The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
155
|
sequence length |
453
|
structure length |
439
|
Chain Sequence |
MILKTNLFGHTYQFKSITDVLAKANEEKSGDRLAGVAAESAEERVAAKVVLSKMTLGDLRNNPVVPYETDEVTRIIQDQVNDRIHDSIKNWTVEELREWILDHKTTDADIKRVARGLTSEIIAAVTKLMSNLDLIYGAKKIRVIAHANTTIGLPGTFSARLQPNHPTDDPDGILASLMEGLTYGIGDAVIGLNPVDDSTDSVVRLLNKFEEFRSKWDVPTQTCVLAHVKTQMEAMRRGAPTGLVFQSIAGSEKGNTAFGFDGATIEEARQLALQSGAATGPNVMYFETGQGSFGVDQVTMEARCYGFAKKFDPFLVNTVVGFYDSKQVIRAGLEDHFMGKLTGISMGCDVCYTNHMADQNDVENLSVLLTAAGCNFIMGIPHGDDVMLNYQTTGYHETATLRELFGLKPIKEFDQWMEKMGFSENGKLTSRAGDASIFL
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
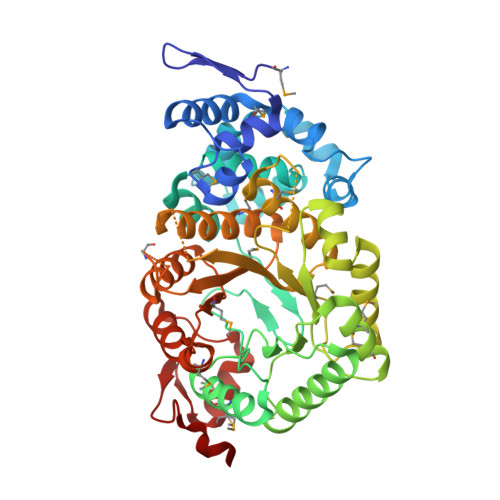
Crystal structure of ethanolamine ammonia-lyase heavy chain (YP_013784.1) from Listeria monocytogenes 4b F2365 at 2.15 A resolution
rcsb |
| molecule keywords |
Ethanolamine ammonia-lyase heavy chain
|
| molecule tags |
Lyase
|
| source organism |
Listeria monocytogenes serotype 4b
|
| total genus |
155
|
| structure length |
439
|
| sequence length |
453
|
| chains with identical sequence |
B, C, D, E, F
|
| ec nomenclature |
ec
4.3.1.7: Ethanolamine ammonia-lyase. |
| pdb deposition date | 2007-06-26 |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Alpha | Orthogonal Bundle | Annexin V; domain 1 | lyase | ||
| Mainly Beta | Roll | Ribosomal Protein L24e; Chain: T; | ethanolamine ammonia-lyase heavy chain domain like | ||
| Alpha Beta | Alpha-Beta Barrel | TIM Barrel | Aldolase class I |
#chains in the Genus database with same CATH superfamily 3A21 A; 3B0V C; 4IXX A; 4JTG A; 2DZX A; 1K7F A; 4O17 A; 2CU0 A; 4QAI A; 5D2T A; 4PH6 A; 5I3J A; 2I1O A; 2J6X A; 4B5N A; 5FI8 A; 2QUT A; 3ZWT A; 2EGZ A; 5G4E A; 3LBM A; 4J9J A; 3M9Y A; 3P80 A; 2BXV A; 1LOS A; 1YYA A; 3KVK A; 3Q2D A; 1KTB A; 1Q3N A; 4LVB A; 4HNV A; 1UBS A; 3UWZ A; 4MYA A; 3MJY A; 4JGD A; 2DJX A; 3G1H A; 2Z2U A; 3HQ1 A; 1LBF A; 1KA9 F; 3W2M A; 3BG5 A; 5BOO A; 3N9R A; 3F1Q A; 3EWW A; 5DY2 A; 2YYU A; 5I3H A; 4UTL A; 4OE7 A; 4ONV A; 2TMD A; 3S5Z A; 3KXQ A; 3MI3 A; 3STC A; 2ZRV A; 1TQJ A; 2AAM A; 1MEW A; 1R2S A; 3W7Q A; 4JTT A; 2QUV A; 2GVG A; 8TIM A; 3N2X A; 4N02 A; 3LTP A; 1H50 A; 2WNQ A; 1JCL A; 3STE A; 3EKZ A; 1LTD A; 5G2H A; 2C6Q A; 2O7Q A; 4NJK A; 4AHO A; 3ZR4 A; 4QIU A; 4JTJ A; 5MHB A; 2WOQ A; 1S5W A; 3Q58 A; 1YPI A; 3W7G A; 2VXN A; 1XBX A; 5C54 A; 4OHQ A; 1FDJ A; 3M44 A; 5D33 A; 2CLO A; 1OFP A; 3SGU A; 3LCL A; 1R30 A; 1UUO A; 2A0N A; 1RZM A; 4JN6 A; 4JTF A; 1D3H A; 1R47 A; 3EX2 A; 2ATS A; 2DU2 A; 2VOM A; 3DFT A; 3HG2 A; 1OYC A; 5EMU A; 1DQW A; 1WXJ A; 3NQ6 A; 1SW7 A; 1KFJ A; 1YDV A; 4MZ1 A; 3DFQ A; 4K37 A; 3WJS A; 3QC3 A; 4WXZ A; 4POD A; 3LCW A; 4AF0 A; 4DF2 A; 1K03 A; 4O12 A; 3LBC A; 3FYP A; 4RZ4 A; 4A2R A; 4H6K A; 3IIP A; 2A6L A; 3NQD A; 2YYT A; 1LCO A; 5FF7 A; 3W07 A; 4XTD A; 2OT0 A; 4IMD A; 4HNN A; 5D2W A; 5G2W A; 1VHC A; 4S1F A; 1GQN A; 3UZJ A; 3TQK A; 3NQ8 A; 4N4P A; 4ZQM A; 1ZAH A; 3LM7 A; 1KM5 A; 3HGO A; 1F6K A; 4ICN A; 2CZD A; 1IIG A; 4POC A; 3NPV A; 4FNP A; 1SO5 A; 2WSQ A; 3C56 A; 1EP2 A; 3R5E A; 4O57 A; 1OYB A; 3G8E A; 1NEY A; 3DFN A; 4OBT A; 4E2B A; 4MIM A; 2ZZ2 A; 5HWN A; 3RLU A; 1Q6Q A; 1PS9 A; 2J27 A; 3ATY A; 3KRS A; 5LKY A; 5K4X A; 3T55 A; 1ZNN A; 2WNZ A; 2G95 A; 3GKF A; 3I65 A; 2PRH A; 4Z1C A; 5DBU A; 4Y96 A; 4LVF A; 1EA0 A; 1EPX A; 4KWW A; 1WOA A; 1H63 A; 3QMT A; 2FLI A; 5H2Z A; 4AAJ A; 5D04 C; 2YR1 A; 3UWU A; 2H3B A; 4AHP A; 5AYZ A; 4UFM A; 2V5L A; 3NNT A; 3P74 A; 5BW6 A; 2J9X A; 1SSG A; 1F73 A; 3IIX A; 4GFS A; 4MZ8 A; 4DEL A; 1A5A A; 4R34 A; 3VE9 A; 2QAP A; 2RDW A; 1PV8 A; 4XTI A; 1M5W A; 1SZE A; 2IQT A; 4LC8 