; 1J5O B; 1S6Q B; 1YRT A; 4ZVH A; 5FD1 A; 2OFG X; 4ZDN A; 1VQN S; 4NYZ A; 4KHP F; 4IH5 A; 2LEA A; 2F1F A; 1NKB A; 4K4W A; 2YX1 A; 2B3T B; 2DGT A; 1X5O A; 3DCA A; 5DDQ C; 2VFV A; 4OJ1 A; 2PT0 A; 7FD1 A; 3N9U C; 3GIJ A; 3KLF B; 2YNG B; 1X0F A; 5B0A A; 1N33 F; 2MKH A; 5BYQ A; 4QWE A; 2RRN A; 5GVQ A; 4RUH A; 4OHY A; 2IRU A; 3KHH A; 2D74 B; 3OR1 B; 5LSO A; 2YNI A; 4I2P B; 4AUK A; 3SYZ A; 1S99 A; 3SQG B; 4AL7 A; 3FFI A; 3HVA A; 4QCL A; 2YKA A; 1SC6 A; 5GAG U; 2BYC A; 2CJA A; 2IC3 A; 4K5R A; 1TZ0 A; 4WFZ A; 3SCI E; 2KJW A; 1B0T A; 1MNE A; 3ISZ A; 2BQU A; 4NLS A; 2F8M A; 4F4W A; 4RW4 B; 2EFV A; 1U1Q A; 1WHX A; 2W9B A; 2QA1 A; 2ROE A; 3QAU A; 2R7X A; 3CUL A; 1JNR B; 4DV6 F; 3PKM A; 4X64 F; 1KRP A; 1UEU A; 4CO5 A; 5CYM B; 4OQJ A; 4IOC Q; 1RYS A; 3GSD A; 2PMH A; 4EZ9 A; 2FD2 A; 4E0D A; 4OJ3 A; 2CQ2 A; 2IMO A; 1JX4 A; 4RLE A; 1ZM2 A; 2YKM A; 5F8H A; 2JEF A; 3KKF A; 2FTR A; 2GML A; 2HGN A; 4MIA A; 4ZMM A; 3QWU A; 1N5T A; 4XIU A; 3T1H F; 3KJV B; 1VI7 A; 2V8G A; 3R1L A; 4YB7 A; 2J5A A; 4F50 A; 4NLX A; 4DF4 A; 2AYE A; 5FJ4 A; 1H7W A; 4CI0 B; 5KT6 A; 1FHT A; 3QLH A; 5T7L B; 4EZ6 A; 1X4F A; 2VFU A; 5L9N A; 2PSZ A; 2I5J B; 1UFW A; 3M8P A; 3UD4 P; 1RT3 B; 3PXG a; 2CQH A; 2KZZ A; 1JJH A; 5A8K B; 4QU6 A; 1OWX A; 4Q43 A; 2BQ3 A; 4TLQ A; 2NZ4 A; 4NZ0 A; 4CTA A; 1K1Q A; 1GTH A; 2LXU A; 3QZ8 A; 5K8M A; 2PMZ D; 1MW7 A; 2EBE A; 2Q3P A; 3JAM Y; 4W90 B; 3GII A; 4IRC A; 5GAE U; 2A3J A; 2WOM A; 1DTT A; 2J0S D; 2RIJ A; 4K4Y A; 3PY8 A; 1JLB A; 1MMN A; 5J2M A; 3H40 A; 2DNM A; 2QO3 A; 5HMX A; 3HDS A; 1N5Y A; 4QU7 A; 1WG1 A; 4EYI B; 1FKO B; 4WK1 A; 5F3Z A; 3V81 B; 5I3U A; 3IRX A; 1S6P B; 1XR7 A; 5E3A A; 4F26 A; 3KLH A; 5I3P A; 1XC9 A; 4FNI A; 2XY6 A; 2ZIT A; 1FRK A; 4X4S A; 2DCL A; 2C3P A; 3IR5 B; 1DQA A; 3OXF A; 4ES3 A; 2LXI A; 2V0N A; 1PC5 A; 2ZM6 F; 2HND A; 1RKI A; 4O3N A; 3U1N A; 2NR0 A; 4IUF A; 2RB7 A; 3IWX A; 3L95 X; 3FRZ A; 4IH7 A; 2KH9 A; 5IT9 Y; 1Y4Z B; 4ECU A; 2FIU A; 2G4B