A; 5AHE A; 3TUU A; 1YLV A; 2NUW A; 3NDO A; 1IF2 A; 2NXG A; 3C61 A; 3L68 A; 1TPF A; 3W7E A; 3DXI A; 1WDW A; 3N14 A; 3P5Y A; 3W22 A; 4AH7 A; 4UXD A; 1OVD A; 4GWE A; 5FEX A; 4RXF A; 4M6V A; 1VRD A; 4FXS A; 3S1X A; 1FY6 A; 1K7E A; 3SZ8 A; 2Y70 A; 4O28 A; 4Z0J A; 3PSW A; 3LB0 A; 3E12 A; 1TPV A; 2Q8L A; 3WJZ A; 4ORI A; 1KFE A; 4UCG A; 1FBA A; 2ZYF A; 1JCX A; 4HT3 A; 4IPI A; 4XBK A; 1BKS A; 1R2T A; 2CFF A; 5F2M A; 4P2V A; 3MI2 A; 5LNW A; 1FDY A; 3FA5 A; 3ELF A; 4WY0 A; 3UZ5 A; 2E09 A; 3GDL A; 1T99 A; 4YCP A; 4PC8 A; 4M6P A; 4LS2 A; 4UTJ A; 3W6Y A; 5AHN A; 1A53 A; 4LUJ A; 3FEM A; 3L65 A; 3R89 A; 3WK1 A; 1X1O A; 3HG4 A; 5FEW A; 4N2Y A; 3C6C A; 3NL2 A; 2CW6 A; 3W88 A; 1UAS A; 1W5O A; 3NQ2 A; 4IX2 A; 3EWY A; 1I2O A; 4N9B A; 1K02 A; 3CIW A; 3HO8 A; 3LTY A; 3GAK A; 2QCF A; 1P5B A; 4UUI A; 4MJM A; 3BVJ A; 2QAF A; 1JQV A; 5GVJ A; 1QO2 A; 1OAB A; 3CQ0 A; 2C0A A; 4LUI A; 3C2V A; 1EB3 A; 3N2M A; 1QAP A; 5HR6 A; 1FWS A; 1X7F A; 4OV4 A; 3G0X A; 3KX6 A; 1I2P A; 3GND A; 1WAU A; 4LC6 A; 4UTK A; 4QJ1 A; 4UMB A; 3LHZ A; 1VCF A; 1V7Y A; 1VPX A; 2Y85 A; 2NX9 A; 3PM6 A; 5LNR A; 6ALD A; 1DBT A; 3PC0 A; 4IX0 A; 4PFM A; 7TIM A; 2RDT A; 1I4N A; 2NLI A; 3EWX A; 3EWZ A; 3MP5 A; 3VND A; 1G4P A; 3IGS A; 1JR1 A; 2JKE A; 2QDG A; 1AL8 A; 3HGS A; 2X30 A; 3KOF A; 5HMS A; 4GE8 A; 3DFO A; 1QO5 A; 5D38 A; 5D2X A; 3TSB A; 1GVF A; 2Z6I A; 4Q4K A; 3W1M A; 1JUB A; 1G4T A; 3EX6 A; 2VC6 A; 1DL3 A; 3P8J A; 1JQX A; 3B06 A; 4NNC A; 3QSR A; 2ISV A; 2E68 A; 4C1L A; 4Z1A A; 4EWN D; 1LRQ A; 4PA8 A; 3G1F A; 3M0Z A; 4W9T A; 2OCZ A; 5AHM A; 5FF2 A; 4JYE A; 1QDS A; 4X3Z A; 3QF0 A; 2R8W A; 4UFL A; 3W86 A; 4TIM A; 5G4W A; 1W3I A; 2BWG A; 2A5H A; 4NJG A; 4E2D A; 5HUL A; 3W7O A; 2NXH A; 4DPP A; 2VEK A; 3LLD A; 3PBW A; 4A22 A; 4EIV A; 2YFN A; 2WKJ A; 3RK8 A; 3O05 A; 3QRH A; 1AW5 A; 2E6F A; 2HS6 A; 3GXN A; 1YXD A; 2QCG A; 3G0U A; 3W76 A; 3T78 A; 1OFD A; 1MEI A; 3S8H A; 1OF8 A; 4NQ1 A; 3OEX A; 3UWQ A; 1LBM A; 3N9S A; 5LZL A; 3PG9 A; 3U6W A; 4NU7 A; 3CB8 A; 1TTJ A; 4JTH A; 2W6R A; 1EEP A; 2ZRZ A; 3EKL A; 3L66 A; 4MFD A; 2GPT A; 2QEZ A; 4MUZ A; 2V2D A; 5LNV A; 1TPB 1; 3TX9 A; 2V81 A; 3BG9 A; 3OA3 A; 4GXM A; 3G3M A; 1A5B A; 5LNS A; 1SFL A; 3OBK A; 4EOU A; 1DVJ A; 5J5D A; 2ZBT A; 2A6N A; 1ICP A; 1IGS A; 1FCB A; 4N9E A; 5KIN A; 1OFO A; 4O19 A; 1KTN A; 4ZZ9 A; 4JD4 A; 5FF3 A; 3R13 A; 3W73 A; 2E5B A; 2XFW A; 1P1X A; 1F2J A; 4Y90 A; 2C3Z A; 5EKY A; 2WSR A; 3S42 A; 4A21 A; 3P61 A; 3DEN A; 4CNP A; 4Q32 A; 4O1A A; 3U8G A; 1DOS A; 1YDN A; 2QCC A; 3TML A; 5CZT A; 5BNK A; 2Z0I A; 2VFI A; 3GDK A; 4TO8 A; 4R33 A; 2FIQ A; 3TNO A; 1BWL A; 1OFA A; 2C15 A; 4HSO A; 4EF9 A; 2QCH A; 3BLF A; 1A50 A; 5D09 A; 2Y7F A; 3DKJ A; 2JGY A; 1T96 A; 4JYF A; 1TPD A; 2PRM A; 1LLZ A; 1H62 A; 4ZQN A; 3W2U A; 4AHQ A; 1B4K A; 3NM3 A; 4O0Z A; 5T26 A; 3MHG A; 3JR2 A; 1LOR A; 3RMJ A; 3S5O A; 3P84 A; 2Y63 A; 2EAW A; 1WV2 A; 3KRZ A; 5SWU A; 4UR8 A; 2ZA1 A; 1UUM A; 5K9X A; 3GX9 A; 3RND A; 3MBD A; 1VH7 A; 3CC1 A; 3ZS4 A; 2P0O A; 2NWS A; 4K8H A; 3LOT A; 4MYX A; 1BEU A; 3W1P A; 4GRS A; 1FWN A; 3QPY A; 5HWM A; 2YXG A; 3IV3 A; 3TCE A; 2A7R A; 3MYP A; 1SUX A; 1NAL 1; 3BV4 A; 1QPR A; 3LHY A; 3ZFH A; 1VGA A; 4N9C A; 5K9C A; 4HNU A; 4DO6 A; 1UB3 A; 3DI1 A; 4IME A; 5KZD A; 2AGK A; 4M19 A; 1TV5 A; 4FXR A; 1VZW A; 1KBI A; 3LXB A; 4CLM A; 5AC8 A; 3M7W A; 4ORM A; 1T0O A; 1VQT A; 1LRN A; 1HO4 A; 1CW2 A; 1TTQ A; 1WOB A; 1LM1 A; 3O07 A; 4JTS A; 3NO5 A; 4ZQO A; 4XKY B; 5A5W A; 1FWW A; 3O1N A; 3E96 A; 4HPX A; 3W7H A; 2W0U A; 3PB0 A; 3W83 A; 5KTL A; 4ALD A; 3A22 A; 3INP A; 5E93 A; 4UTW C; 3HG3 A; 4X2R A; 1RQE A; 3W23 A; 3LGE A; 1Q6R A; 3W7R A; 3LV5 A; 1TTI A; 4JY9 A; 2GQ8 A; 4FO4 A; 3V1P A; 3UPB A; 1W0M A; 4ZVJ A; 5DBT A; 3LXA A; 5KSW A; 4MY8 A; 2J24 A; 4FF7 A; 3LTS A; 3M5X A; 1RQH A; 4IMC A; 3HIJ A; 5L9F A; 2JKP A; 1NF0 A; 4ADT A; 2FPV A; 4YLW A; 2E6A A; 4IQL A; 3DBP A; 2C16 A; 3NZ1 A; 1OX4 A; 3W1Q A; 1GZG A; 1LRO A; 1YAD A; 4GUJ A; 2C1H A; 3ATZ A; 3NQM A; 1LLW A; 1SZG A; 2PUR A; 3GDT A; 4CQ8 A; 2EPH A; 4UTI A; 5D05 A; 1OX6 A; 1EWD A; 3CM4 A; 4JTI A; 2X16 A; 3R8R A; 2WV8 A; 4YWI A; 3BW3 A; 4NVT A; 2VEI A; 1ONR A; 5IM5 1; 3SR7 A; 3DFP A; 4O15 A; 3C8F A; 2V5B A; 2HS8 A; 2Y7D A; 4FHA A; 1AW1 A; 2UV8 G; 4KFN A; 3FS2 A; 5TBO A; 1LBL A; 1WA3 A; 3N34 A; 4JX5 A; 2Y61 A; 5F2I A; 4BFA A; 4MVA A; 1LDC A; 1L6Y A; 1ZHA A; 2I14 A; 5FEZ A; 3TC6 A; 1ZFJ A; 3DZ1 A; 2E1D A; 5DEL A; 3HPX A; 4DXV A; 1MO0 A; 3P5Z A; 4IXH A; 4JXC A; 3MUX A; 2C14 A; 3CIX A; 3LHT A; 2Y6Z A; 2Q3R A; 4CCC A; 2ZDR A; 3IGX A; 1IIH A; 3CTL A; 3K30 A; 3HPZ A; 1HG3 A; 3F4W A; 3LLF A; 2TYS A; 5K9D A; 3QH2 A; 1PHQ A; 3TXV A; 2B7P A; 1YTK A; 1OX5 A; 1TSI A; 1W54 A; 1H1Z A; 4NX5 A; 4C1K A; 3GR7 A; 2QUU A; 2DP3 A; 1TJP A; 3NQG A; 4JIC A; 3PBV A; 2BLE A; 4GC4 A; 1QML A; 5DN1 A; 4TR9 A; 3GR8 A; 3GXT A; 3GYE A; 3DU0 A; 4AMA A; 1TPE A; 2I9E A; 1KTC A; 1FWR A; 3SIZ A; 2JKA A; 3E49 A; 3HBL A; 4NXK A; 1H7O A; 4OT7 A; 1CI1 A; 2ABB A; 2A1Y A; 4MKN A; 5EYW A; 5C55 A; 1O94 A; 3CPR A; 3FJ6 A; 2X1U A; 1FX6 A; 2HTM A; 4Z08 A; 4RZ5 A; 3HPS A; 4H4I A; 4DO5 A; 3CU2 A; 1H7R A; 1Y0E A; 3TSD A; 1QPQ A; 2FQI A; 4HHP A; 3QPZ A; 3T7V A; 3Q94 A; 1VR6 A; 4QM1 A; 4OV9 A; 5FF0 A; 4WJE A; 1LZO A; 2CLK A; 1FXP A; 2JBM A; 3T40 A; 2VFE A; 4IJB A; 4ZQR A; 2PRL A; 5LHF A; 2ZA3 A; 4GJ1 A; 1LP6 A; 5F4X A; 1ZEN A; 3QFE A; 3BAR A; 4JX4 A; 4O54 A; 5CZJ A; 4NNB A; 3CEU A; 1F76 A; 2QJG A; 2OMA A; 2A85 A; 3W1X A; 3TIM A; 2ZTK A; 5F4S A; 4XWU A; 2ISS A; 4GUF A; 3UY8 A; 5IBX C; 5LHE A; 3SJ3 A; 3W3O A; 5J5R A; 2B7N A; 3G3D A; 1JUL A; 2DZS A; 3C3N A; 3S1U A; 2V2H A; 4O10 A; 3M5V A; 2BTM A; 5CGQ A; 3FYO A; 3MWA A; 3W70 A; 4JTU A; 1FCQ A; 2X1T A; 3G18 A; 2QCN A; 1F7B A; 3W7L A; 3GXP A; 1GTE A; 2QCE A; 1VLI A; 3TW6 A; 3SY5 A; 4IGH A; 3W2N A; 1THF D; 5DY3 A; 3LI0 A; 4WUI A; 3SSJ A; 3I7Q A; 5G1Y A; 3TH6 A; 2P1F A; 1SO6 A; 4YMZ A; 3LCX A; 2GQA A; 3LRM A; 2BSL A; 3C4U A; 2NUY A; 2WPB A; 3W1L A; 1QPO A; 2DOR A; 3JRK A; 2YDA A; 3L0N A; 4RXG A; 2QCM A; 3UQN A; 1SQ7 A; 2PE4 A; 3IVS A; 3LXC A; 3O16 A; 1ME9 A; 2NX3 A; 3BPW A; 1PE1 A; 5D32 A; 2WQP A; 4FIQ A; 4O1C A; 1ME8 A; 3A5F A; 4NJJ A; 3FJL A; 3W9Z A; 4XQ6 A; 4I7V A; 1L6S A; 3F4N A; 1N8F A; 3ABO A; 4LVG A; 4PTN A; 4LNY A; 4EF8 A; 2QR6 A; 1GYN A; 1KM6 A; 2Y88 A; 3MYO A; 1W3T A; 1W5N A; 1AL7 A; 3CEP A; 4YCO A; 1QR7 A; 3C2O A; 3W85 A; 4LVD A; 4ZL1 A; 5D2Y A; 1QNV A; 4MPQ A; 1XUU A; 1TO3 A; 5BMX A; 1VLW A; 2Y62 A; 4TMB A; 3KRU A; 3A23 A; 3L5A A; 3S5Y A; 2AQW A; 4K38 A; 4OU1 A; 3TJX A; 3BLE A; 5LNU A; 5K2Z A; 4TMC A; 1PCW A; 3FFS A; 1XFB A; 4FX7 A; 4UC5 A; 4K8E A; 1VJI A; 3OVR A; 3GZ3 A; 2QJI A; 5DCE A; 1W5Q A; 4O53 A; 4XZ9 A; 3ABQ A; 1MEH A; 1L2U A; 4DF0 A; 4O18 A; 1L9W A; 4QIT A; 1TRI A; 3Q37 A; 3G1V A; 3VFL A; 4MY9 A; 1TYG A; 4O50 A; 3NL5 A; 3P81 A; 4KFP A; 5AC7 A; 3NZR A; 1H7N A; 3QW4 B; 3NGJ A; 5CZS A; 1TTP A; 5EY5 A; 5TQL A; 3DHD A; 3PBY A; 3C0J A; 2ZRX A; 4M7S A; 3NG3 A; 1J2W A; 4LWW A; 3KAO A; 1FXQ A; 1BWK A; 2ZZ5 A; 3ABS A; 3VKJ A; 3W1A A; 3HJZ A; 1ADO A; 4O52 A; 2NXI A; 3WJW A; 1SZN A; 4K6A A; 3THA A; 1QWG A; 1H7W A; 5D2V A; 5H91 A; 3RFA A; 3NEV A; 3WJY A; 3HMJ G; 3UWV A; 3UYC A; 1N55 A; 1PHW A; 4DBE A; 3M5Y A; 3PAJ A; 3EX1 A; 3P8I A; 2ABA A; 1DQX A; 3EXS A; 4L4L A; 2ISW A; 2V2C A; 1GOX A; 1W5P A; 1F6P A; 1A5C A; 3NQV A; 4LT9 A; 1GVR A; 1UJP A; 1C29 A; 2E77 