A; 4IR9 A; 4X4V A; 1KHW A; 3KLE A; 3PEH A; 1LW2 B; 1REV B; 3HKZ D; 4WYW A; 4Y0F A; 4NLU A; 3LP0 A; 3QAE A; 4LIR A; 3CC2 S; 1VBY A; 3M30 B; 5EHH A; 1O51 A; 2GMJ A; 1NEE A; 2I0X A; 2OPP A; 3JB9 k; 2IVY A; 1JLG A; 1YJR A; 3J7Z T; 4YB5 A; 1HWK A; 1M5S A; 4LQ3 A; 1TK8 A; 3UW1 A; 3TI0 A; 4KAI A; 1ZET A; 3THT A; 2HIQ A; 5IM0 B; 3O1L A; 1X4G A; 2DO0 A; 1SIZ A; 2BJE A; 4ZA1 A; 2ZZM A; 3IO1 A; 4DFK A; 4PPZ A; 2DNG A; 2BXJ A; 2JVR A; 4YDD B; 3DN9 A; 4DV3 F; 4LSN A; 3ZZU A; 3NMR A; 2F7V A; 4IG3 B; 4WXW A; 4FXD A; 2R7R A; 2QYC A; 2ZFH A; 3DLJ A; 3BGU A; 3OUY B; 3H98 A; 4J9O A; 3T5H A; 1VQL S; 1NZA A; 4MAK A; 2JRS A; 4LQ9 A; 5A8R A; 4I6Y A; 2CJK A; 1RT6 B; 4O4G B; 1O8B A; 5GAG I; 3HVW A; 3I4P A; 1NK6 A; 2MKJ A; 4NCA A; 4ECT A; 5KFM A; 1ULR A; 2LEP A; 2OML A; 4NLV A; 4ZVF A; 2K7J A; 3F56 A; 4KC5 A; 3I55 S; 1U09 A; 3V6D B; 3TAM A; 4ATN A; 1QSL A; 2GO9 A; 1D9D A; 4WTY A; 1UMJ A; 2BV3 A; 2W7P A; 4M94 A; 2CPJ A; 5KT5 A; 3MEG A; 2W9C B; 3NBP B; 5DM6 Q; 4A8F A; 1LQ9 A; 4TN2 A; 2X1B A; 3KG4 A; 4C11 A; 3CJK B; 1YJN S; 2BQR A; 1FTC A; 2MGZ B; 4F4K A; 2HQ3 A; 4X66 F; 2HS0 A; 4URQ U; 2AD9 A; 5KFL A; 4B9U A; 2SXL A; 2YNH B; 3HLU A; 4R5P A; 4QW9 A; 3ZFV A; 1SBR A; 2C42 A; 4Y2C A; 1KQF B; 1R8B A; 2DNH A; 1R3N A; 1RGV A; 1Y5I B; 2UXC F; 2LDY A; 5K8P A; 1D3W A; 3I9Z A; 4MKY A; 1FJC A; 4U7C A; 2YTC A; 1FJG F; 2PI5 A; 2BOP A; 1RTH A; 3H4D A; 2VG7 A; 3I6P A; 1LOU A; 1X5T A; 4XPE A; 2BAC A; 1N6Q A; 4I4B A; 4FYD A; 5CH7 B; 2V9W A; 2NSB A; 3QYY A; 2HYI B; 4JYA F; 1F0X A; 1S1U B; 2ZD1 A; 3N0V A; 4CNY A; 3CCL S; 1TKX B; 1P27 B; 4QR1 A; 1BQX A; 1JKH A; 3T9Z A; 1VFJ A; 1HI8 A; 3M4X A; 5A0Y B; 1WEZ A; 3SCL E; 5IS2 A; 3PJX A; 1OPZ A; 1YRU A; 2C2D A; 2FPH X; 2J7K A; 3NLC A; 4HH1 A; 4IFY B; 4QYR A; 4A8Y A; 4KHM A; 3DRS B; 3FDS A; 2FVC A; 3T3F A; 3RRG A; 4X4O A; 4F2R A; 2XSF A; 1JLF B; 1R7I A; 4B9T A; 4AFF A; 1Q81 T; 3O8W A; 1S10 A; 1VRU A; 3HVR A; 1UET A; 2JL9 A; 5GAD U; 1FXD A; 4I64 A; 