A; 4AB4 A; 3B8D A; 4NXS A; 5C6M A; 3M16 A; 1P4C A; 5KMY A; 5EBU A; 5F2G A; 1G7V A; 1KBJ A; 4ML9 A; 4JDY A; 2RH9 A; 3OVQ A; 4UTT A; 5C5Y A; 2G97 A; 3PUE A; 2CLF A; 4I9A A; 3L67 A; 2ZRW A; 1SO3 A; 1V5X A; 1MZH A; 3PUD A; 3W2J A; 1DJQ A; 1F74 A; 4JIP A; 4Z0S A; 3W7M A; 5LSM A; 3QZE A; 6TIM A; 5HIN A; 3NL6 A; 4BI5 A; 1PVN A; 1AW2 A; 4FIR A; 3KFT A; 3TMQ A; 1KFK A; 1TB3 A; 4CQ9 A; 1GEQ A; 3O6C A; 3CLM A; 3LDV A; 4IWW A; 1O5K A; 4TU1 A; 3VCR A; 5KIT A; 3PB2 A; 4LRT A; 5E0K A; 4G1K A; 1RD5 A; 3QW3 A; 1I8J A; 3ABR A; 1YPF A; 1SFJ A; 2BZN A; 5CSR A; 3FKR A; 1KM0 A; 4MLJ A; 2Z1B A; 1VC4 A; 4TX9 A; 3BO9 A; 3S5N A; 3NPW A; 5AB3 A; 3NQC A; 4R7J A; 3UXD A; 3B03 A; 3C2F A; 3G24 A; 3T4C A; 4QNW A; 4JY8 A; 1H5Y A; 5CZ0 A; 2ZZ7 A; 1E51 A; 1OX4 B; 1Q6O A; 4JR5 A; 1QCW A; 2V8Z A; 3WK2 A; 1YTD A; 5AOU A; 1GWJ A; 3UND A; 3BK0 A; 2RHG A; 1DOR A; 3I7R A; 1KFL A; 5AN7 A; 4O14 A; 4JTE A; 4Z1D A; 5D02 A; 1OFB A; 4RCX A; 2ZFA A; 4LS0 A; 2RKX A; 3RLV A; 3M43 A; 3NQE A; 1VCV A; 1WBH A; 3USB A; 4U28 A; 1FUY A; 1RV8 A; 3PSV A; 5FEP A; 1BTM A; 4FEZ A; 1OK6 A; 1O95 A; 4UFI A; 3GK0 A; 1ZCO A; 4IMG A; 2OJP A; 3OVP A; 3TRO A; 1SU5 A; 5BRB A; 3PY2 A; 4IPJ A; 4O13 A; 1W8S A; 5ABT A; 3NPX A; 2CZF A; 1JRC A; 3O63 A; 1GVS A; 4K7Y A; 1D3G A; 3S1V A; 5F4W A; 2FB3 A; 2J9Y A; 4UFJ A; 5EPD A; 3G1X A; 1JCJ A; 1ZAJ A; 4JTK A; 4Q33 A; 2F84 A; 4RJE A; 1Z41 A; 3L5L A; 3G1A A; 3H55 A; 4UMC A; 3W2L A; 4PEK A; 1LRT A; 3EWB X; 4X9S A; 1PII A; 1DV7 A; 4M6Q A; 3KVJ A; 2F7F A; 1XUZ A; 3LCI A; 3A9I A; 2VEP A; 2ZCG A; 3TJL A; 1LOL A; 5CPO A; 1F05 A; 3DKL A; 2ZRY A; 4LRS A; 3QQ0 A; 1U83 A; 1JCY A; 4O1D A; 5G2I A; 2E6D A; 3EXT A; 3MHU A; 2ZZ6 A; 1ZAI A; 4AF0 B; 1U5J A; 1Z44 A; 5CKS A; 4I7U A; 1TIM A; 4YXG A; 1M6J A; 4WOZ A; 2D73 A; 3PR2 A; 1G69 A; 1GG1 A; 2WNN A; 1IXO A; 3EEG A; 4QSH A; 2TPS A; 3QMS A; 5HNR A; 2J88 A; 3IGU A; 1TPC 1; 4UTM A; 1XC4 A; 3W7K A; 4NNA A; 3B05 A; 3TFX A; 1O5X A; 2QCD A; 3LCH A; 3NQ7 A; 2YW3 A; 3W7C A; 1KM1 A; 4BK9 A; 1YXY A; 2YW4 A; 2VFG A; 3SGZ A; 1I2Q A; 2HMC A; 1R2R A; 3A5V A; 1W31 A; 2NV1 A; 2CZE A; 3W7I A; 2V9D A; 4WY0 F; 1EP3 A; 2YFO A; 1I2N A; 3I68 A; 2H3D A; 3NOE A; 3GDR A; 1KV8 A; 3L2I A; 2C13 A; 1EWE A; 3WJX A; 4UFH A; 2VEL A; 4AWU A; 3EB2 A; 4M7T A; 4WZH A; 3O6Y X; 3O15 A; 1JCN A; 5C2X A; 1N7K A; 1W37 A; 2ALD A; 1M7O A; 2QDH A; 3DI0 A; 3TAO A; 4GUG A; 1RR2 A; 1H1Y A; 1VYP X; 2V0T A; 1I2R A; 2GJN A; 2Y7G A; 1JRB A; 4RX0 A; 3EX4 A; 3N3M A; 1EX5 A; 1OK4 A; 3W74 A; 5E1Q A; 1X6U A; 3PBU A; 2B7Q A; 4RR4 A; 1KM3 A; 3P60 A; 3LX9 A; 3MMT A; 3G1D A; 3RF9 A; 5I3G A; 1P0K A; 1XDL A; 5HJL A; 3UD6 A; 5I3F A; 3YPI A; 1EIX A; 3G1Y A; 2NWR A; 1O60 A; 4S2B A; 4UNL A; 4RZ6 A; 3TAK A; 3BG3 A; 4NT0 A; 3T44 A; 1GJP A; 3HGJ A; 2C18 A; 2YPI A; 3FOK A; 3B04 A; 3PF3 A; 4LW7 A; 3W7N A; 1TV7 A; 5CG7 A; 1SW3 A; 1JVN A; 3H54 A; 5I3I A; 1JUE A; 2H6R A; 2R94 A; 3TQV A; 1A5S A; 1XXX A; 1SZF A; 4UMA A; 3E9A A; 4R53 A; 3W7J A; 4IMF A; 2H8Z A; 3E02 A; 3AJX A; 3HB9 A; 4WOQ A; 5AHI A; 3E0I A; 3MP4 A; 1FCU A; 1Z48 A; 2A7N A; 1DJN A; 1TRE A; 3C2R A; 5K4Z A; 1HTI A; 4UNK A; 3RU6 A; 5EA9 A; 4ZMG A; 5LGX A; 1YTE A; 4A29 A; 2GUU A; 2X2G A; 2DZT A; 4ZQP A; 1K8X A; 2YGY A; 2ZTJ A; 1XM3 A; 5TIM A; 5HR7 A; 3LI1 A; 4WY0 B; 1SPQ A; 4A2S A; 1Q45 A; 2GVJ A; 4H3D A; 1VYR A; 2B7O A; 4LV9 A; 4BI6 A; 5K3V A; 1MXS A; 5HQE A; 3TU9 A; 3N19 B; 3W7P A; 3GIY A; 4JDB A; 4D2J A; 1SO4 A; 5CPN A; 1TPU A; 3OHI A; 2ZZ4 A; 2X1S A; 4UTU A; 4LU0 A; 2B4G A; 3W87 A; 4UR7 A; 1WQ5 A; 5G1T A; 2E5D A; 3QYQ A; 3B4U A; 1SSD A; 1QPN A; 4RKA A; 2VEM A; 4FX8 A; 2FTP A; 4WD0 A; 3P62 A; 3FLU A; 3IVU A; 3NYZ A; 1B4E A; 3GVG A; 4ADU A; 4NUW A; 1EP1 