4L3N A; 1VS3 A; 2A18 A; 2VBW A; 4C8L A; 1AFI A; 1HWJ A; 2BZ2 A; 4ED2 A; 2JEI A; 1Z2L A; 4B3O B; 1VIX A; 2HRY A; 1QAI A; 4NXM F; 2RQM A; 1HBM B; 1R8C A; 4NWB A; 4WII A; 4PXB A; 2DGW A; 2MDA A; 2V4J A; 2B9Y A; 5C25 A; 4MIW A; 2K1R A; 1XTZ A; 3CJ3 A; 2C2E A; 1FXR A; 4XK8 C; 3OJU A; 1UWB B; 3DRR A; 5CYM A; 2DR8 A; 2V8V A; 3D28 A; 1UFL A; 2FB0 A; 2AJ0 A; 3K5O A; 2Q1L A; 3J7A 1; 1TAU A; 1YI2 S; 4DN0 A; 4LFA F; 3BSO A; 3MM6 A; 4N8N A; 4DG1 B; 4PG4 A; 3O44 A; 3KJV A; 3Q8S B; 1DTQ B; 5L1I A; 4IOB A; 4RW7 B; 2MKI A; 1LK5 A; 3DRP B; 3VTI A; 2LVW A; 5T7L A; 2ZHA A; 2EVZ A; 1FVQ A; 3V4I B; 3LWM A; 3HK2 A; 4N47 A; 1C1B B; 2MZT A; 2WK4 A; 2ZHO A; 4LF9 F; 3OXL A; 1GXT A; 5B0F A; 1XMQ F; 4QIG A; 2IYG A; 3G8S A; 1V6H A; 2LQ5 A; 3MMC B; 4UUX A; 5A8W B; 2QSF A; 3CDE A; 3VQT A; 2AYM A; 4DSL A; 3H5X A; 3M32 A; 3M8R A; 4R0E A; 2Q79 A; 3IR7 B; 1CG2 A; 1S6U A; 1SKS A; 2PFF B; 4Q8F A; 1L3U A; 3BZQ A; 4R8U B; 2DPJ A; 1HD0 A; 2BRL A; 2KVI A; 1WEX A; 3W2V B; 4AXJ A; 2FHJ A; 3CD6 S; 2B43 A; 2J0Q D; 2W25 A; 4G1I A; 3F73 A; 2E5G A; 5KFX A; 1OSC A; 3GIL A; 1N48 A; 3MUR P; 3RRH A; 3C3L A; 2P9C A; 5KFQ A; 2RS8 A; 2MZR A; 1IJE B; 5KT7 A; 3BDP A; 3IWL A; 1BY9 A; 3R1H A; 3VAJ A; 4N56 A; 4C9D A; 2OPQ A; 2PC6 A; 3CT9 A; 5IQQ A; 3TAQ A; 5D78 A; 1UEV A; 2F9D A; 1FDN A; 1B64 A; 3M6V A; 5IT4 A; 2AJ4 A; 4MK8 A; 1MML A; 4GMK A; 3KLV A; 3IAM 9; 4A8S A; 1NK9 A; 1BGX T; 4N76 A; 2LQJ A; 1UL3 A; 3M9O B; 3CJ0 A; 3PKY A; 2PN6 A; 1KSP A; 3IWN C; 4KKO B; 3I0S A; 4ALZ A; 3ENW A; 1CQM A; 3OTB A; 1C0T A; 1HPZ B; 5KFS A; 3LW5 C; 3KG0 A; 4CH1 A; 1KQG B; 4LJM A; 5CZX A; 5U47 A; 4IRD A; 3DMJ A; 3NKY A; 5F0W A; 3CJ5 A; 2DNQ A; 2XMW A; 3K0J A; 4RKU C; 1UA1 A; 1H2U X; 4K4V A; 3T5J A; 4IQM A; 1X0P A; 2FGO A; 4ES2 A; 1U47 A; 2XO7 A; 2D9P A; 4KFB B; 5G0R B; 3NGK A; 2CVE A; 3DHX A; 1A6L A; 2X1G A; 4EFA C; 3E2E A; 1X4B A; 5C42 B; 5E33 A; 2VBX A; 3BQ0 A; 3AQO A; 4PXE A; 2V8H A; 4RNO A; 4G1Q A; 3R3T A; 1Y7P A; 3EPG A; 1XR5 A; 5IM0 A; 2OLW A; 5KFH A; 3SFG A; 3M8Q B; 4NZ7 A; 2KN4 A; 2RDI A; 2MZS A; 1HWL A; 2NUU G; 3OFH A; 3S8S A; 1IBK F; 3C46 A; 2PA8 D; 1RJJ A; 4OZ5 A; 3AY0 A; 4U9R A; 2CPI A; 2VBY A; 3G6E S; 1QSS A; 4JI4 F; 2LYV A; 4EO6 A; 1WG4 A; 1U6Z A; 5DJB A; 3PX4 A; 3GWC A; 4B9S A; 4LFC F; 4X4U A; 2UV8 A; 1ZAW A; 5EUH A; 4ETX A; 1R27 B; 3NGD A; 3DM2 A; 4DS5 A; 1SKW A; 3H2V E; 3PIP Q; 1N5S A; 5H1S V; 2KM8 C; 2XHW A; 3PW7 A; 2CZ4 A; 2DHA A; 5GAD I; 1VGY A; 1HL6 A; 1IKV A; 3PDN A; 1ZM4 A; 3D0G E; 3HI9 A; 5AE2 A; 2Z0G A; 3CCR S; 4K4X A; 1P6T A; 3KWM A; 4HEA 9; 3GFY A; 2DR9 A; 4RNF A; 5KFV A; 4IR1 A; 1N54 B; 2EFG A; 1Q8I A; 3W2W B; 3DOK A; 1LWE B; 5A6W A; 4Y65 A; 2MMY A; 1E6V A; 2KJV A; 4HT5 A; 2CYY A; 3CDA A; 3NBP A; 5GAH U; 3HHN B; 1T94 A; 2ABY A; 3QGP A; 1T03 A; 2E9T A; 2YWH A; 3CJK A; 2MGZ A; 5KFT A; 1AXQ A; 3MM7 B; 3MUM P; 2CQI A; 3IC1 A; 4XDD A; 3IFE A; 2JEJ A; 2IRY A; 1C0U B; 1DFD A; 2DNO A; 4RUC A; 1IKX B; 3HHL B; 4LVO P; 4I2Q B; 1QAJ A; 1RT1 B; 2N4D A; 1YQH A; 4ECZ A; 2V9K A; 1TUV A; 3QGH A; 4ZVE A; 1F60 B; 2OD4 A; 2JSX A; 2O5D A; 2DGQ A; 2J0W A; 4X4Q A; 2QIF A; 4DL6 A; 4ZS6 A; 4DL3 A; 1JWW A; 2C22 A; 1Q4R A; 3ISN D; 1F5C A; 3SEO A; 3MTK A; 5KFF A; 1S1U A; 4Y3C A; 4KR0 B; 2XLI A; 2ZH3 A; 4NYP A; 4A8Q A; 1VOM A; 1H6K X; 1KLM A; 4U6P A; 3OXG A; 3FSI A; 3QGI A; 3IS9 A; 2JC0 A; 2A1B A; 4WFY A; 4R25 A; 1HNX F; 4DDF A; 2RQ4 A; 4USJ C; 1Y3J A; 1S48 A; 2GQC A; 4FZV A; 4WU2 A; 3CCQ S; 2DR5 A; 2X3G A; 2FJD B; 2XMT A; 1SJF A; 4P7V A; 2MZJ A; 1GXL A; 1JLF A; 5HP1 A; 2W9A A; 1QUV A; 3MGJ A; 4HL9 A; 2AYG A; 2CG4 A; 3EXC X; 3VWS A; 3TA2 A; 2J7U A; 3LO3 A; 1RT7 B; 4OI2 A; 1UVJ A; 1NKE A; 2M52 A; 4B3O A; 2HQI A; 3LUY A; 2I9S A; 3E8O A; 3IN5 A; 1HBM A; 1Y00 A; 2E5I A; 2J0X A; 2XNR A; 5E3Q A; 3VZI A; 1VHF A; 3TO8 A; 2HVZ A; 2J6T A; 2CQD A; 2CQP A; 2DHG A; 1L5U A; 3HZE A; 2HHV A; 4O3S A; 2ZVS A; 5E5N A; 3MAH A; 1LWF B; 2A2D A; 3THP A; 4QZV B; 5HRO B; 1HQE B; 1ZOT A; 1JLC