A; 1EUA A; 2OX1 A; 1ZJI A; 3SEC A; 2FFC A; 3IEB A; 3S0C A; 1VCG A; 2RDU A; 4WQ6 A; 5D37 A; 1TMH A; 3W72 A; 5U4N A; 5A8G A; 4XKY A; 1D9E A; 3KHJ A; 5AFD A; 1NF7 A; 2EHH A; 1GYL A; 2DJL A; 4DO4 A; 5D30 A; 1TV8 A; 5FF4 A; 3W1T A; 1LOQ A; 4CNO A; 3NXF A; 2ATM A; 2H90 A; 1UCW A; 4NJH A; 2YZR A; 4Y8F A; 1GVO A; 4BWL A; 3FIG A; 4RV8 A; 1FWT A; 3PS7 A; 1GVQ A; 5BMW A; 4UTT C; 2TRS A; 3HF3 A; 4X22 A; 3QST A; 2NX1 A; 1T8X A; 4LTS A; 3TC7 A; 1H51 A; 2E5C A; 2XN0 A; 1P0N A; 3PG8 A; 4QNE A; 3IIV A; 3LCG A; 1WYI A; 1S3H A; 4UTW A; 1FCV A; 1TPH 1; 1FQ0 A; 1XDM A; 1XKY A; 1H7X A; 2H8X A; 4LVA A; 2CZ5 A; 2CLE A; 1C9D A; 2OZ0 A; 3GLC A; 4QQ3 A; 3DGR A; 3LV6 A; 2P10 A; 3G8R A; 1KV5 A; 1OHL A; 4OWG A; 1TRD A; 1RVG A; 1H60 A; 4WXY A; 1ME7 A; 2VFF A; 3STF A; 1NVM A; 3W71 A; 3DFS A; 2NUX A; 3LHW A; 3TE9 A; 2VEN A; 2DZV A; 5DXX A; 2QKF A; 2Y7E A; 1TJR A; 4HPJ A; 2ZZ3 A; 1AG1 O; 3THQ A; 1ALD A; 3G22 A; 2E6Y A; 4BF9 A; 4CNN A; 3KVL A; 4PCF A; 3ZWS A; 2ZRU A; 3P7Y A; 1W1Z A; 5AHL A; 1KW1 A; 2G5W A; 1VHN A; 1DHP A; 4MLR A; 3CWN A; 1RPX A; 3PUL A; 5AYX A; 3GB6 A; 3TXZ A; 1XI3 A; 2YSW A; 4L4M A; 4Y6G A; 1B57 A; 1KLY A; 4K7V A; 5FES A; 1O4U A; 2YX0 A; 1IXQ A; 5F2L A; 3NQA A; 2FJK A; 3VE7 A; 5G5I A; 4IOT A; 3VI2 A; 1HUV A; 1K8Y A; 1W3N A; 4HN4 A; 3C2E A; 2V30 A; 4O4V A; 3BW4 A; 3HOJ A; 3TR2 A; 3LHV A; 2VFD A; 4PL2 A; 2J9Z A; 2R91 A; 2G96 A; 3F03 K; 4AXK A; 3G1S A; 2QF7 A; 5D03 A; 3LCF A; 1WBJ A; 5KZE A; 1AK5 A; 1KFC A; 1QOQ A; 4HIB A; 3O6D A; 3NVT A; 4LV4 A; 1SW0 A; 4GNJ A; 3BGG A; 1YDO A; 1GG0 A; 1KM2 A; 3BLI A; 3IRD A; 4FNT A; 3NPU A; 1TPW A; 2FPT A; 3STG A; 2YCE A; 4NJI A; 4OGZ A; 4N9D A; 5HUO A; 1F5Z A; 2R14 A; 2C19 A; 5F1U A; 3EX3 A; 3S6D A; 4N4Q A; 2CLM A; 5HWJ A; 3NL3 A; 4RLI A; 1SR9 A; 4U4M A; 2GQ9 A; 2O7S A; 4JNM A; 5KZM A; 3H53 A; 2ZZ1 A; 1QFE A; 4LOC A; 1R46 A; 3M6Y A; 1FDZ A; 3MO7 A; 3GKA A; 4MY1 A; 2GOU A; 3IIZ A; 4FNQ A; 4KFO A; 1G6C A; 3N73 A; 3DAQ A; 2B0M A; 3HG5 A; 2QJH A; 3LRL A; 2DZW A; 5L6U A; 4ADS A; 1VYS X; 3QQ1 A; 3GNN A; 1X8F A; 4BR1 A; 5U7S A; 3R12 A; 4OQV A; 1RQB A; 3L0K A; 2LLE A; 2NZL A; 3WK3 A; 1L6W A; 1JJK A; 3L0G A; 3WK0 A; 3ANY A; 5DCD A; 5F1V A; 1MSS A; 4AEO A; 4O1B A; 1H7P A; 3S9Y A; 1OJX A; 3S1W A; 3BGJ A; 5LGZ A; 2CLI A; 2ZA2 A; 1WX0 A; 2PC4 A; 1G67 A; 2WSY A; 2QCD B; 2X1R A; 4K36 A; 3W2K A; 5IBX A; 4UIO A; 3O8A A; 3I7S A; 3TW7 A; 4ZQC A; 3TKF A; 1JCM P; 2V82 A; 4HNT A; 3UWY A; 4PL1 A; 3TK7 A; 5DLC A; 1PL9 A; 3FKK A; 1GPW A; 2NV2 A; 1PCK A; 4DBD A; 4E0C A; 3PWA A; 4MOZ A; 2VFH A; 2WO5 A; 1XBY A; 5I3K A; 3TV8 A; 3UWW A; 3CWO X; 4A3U A; 4YL2 A; 1OG0 A; 4O11 A; 3U2O A; 2DZP A; 4DEF A; 4GUH A; 3QEZ A; 3W1N A; 3V75 A; 3L5M A; 4E38 A; 3AO0 A; 2CWN A; 1HFB A; 3OG3 A; 5CPM A; 2OT1 A; 1G4E A; 4M5P A; 4XBS A; 3P82 A; 1IXN A; 2W79 A; 5CPL A; 4CQA A; 5DXY A; 3TFC A; 4AWS A; 3MW7 A; 4S2C A; 3SI9 A; 1IXP A; 1K3U A; 3M1Z A; 3W75 A; 2A21 A; 1ZAL A; 2DZU A; 3L9C A; 4MFE A; 1GT8 A; 1KLZ A; 4N6E A; 2Q8Z A; 4RK8 A; 4O0K A; 3I6R A; 2FDS A; 1J5T A; 3A24 A; 3C52 A; 1EUN A; 2Q3O A; 2WJZ A; 1XBV A; 4NZJ A; 3EWU A; 3TQ0 A; 1W5M A; 5AC6 A; 3QM3 A; 1OFR A; 1S5V A; 4AVF A; 1ZY9 A; 4XUG A; 4FB7 A; 1ICS A; 2XN1 A; 2Z6J A; 3GDM A; 3HGR A; 2KZH A; 4P61 A; 2RFG A; 2A2I A; 3NA8 A; 3LRK A; 1XCF A; 4JEQ A; 3P67 A; 4O4W A; 2FPY A; 3TDF A; 3W1U A; 4FNR A; 1XBZ A; 3PUO A; 1LYX A; 1JUK A; 1AMK A; 3NAV A; 4JYD A; 1OFE A; 1B3O A; 2CLL A; 3R2G A; 1HO1 A; 4DQW A; 4Y9A A; 4IUO A; 2ZQ0 A; 4MZY A; 1QOP A; 4NX0 A; 1DKW A; 1X1Z A; 1NSJ A; 3TA6 A; 4O16 A; 3TSM A; 1I45 A; 1S5T A; 3D0C A; 4EVZ A; 3DH7 A; 3CHV A; 2FB2 A; 2JK2 A; 4Z1B A; 4GUI A; 1Z42 A; 2HSA A; 3EXR A; 4UTH A; 3UPW A; 2EF9 