B; 2R2S A; 1TVR B; 2O1P A; 4OLP A; 3QZ7 A; 2XMV A; 3V6H A; 3TVZ A; 4G3I A; 2B6A B; 3RB3 A; 2AX0 A; 4F3O A; 5KOX A; 4EO8 A; 4RW9 B; 3OTO F; 2J6U A; 2FLL A; 5BYS A; 1S9F A; 1GAO A; 4RW7 A; 4WE1 A; 3DRP A; 3N79 A; 2DH8 A; 1YIJ S; 4LMZ A; 4IO9 Q; 2J9D E; 4PQU A; 4BWJ A; 3CF5 Q; 3R5D A; 1DHM A; 4RW6 B; 3OL8 A; 3BSC A; 2EBG A; 5KFR A; 2DGS A; 4H54 A; 4KBI A; 5FLX Y; 2CVI A; 3LP2 B; 2RJZ A; 1H7X A; 4M1K A; 3U02 A; 1HBO A; 3MMC A; 5E2Q A; 4JI6 F; 5A8W A; 2QLW A; 2JFD A; 5I42 B; 4WCE Q; 2GQ2 A; 2RF2 A; 3RW6 A; 3PZP A; 3GYX B; 3UDC A; 2NQP A; 2FVR A; 3DRX A; 3W2V A; 1NJI T; 2FHM A; 1Q86 T; 3EXY A; 4Q2L A; 3DOM A; 5DM7 Q; 1XR6 A; 5SUH A; 2KXH A; 5SZT A; 3HP6 A; 1FDD A; 2M8D B; 2OH2 A; 4QIV A; 4EUV A; 3MUT P; 3UCZ P; 5JXS A; 2POK A; 2DIT A; 3IR6 B; 2VQE F; 4JOU A; 2M70 A; 3UR0 A; 5FII A; 4ED8 A; 3DOL B; 3C2P A; 1Z4U A; 3BR9 A; 4N41 A; 1HQU B; 1DUR A; 4OI3 A; 2VPX B; 2ADC A; 3HM9 A; 4JV0 A; 4F4X A; 2DIS A; 2E1C A; 5ITH A; 1CVJ A; 2AGP A; 3JYT B; 3OJS A; 1SXL A; 1HPZ A; 1S9E B; 4URS A; 3E01 A; 5L38 A; 2KXN B; 2XS5 A; 4B3S F; 4AL5 A; 1Q78 A; 1T3O A; 4Z2X A; 3BDE A; 2HFN A; 3M1V A; 1MRO B; 1VMB A; 2BAB A; 3PAC A; 3OBI A; 4IFV A; 1IM4 A; 1U4B A; 5K14 B; 4PRF A; 2I1R A; 5F8L A; 2VOP A; 3M2R A; 1LXJ A; 1TR0 A; 1X4H A; 2GO8 A; 2HGL A; 3G86 A; 1ZTT A; 2JC1 A; 3JSR A; 2CPY A; 3NCQ A; 4RX6 A; 3C66 A; 1DBD A; 3UR3 C; 4DV7 F; 4HYK A; 1N0V C; 4WF9 Q; 3RB6 A; 4ILR A; 1HHT P; 1PC4 A; 3JTP A; 1SL1 A; 2E9Z A; 2R8I A; 1SJQ A; 2QE5 A; 1H38 A; 3H5S A; 3QJL A; 4PWD B; 1QUP A; 1HMV B; 3WDO A; 3DXS X; 5B7W A; 1NJZ A; 1RKJ A; 3THV A; 4UOC A; 2XWX A; 1VC6 A; 3M2V A; 5C42 A; 1LXN A; 1LW0 B; 5D8H B; 4J37 A; 2UUB F; 4QOT A; 5B0D A; 1SQE A; 3MM9 B; 4YR2 A; 1WF1 A; 1Y5L B; 1EET A; 5CHC B; 4LF5 F; 1S1W B; 2DCU B; 4K0K F; 2HHW A; 1NU4 A; 3LGN A; 1VQ4 S; 2RQK A; 1U1M A; 4P7T A; 2NXU A; 4ID8 A; 5J1E A; 3CCS S; 2LEC A; 4GC6 A; 2WSC C; 2VG6 B; 1WHY A; 1VQK S; 1WTF A; 2P6T A; 3C6T A; 3AH6 A; 4MJK