A; 2PCQ A; 3B0U X; 1B9B A; 1GTH A; 3M41 A; 3LHU A; 3MP3 A; 4FX6 M; 2A7P A; 4KKX A; 4HKP A; 1TCD A; 4N6F A; 1Q6L A; 1G7U A; 3B0P A; 4I7W A; 4DF1 A; 5AYY A; 4O8R A; 4FNU A; 4GBU A; 5AHF A; 1ML1 A; 3LER A; 2CLH A; 2Y89 A; 3JS3 A; 3W84 A; 4CCD A; 3BW2 A; 3WFA A; 1J4E A; 3L21 A; 2EKC A; 4QIS A; 1TQX A; 3NR0 A; 1W56 A; 5EL1 A; 2QCL A; 1OYA A; 3IIO A; 3MI6 A; 1OFQ A; 4K39 A; 3W7D A; 1VS1 A; 1M7P A; 3M47 A; 3OIX A; 4AWT A; 2JGQ A; 5DCB A; 4MJZ A; 4JS3 A; 1HL2 A; 3W1R A; 2GVL A; 3M5Z A; 1NFB A; 4DPQ A; 5HUP A; 4JIQ A; 4NZF A; 3SW6 A; 1CX9 A; 1KFB A; 3IVT A; 3MBF A; 1H61 A; 3DHF A; 2VKZ G; 5K7J A; 4BWL C; 1XL9 A; 4LS1 A; 2BX7 A; 1C8V A; 3UY7 A; 3CT7 A; 4JTL A; 1OF6 A; 2A4A A; 3GAY A; 3MHF A; 4BI7 A; 3NM1 A; 1G4S A; 1ICQ A; 4JX6 A; 1YXC A; 5CSS A; 3KVM A; 4UFK A; 4WX2 A; 2GJL A; 1KM4 A; 2TSY A; 4HSN A; 3G0S A; 3PVF A; 4FNS A; 3O06 A; 4KWV A; 2XN2 A; 4LY8 A; 3NQF A; 4CCE A; 3QJA A; 3QMR A; 1K8Z A; 1K7X A; 3SFK A; 3UXA A; #chains in the Genus database with same CATH topology 2PWH A; 2GWN A; 4I9T A; 1X96 A; 5DRC A; 4I3G A; 5I3J A; 3E0L A; 1H41 A; 2EPK X; 2J6X A; 3GIP A; 4B5N A; 3SSZ A; 2QUT A; 5G4E A; 4GCX A; 3LBM A; 3I3E A; 1SMD A; 1AFS A; 1LOS A; 1YYA A; 1JPD X; 3WDH A; 4ZEP A; 1MW1 A; 2XZ9 A; 2G4J A; 4R7W A; 2PZ0 A; 4MYA A; 1YBQ A; 3MJY A; 4JGD A; 2J8F A; 4XKM A; 1BC1 A; 4AZ7 A; 1GKQ A; 4LAN A; 1W6T A; 3GC5 A; 3WFL A; 4WDX A; 3AS0 A; 4QHR A; 4LCQ A; 4RZB A; 3M0J A; 1IEX A; 4OE7 A; 1EHA A; 2UY4 A; 3STC A; 1ZSX A; 4HU8 A; 1VQO U; 1TX0 A; 4MB4 A; 2GVG A; 1PZ2 A; 4HJW A; 5IDI A; 3WD1 A; 8TIM A; 2AL2 B; 4GIY A; 4LX4 A; 4QLL A; 1FWI C; 4XVE A; 2TOD A; 4YTQ A; 3AU6 A; 2IPG A; 3H7R A; 5MHB A; 1VIW A; 3ES8 A; 4QSF A; 3W7G A; 1UR0 A; 1XBX A; 1E4L A; 2IPJ A; 1FDJ A; 5GPR A; 3URB A; 3V5O A; 4IGN A; 1OFP A; 3SGU A; 1KXQ A; 2A0N A; 2GGH A; 4JN6 A; 1JAE A; 2IQ0 A; 2DU2 A; 2VOM A; 1V8E A; 1IT0 A; 4AD2 A; 4KGJ A; 4RDZ A; 5TNV A; 2VM8 A; 1RP8 A; 4C7G A; 4MZ1 A; 1FHD A; 3DFQ A; 3OW2 T; 1MDR A; 4WXZ A; 4B3Z A; 2PP1 A; 3CCS U; 5CC3 A; 1XF0 A; 3CMA U; 3WQF A; 1JPH A; 5ECV A; 4RZ4 A; 4A2R A; 3CXO A; 4H6K A; 3IIP A; 1AUS L; 4U33 A; 3WQW A; 2YYT A; 3W07 A; 4XTD A; 4IMD A; 4GVF A; 2AL1 A; 5G2W A; 4S1F A; 3NQ8 A; 2DDX A; 4JTD A; 4L6O A; 2PD9 A; 3HGO A; 2Q4C A; 1OGS A; 2CZD A; 2I7G A; 3NPV A; 1C92 A; 3KUM A; 5T9B G; 3WQG A; 1R3S A; 3C56 A; 1OYB A; 1NEY A; 4E2B A; 2DSV A; 3BLL A; 2QUM A; 1M7X A; 1PS9 A; 1US3 A; 3SOH B; 2HK1 A; 5K4X A; 1V3J A; 3GKF A; 2H0L A; 1W9P A; 3R10 A; 4Y96 A; 3T8L A; 1EPX A; 1ODZ A; 3PJT A; 3B8U A; 3CHC A; 3M0I A; 1OCQ A; 1H63 A; 2ZVI A; 4LUT A; 2PB1 A; 3K4W A; 2H3B A; 4AHP A; 2OOF A; 4UFM A; 3S2L A; 3ZJ7 A; 4JPG A; 2J9X A; 2BBF A; 1A5A A; 5XIN A; 2YCL B; 4JFT A; 3GSM A; 3NJ3 A; 5HNT A; 1SZE A; 4AQL A; 2IQT A; 1XH0 A; 4LC8 A; 2WKL A; 3O2L A; 1XIF A; 3GXI A; 1MFU A; 3W7E A; 4GXP A; 5JVL A; 1NFP A; 2XH2 A; 2ISF A; 3AMC A; 1YIX A; 1C9W A; 4WSK A; 1XLA A; 1EFZ A; 3CAQ A; 3QVQ A; 4M6V A; 1VRD A; 1DXI A; 4BQ3 A; 2CBV A; 1YX1 A; 1JF6 A; 3B8I A; 2VGI A; 3LB0 A; 3E12 A; 3PSW A; 3WQV A; 3JVA A; 1FBA A; 1A47 A; 1R2T A; 3GTF A; 3IJD A; 4JK1 X; 4PC8 A; 2WSK A; 3WF2 A; 3W6Y A; 3R6I A; 2OTL U; 3AMD A; 5LYR A; 4C1N A; 3VKK A; 3R89 A; 3WK1 A; 3CV2 A; 3HG4 A; 3C6C A; 3POH A; 1VQM U; 3NL2 A; 1S1P A; 4GZJ A; 1X6L A; 3NQ2 A; 1ZJH A; 3EWY A; 4ZXO A; 1X7I A; 1AZ1 A; 4UUI A; 4MJM A; 3B0X A; 5C8B B; 1W3H A; 4R3U A; 3CH0 A; 5HQL A; 2RGM A; 1QAP A; 5HR6 A; 1LQ2 A; 1X7F A; 1V3H A; 1Y5W A; 1I2P A; 1WAU A; 1UBP C; 1PEZ A; 3LHZ A; 2XII A; 3VIF A; 1FFR A; 3U48 A; 1LL6 A; 3WL0 A; 3PC0 A; 4D8Z A; 3ZLG A; 1CLV A; 3WF3 A; 2RDT A; 2X05 A; 1I4N A; 3EWX A; 2J7H A; 3MP5 A; 3VND A; 1G4P A; 8XIM A; 