A; 3KK2 B; 2Y1J A; 3M2U B; 2RD5 C; 2WB1 D; 1Q16 B; 4EYH B; 2OPR B; 4XAK A; 4TXS A; 1CC8 A; 1HWI A; 1HNW F; 1LK7 A; 1LFP A; 1S9G B; 1SKR A; 4ED7 A; 3DDN A; 4XO0 A; 2ZH8 A; 3G8Q A; 2ADB A; 2XCP A; 1NK5 A; 3OVS A; 2E29 A; 4DUY F; 1IBM F; 2N7Y A; 5J1N A; 2OO4 A; 2YKQ A; 2IBO A; 2M2B A; 3A1Y G; 3GQC A; 1ZTW A; 1S72 S; 5IFM A; 1QE1 B; 2BR0 A; 1P8G A; 2KFZ A; 1KFS A; 2AP6 A; 4LF6 F; 4ECX A; 4X2B A; 1SI9 A; 5FDL B; 1TL1 B; 3ETO A; 2BE2 B; 5KAK A; 3I0R A; 4JI3 F; 4ECW A; 3RB0 A; 2DNP A; 1WI8 A; 2LKZ A; 2XC9 A; 2YKN A; 2CKW A; 3MM7 A; 4LUB A; 2KZM A; 2IJN A; 4CT8 A; 4OI0 A; 2D9O A; 2PA3 A; 3ZZT A; 2ILZ A; 1RTD A; 1RYR A; 3R5B A; 2GV1 A; 4G6V B; 4NLO A; 2XMJ A; 3ZZ0 A; 5HBM A; 4DR3 F; 2Z2L A; 3QWP A; 2KO1 A; 5KFU A; 3AHP A; 1IKX A; 2VA3 A; 1SUQ A; 2DBB A; 4I2Q A; 1WNE A; 4DV1 F; 4NLP A; 5KFA A; 2BUN A; 1HBN B; 2RSQ A; 1XMB A; 4FF1 A; 4WFX A; 3I56 S; 4BC7 A; 3RAX A; 2B9W A; 2HLU A; 3TUJ C; 4ELU A; 1FCA A; 4ZHR B; 1M5H A; 2PI4 A; 4Y2I A; 3LAK A; 2RIL A; 2VPW B; 1H2T Z; 2YX4 A; 3DLG B; 5HQ8 A; 2KFY A; 4EBC A; 2QV6 A; 4JI5 F; 3VF0 B; 3VAK A; 1HI0 P; 1YG0 A; 5A2Q Y; 2MHN A; 1X9S A; 4C2M A; 4J9L A; 1YIT S; 1HD1 A; 3NL0 A; 3OL6 A; 2DGV A; 2R7W A; 2DTJ A; 3MMA B; 5K5M A; 1X9W A; 1XNR F; 1YJU A; 3MEC B; 1WHW A; 3N5F A; 2UUW A; 2JOQ A; 1JLQ B; 2XS7 A; 4CNZ A; 1DZ5 A; 4PPD A; 2MST A; 4I7F A; 2OMO A; 5LT5 A; 2RKI A; 1VQ7 S; 1HBU B; 4LFB F; 1IKW B; 4QBU A; 2WB4 A; 2E9R X; 1RT7 A; 2R8H A; 1M1K T; 2L48 A; 2R7Q A; 1GH8 A; 2E7X A; 2BAN B; 2C3Y A; 2RNE A; 4LSL A; 4YUD A; 2XMK A; 2VPY B; 1N5Q A; 2GC8 A; 2L3M A; 1TFY A; 4TU9 A; 2AX1 A; 4J9K A; 3OHA A; 4E8U A; 4C8Y A; 1CEZ A; 1UJ6 A; 1FA0 A; 2VUG A; 5IXU A; 2WRM A; 2WSE C; 5M2P A; 1QSY A; 4WZQ A; 2LLZ A; 1HHS A; 2RUG A; 3DLK A; 3MR5 A; 3LAM A; 3N28 A; 4MFB A; 3S01 A; 2B4P A; 1V3S A; 5LJ5 Y; 5J2P A; 2XY7 A; 4DR7 F; 1Y0H A; 3H2L A; 3FJ2 A; 4AXI A; 1U7L A; 3IA0 A; 3PJW A; 1RT5 B; 1HQE A; 2GIR A; 3AB4 B; 1TVR A; 3MEE B; 5KFO A; 4RUA