1YCN A; 2QDG A; 1BGG A; 1A0C A; 1QO5 A; 1V79 A; 5D2X A; 3TSB A; 1GVF A; 2AMG A; 1E3X A; 2W63 A; 4JN7 A; 2Z6I A; 4Q4K A; 3W1M A; 1DJG A; 1NJJ A; 3AYV A; 1DL3 A; 3P8J A; 3F4D A; 5CBB A; 3R11 A; 1IGW A; 1OGB A; 4JHZ A; 3S2C A; 3UP8 A; 1LRQ A; 4PA8 A; 2OCZ A; 4JYE A; 2DV0 A; 1OY0 A; 2YCJ A; 3QZA A; 3NDZ A; 4X3Z A; 3QF0 A; 1ENU A; 1O5R A; 2VVN A; 4UFL A; 5RUB A; 1BYB A; 1RAL A; 2BWG A; 2Z1W A; 4MWA C; 4AC7 C; 2DFA A; 3LZ3 A; 2W27 A; 3R43 A; 4DPP A; 2VEK A; 1NDY A; 1RD6 A; 3PBW A; 1E0T A; 2OTD A; 2YFN A; 1R3Q A; 3RK8 A; 3QRH A; 2CIT A; 1AW5 A; 2E6F A; 3GXN A; 1YXD A; 3W76 A; 3T78 A; 3EPN A; 3CL7 A; 3S8H A; 2XZ7 A; 1OF8 A; 3DEQ A; 2VC5 A; 4JMI A; 2RGL A; 3N9S A; 5C2C A; 2PU1 A; 3CUJ A; 4NU7 A; 1TTJ A; 2W6R A; 4OIF A; 2GPT A; 2QVH A; 3MSR A; 5LNV A; 1NIU A; 5IFP A; 1W3L A; 5CPT A; 1R5Y A; 1RCO B; 1TE6 A; 1A5B A; 3M7S A; 1R7O A; 1W2P A; 3AIU A; 3QY6 A; 4EOU A; 1DVJ A; 3UU0 A; 1XID A; 1FCB A; 4H83 A; 3GO2 A; 4N9E A; 3A4J A; 4ZZ9 A; 4JD4 A; 3CZJ A; 2XFW A; 1P1X A; 4A8N A; 2C3Z A; 3F4C A; 3L4U A; 1GJU A; 1TVP A; 4A21 A; 3DEN A; 4CNP A; 4HOZ A; 4Q32 A; 5GQB A; 3U8G A; 1YDN A; 1LWJ A; 3TML A; 4DM1 A; 1QVF T; 1G0C A; 2FVM A; 1ZBW A; 3TNO A; 1BWL A; 1OFA A; 1VBU A; 1TWZ A; 2C15 A; 5CPS A; 3BLF A; 4D72 A; 2RJG A; 4PSP A; 1A50 A; 2F38 A; 4UBP C; 3QZ6 A; 2JGY A; 4W8B A; 2PRM A; 1S3T C; 1HKK A; 1JG9 A; 4JHO A; 1H62 A; 4NZ5 A; 4ZQN A; 4AHQ A; 4KBX A; 1E40 A; 5CBI A; 3JR2 A; 3GG7 A; 1SMA A; 2QQ6 A; 2BVY A; 2YDQ A; 3RR4 A; 1LQ0 A; 4RPQ A; 3CJP A; 3L8N A; 2ZA1 A; 4AEE A; 1UA3 A; 2Z2A A; 3NWR A; 3RND A; 1OC5 A; 3MBD A; 4GKL A; 4IQE A; 2GWG A; 3TN4 A; 2NWS A; 2VZO A; 2BY0 A; 4JWL A; 4MYX A; 1V6W A; 3QPY A; 2YXG A; 1NAL 1; 1RSC A; 3RDK A; 4HNU A; 2WCA A; 4IME A; 4JOM A; 1VZW A; 2FYM A; 3LXB A; 4CLM A; 1W1Y A; 1A80 A; 4ORM A; 1T0O A; 1VQT A; 1GKP A; 4D8A A; 1WOB A; 3CYJ A; 2XIB A; 1V3M A; 3NO5 A; 3WIJ A; 4ZQO A; 1LG2 A; 3TON A; 3W7H A; 1F8I A; 4Z17 A; 1PB0 A; 1IQ8 A; 3HG3 A; 2HQA A; 5E0F A; 4PBE A; 3BCF A; 1RQE A; 3W23 A; 5DQP A; 4JFV A; 2WXD M; 2YZ5 A; 3LV5 A; 1TTI A; 4FO4 A; 3V1P A; 4HRE A; 1W0M A; 1Q7Z A; 5DBT A; 3AXH A; 1G1Y A; 3HU7 A; 3POC A; 1OM0 A; 4MY8 A; 1JDE A; 1RQH A; 2INF A; 4H1Z A; 3HIJ A; 3VNZ A; 2INZ A; 1VQ5 U; 1NF0 A; 2NVA A; 1FP9 A; 3AIB A; 4MNL A; 3G4S U; 3KL0 A; 2PTZ A; 4BQ2 A; 4L6D A; 1A5M C; 1A8A A; 3ATZ A; 3NQM A; 1SZG A; 2PUR A; 3TYA A; 3GDT A; 1OC7 A; 1OX6 A; 1EWD A; 2A3B A; 3WD3 A; 4JTI A; 1KCL A; 2F2H A; 2VEI A; 4GZE A; 3SR7 A; 1ISY A; 3HV9 A; 4DHG A; 4XUY A; 3CMF A; 2J79 A; 4FHA A; 9XIM A; 4I8D A; 1AW1 A; 3FCP A; 3FS2 A; 1BRL A; 3N34 A; 4JX5 A; 2Y61 A; 4Z2L A; 3OP2 A; 4E3V A; 1ZHA A; 2I14 A; 1FA2 A; 5FEZ A; 2E1D A; 5D9P A; 3FN9 A; 3P0X A; 3LHT A; 2QMJ A; 3UBP C; 4WGX A; 2Q3R A; 2X7V A; 2Y8V A; 4CCC A; 1GQL A; 1BYD A; 3GH7 A; 3UEQ A; 3CTL A; 1HG3 A; 5CLW A; 1CWY A; 4KEZ A; 1PHQ A; 2B7P A; 4FAL A; 1TSI A; 1KBB A; 4NX5 A; 3WDQ A; 3GR7 A; 1TJP A; 3NQG A; 3PBV A; 4GC4 A; 3LBO A; 3GR8 A; 4IIE A; 5ABF A; 1GVE A; 2I9E A; 3L2M A; 2IE7 A; 3SIZ A; 3WNM A; 2ZHJ A; 1DWG M; 2V68 A; 3HBL A; 4OT7 A; 1UPP A; 5C55 A; 3EOS A; 3CPR A; 3FJ6 A; 3N3T A; 3I10 A; 3HPS A; 5E5C A; 4PMV A; 3AC0 A; 3AFB A; 5FJR A; 3H70 A; 1Y0E A; 4R27 A; 1QPQ A; 2CEQ A; 3OA5 A; 4XX6 A; 4H7C A; 3T7V A; 3Q94 A; 1VR6 A; 4JNZ A; 2R09 A; 1PPI A; 1UWI A; 3BAJ A; 5FF0 A; 1LZO A; 3LM3 A; 3NZP A; 2VFE A; 3KWX A; 3ZIZ A; 1CXL A; 1LP6 A; 5I09 A; 3ARV A; 2W92 A; 3CLW A; 1UH4 A; 1ZEN A; 2J7G A; 6XIM A; 1XRF A; 4FOU A; 1UC5 A; 2A3A A; 2G3M A; 3BAR A; 4JX4 A; 4O54 A; 4GAC A; 2HE8 A; 2OG9 A; 2QJG A; 3G77 A; 2PDQ A; 2MUC A; 1AD1 A; 2FTY A; 4IT1 A; 3W1X A; 1P0B A; 2CC0 A; 1I9C B; 1Z9A A; 2D2H A; 2WVT A; 