A; 3DYA B; 5KFJ A; 3OR2 A; 4EEY A; 1C1C B; 4OZ0 A; 1UW4 A; 3HKW A; 3I5B A; 5D77 A; 3MM8 A; 2RE1 A; 2L9N A; 3GFH A; 2Q6C A; 2E5H A; 3AAW B; 4U67 Q; 3TX8 A; 3ICL A; 1SV5 A; 1ZPW X; 1S1V A; 2C28 A; 1CSJ A; 2X51 A; 5BYR A; 4EU0 A; 4DLE A; 4NLR A; 3CSK A; 4DS4 A; 4J9R A; 1M5P C; 3VQS A; 3KLI B; 3OLB A; 4PXD A; 4I0W A; 5H9F J; 3OFE A; 2IRX A; 2FJX A; 2UVU A; 3LP2 A; 2DNR A; 5JJB A; 2LU2 A; 1HWU A; 2JHE A; 1S1X A; 3M05 A; 4YME A; 1FNX H; 3DLH A; 2K8G A; 4OLO A; 4DL0 C; 2B7B B; 2GMH A; 4BHQ A; 4Y2K A; 4N5S A; 1F5B A; 2KYZ A; 5JUM A; 4JV2 A; 1LWC B; 3NNA A; 4MIB A; 1I94 F; 2B7C B; 2HPU A; 5CA5 A; 5KFD A; 1Y3K A; 2VAS A; 4Q44 A; 5IWA F; 1U45 A; 4QR2 A; 4X65 F; 3H25 A; 3RR8 A; 1R6Y A; 2GHP A; 3V5R A; 5J2Q B; 1VBX A; 1FK9 B; 4PG5 A; 3RAM A; 2HNZ A; 4CO0 A; 1VQ5 S; 4QIW D; 1JLA B; 2E1A A; 2QMX A; 1A9N B; 1D9A A; 2QSW A; 1KFD A; 2R8J A; 3M9M B; 3K8V A; 2QT7 A; 2B56 A; 5BR8 F; 3IGV A; 1QTM A; 3OVB A; 4DSE A; 4X4N A; 3JYT A; 2ZE2 B; 5J2N B; 5M2R A; 1MU2 A; 3ITH A; 2LA6 A; 3W7B A; 3FSY A; 1QY7 A; 4F67 A; 3TAN A; 2B4U A; 1VQM S; 4QW8 A; 4DQS A; 3WC0 A; 4Q7A A; 2PO4 A; 5JVH Q; 3J7Z 5; 1S76 D; 1R43 A; 1UJ4 A; 2CFX A; 2IJF A; 2MXY A; 5DET A; 1FRJ A; 1T02 A; 2XLK A; 1MRO A; 5D4P A; 5KG1 A; 2J8A A; 2UXB F; 3GFZ A; 4K2X A; 3KLG A; 3KG1 A; 3CIM A; 2DO4 A; 3DI6 B; 5CXT A; 3BSN A; 2MPU A; 2QMW A; 1L3T A; 4RWX A; 3VAI A; 1S0M A; 2VG5 B; 3SDE B; 3TP2 A; 1FKP A; 4B3T F; 2FHJ D; 1FKA F; 3HVT B; 1YVF A; 3SZ2 A; 4PWD A; 3H2U B; 4TKT A; 1VC5 A; 2GQQ A; 5AE3 A; 2K3K A; 5LSL A; 2R2T A; 1NJY A; 2OPS B; 3CCT A; 4RW8 B; 5LMU F; 2O67 A; 2MQL A; 2MQM A; 3OSN A; 3MM9 A; 4AL6 A; 4DF8 A; 1FEH A; 2VKR A; 1P1T A; 3U27 A; 1HYS B; 2H5E A; 2HRU A; 4QIE A; 1S1W A; 3G6Y A; 2RU3 A; 4Q0B B; 2BM0 A; 5EX0 A; 4OZN A; 1N4L A; 3WC2 A; 1KVI A; 1WG5 A; 1S6Q A; 3ULH A; 4DQR A; 5DQH A; 1J5O A; 2UUT A; 4O44 B; 5HLF B; 4OL8 A; 1HAR A; 2VG6 A; 5E41 A; 1GT8 A; 4O3P A; 3UNG C; 3O2I A; 1YJW S; 1JLE B; 2JX2 A; 1S0N A; 1DQ8 A; 4D3H A; 1BQM B; 1DJ0 A; 4UY8 T; 2FC9 A; 2HHX A; 1GQE A; 2MSS A; 2YNG A; 3LP1 B; 5LL6 d; 3KLF A; 1OPI A; 1RT4 B; 3QGG A; 4GC7 A; 1S9G A; 2DIU A; 4NLG A; 3V6K A; 2LQB A; 3CJ2 A; 3T6B A; 1KVJ A; 3PFO A; 1TKD A; 1TL3 B; 2YH0 A; 1O5J A; 3OR1 A; 5IT0 A; 1K73 T; 4ZP6 A; 3GR1 A; 1TAQ A; 2B4V A; 4I2P A; 4KO0 B; 3CE8 A; 1G7C B; 4OHX A; 3A8E A; 2NRE A; 1X4A A; 2HHP A; 5DS7 A; 3SQG A; 3SXL A; 2IM1 A; 1QE1 A; 2RQC A; 3OPK A; 2Y0S D; 4KPY A; 2JLF A; 2WCX A; 4MYU A; 3SMZ A; 1NH8 A; 2MKK A; 4KTQ A; 2XI3 A; 2FLP A; 3E51 A; 1YTY A; 2R9L A; 4RW4 A; 2GJ2 A; 4B9L A; 3LAN A; 3NRB A; 4JV1 A; 2BE2 A; 2HS4 A; 1JJ4 A; 1NK8 A; 2IM3 A; 4QWB A; 2GGP B; 3K7R A; 5B08 A; 4RBV A; 3EX7 B; 1GNK A; 4M5D B; 1OQ3 A; 5AXK A; 4O23 A; 3SI8 A; 2UVW A; 4GKJ F; 3NCR A; 1Z1Z A; 4I6W A; 1U24 A; 1YGY A; 4A4J A; 1KN6 A; 3PR5 B; 3BSA A; 4ERD A; 2B5J B; 5DG9 A; 4K50 A; 1L3V A; 3JSM B; 2UVR A; 4ILM A; 2M3D A; 5L9X A; 4BBY A; 2ZH1 A; 4ZHR A; 2MKC A; 4HDG A; 1VRT B; 2NPF A; 4DSJ A; 4UY8 5; 2HHT A; 1KQK A; 3MMB B; 5KT4 A; 3NAI A; 1JK9 B; 2EG1 A; 3RRK A; 1HW9 A; 1N5V A; 1QAY A; 1HFE L; 2GHW A; 3MM5 B; 2ZDO A; 4DLG A; 2I5J A; 3AJ1 A; 3HXM A; 1RT3 A; 2ZBC A; 5M2U A; 3POT B; 5A8K A; 4Y6I A; 5L39 A; 3CCU S; 4YFU A; 4AQY F; 3G9C A; 4CT9 A; 2ROG A; 2ZOG A; 2DNK A; 4N77 A; 1SJR A; 4WFN Q; 4CCH A; 4RZR A; 2OD6 A; 6FD1 A; 4J9S A; 2ATL A; 3M6U A; 2VBZ A; 2CQB A; 2XY5 A; 2RDJ A; 2VB6 A; 2UUA F; 2XB2 D; 1T3N A; 2YNF B; 4IOA Q; 5HI7 A; 1TQL A; 3FEZ A; 5ADZ A; 4IQX A; 1T95 A; 3RYL A; 1KTQ A; 3E3X A; 1J2V A; 1HW8 A; 1QM9 A; 1TKT B; 1IXK A; 2DB1 A; 5KFW A; 1FD8 A; 2JH3 A; 2XG8 A; 2ZBI A; 1Q7Y T; 3C6U B; 1M5K C; 1FKO A; 3M6X A; 3Q22 A; 3V81 A; 2YKO A; 1S6P A; 1SJ1 A; 2BAN A; 4B17 A; 4Z37 A; 3G96 A; 4DFP A; 1D8Z A; 4FPR A; 3S6E A; 2DNZ A; 2IFX A; 2FVP A; 4ED0 A; 1VQP S; 2E5L F; 2EFQ A; 1QVF R; 4F4Y A; 1WD6 A; 4DFJ A; 3MI7 X; 2KHD A; 2ASD A; 5L8R C;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...