4XWU A; 1RJR A; 3UY8 A; 3CAS A; 3GXM A; 6TAA A; 3APG A; 1XT0 B; 2VI0 A; 3SJ3 A; 2B7N A; 1AVH A; 1YPX A; 2FKP A; 2DZS A; 2E3Z A; 1AQF A; 1V6Y A; 2V2H A; 1EC8 A; 3M5V A; 3U4F A; 3FYO A; 2HPM A; 1N8R V; 3UZZ A; 4HZ6 A; 2O9R A; 3OLE A; 3N4F A; 1F7B A; 3RN6 A; 3W7L A; 2XYL A; 3SN4 A; 2QCE A; 4Q4R A; 1THF D; 1AMY A; 1E72 M; 3EDE A; 1E1E A; 1YKW A; 1WP6 A; 1SO6 A; 2P1F A; 3LCX A; 4YMZ A; 2GQA A; 3C4U A; 1VJZ A; 1DJY A; 4AZC A; 4UB9 A; 4KH2 A; 2WPB A; 3W1L A; 4ERI A; 3ORW A; 4RW5 A; 4W7W A; 3AIS A; 1FWE C; 3PNU A; 2YDA A; 3TOP A; 3VZZ A; 3ISI X; 1OC6 A; 2W62 A; 2YBV A; 2QCM A; 4LF1 A; 4IIS A; 4ZOB A; 4AU0 A; 3EZ4 A; 1ME9 A; 3T81 A; 1EL3 A; 2P3E A; 1O66 A; 3N4E A; 4GAB A; 2A3L A; 3RRX A; 2WQP A; 3A5F A; 1J0H A; 2P88 A; 1XIM A; 1N8F A; 1OSE A; 3U0H A; 3ABO A; 4CQB A; 5M3W A; 2KAU C; 4K8G A; 4YCO A; 3WEN A; 2EBN A; 5BWF A; 4LVD A; 5D2Y A; 2VUR A; 5T8G A; 1TO3 A; 3TN3 A; 4Q8U A; 5BMX A; 3S2M A; 3S5Y A; 3IUI A; 5A6M A; 1J6P A; 4K38 A; 1TVN A; 3D5H A; 3TJX A; 1J12 A; 3V7P A; 3GYC A; 1VJI A; 3OVR A; 3WLJ A; 2CET A; 1KRM A; 1Q6G A; 1JFX A; 3GH4 A; 3EA3 A; 3IK4 A; 1UR2 A; 1W1V A; 4DAF A; 3KDO A; 4MY9 A; 2DW7 A; 4PMD A; 4O50 A; 3ALG A; 1LT7 A; 3WSU A; 4KFP A; 3NZR A; 3NGJ A; 5CZS A; 3AS1 A; 5TQL A; 4CNT A; 3PBY A; 3C0J A; 3TTQ A; 2BGQ A; 1IWP A; 2MNR A; 3GXF A; 1G01 A; 1CGV A; 3N29 A; 2NXI A; 3WJW A; 3TYD A; 3UA4 A; 3THA A; 1DWI M; 4IQJ A; 2Z1X A; 3HMJ G; 1N55 A; 4RNF A; 3FXG A; 4ENL A; 3UES A; 4Z2J A; 3P8I A; 1DQX A; 3EXS A; 1JZ2 A; 3CZK A; 3BAX A; 2O4M A; 2PBO A; 4HYW A; 3RLG A; 1EBH A; 5CAH A; 3GC4 A; 1PDZ A; 1FWD C; 4ZCW A; 1YJ9 U; 1N42 A; 3PN8 A; 4NXS A; 1R3R A; 4K68 A; 2WJE A; 4NZ1 A; 4K1W A; 3M16 A; 1P4C A; 1P7T B; 2PPG A; 2D0F A; 4ML9 A; 2RH9 A; 3RJY A; 5C5Y A; 2G97 A; 4QX4 A; 2CLF A; 4I9A A; 4MPK A; 4PMX A; 3BXW A; 4HK5 A; 1XIB A; 4J35 A; 3WLP A; 1F74 A; 3LMZ A; 3C0S A; 2HVO A; 1B0I A; 1S72 U; 3TOY A; 4AW7 A; 5JBF A; 1AW2 A; 4LAU A; 4FIR A; 3BLO A; 2POT A; 4YYC A; 4XAZ A; 3CLM A; 2BP1 A; 3U2E A; 1WKY A; 3PB2 A; 4G1K A; 1I8J A; 4KET A; 3QV6 A; 2WSP A; 3GBE A; 1SFJ A; 2BZN A; 2POD A; 1VC4 A; 4TX9 A; 3BO9 A; 4PMR A; 3UET A; 4UY8 X; 3NQC A; 3UXD A; 5EML A; 5HME A; 3C2F A; 1PZ1 A; 3T4C A; 5CZ0 A; 2ZZ7 A; 1OX4 B; 2WZH A; 3VNL A; 4JFU A; 3NSM A; 3BK0 A; 4DBU A; 1YBQ B; 4JTE A; 4OZO A; 5D02 A; 1OFB A; 4RCX A; 2RKX A; 1IYX A; 2WVV A; 1WBH A; 3T09 A; 1J93 A; 1RV8 A; 3WBY A; 1OK6 A; 2FZ9 A; 4B3L A; 2PMQ A; 4JFS A; 2Z1V A; 3V5F A; 5BRB A; 3THD A; 1TKK A; 4I5X A; 5HOS A; 3NPX A; 1JRC A; 3OG2 A; 4LAL A; 4DM2 A; 2HNH A; 2G3F A; 3AHV A; 3S1V A; 2J9Y A; 4L4O A; 4UFJ A; 1R3V A; 1JCJ A; 4Q33 A; 3G1A A; 2QJN A; 3MT1 A; 4AZB A; 4BA0 A; 1FWG C; 3EWB X; 4AFY A; 4DYJ A; 2GSH A; 2PDB A; 3KVJ A; 2F7F A; 3EPM A; 3WFZ A; 3A9I A; 3CT2 A; 4GBD A; 4GZ7 A; 3AMK A; 3IAQ A; 2C1G A; 5EBA A; 4AMC A; 1Q65 A; 1A0E A; 1DXF A; 4HA1 A; 5CLT A; 3I4K A; 1F05 A; 4D6F A; 4FXF A; 1JCY A; 2ZKM X; 5G2I A; 1XH2 A; 3EXT A; 2GGG A; 1HKW A; 3JZE A; 4HYV A; 3AZS A; 1TIM A; 4YXG A; 3N25 A; 3PR2 A; 1G69 A; 4QSH A; 3ARO A; 2TPS A; 5XIA A; 4H41 A; 2J88 A; 3IGU A; 1TPC 1; 4DJD A; 2VL4 A; 3GW3 A; 2HYV A; 4NNA A; 3VD5 A; 3W7C A; 3LCH A; 1A5K C; 2YW4 A; 1C93 A; 2VFG A; 4J40 A; 3CAK A; 1R2R A; 3B4Y A; 2EJ1 A; 2NV1 A; 2CZE A; 3W7I A; 4WY0 F; 3FA3 A; 2YFO A; 1MVY A; 1KV8 A; 1UR4 A; 1ZPT A; 4IJC A; 3E2V A; 1KU1 A; 4LH8 A; 3EB2 A; 4M7T A; 4DJD D; 5CC7 A; 4FLS A; 5C2X A; 3T05 A; 3IAR A; 5BRP A; 1PX3 A; 3DI0 A; 3W29 A; 3WF0 A; 4FR6 A; 3B9D A; 2QYG A; 2V0T A; 1I2R A; 1WPC A; 3N3M A; 5CGM A; 3AUJ A; 1EX5 A; 3W74 A; 5FAG A; 3N9K A; 2OWX A; 2PI6 A; 4LUT B; 3B3D A; 1KM3 A; 3P60 A; 4EP8 C; 1P0K A; 1XDL A; 5HJL A; 1XLK A; 3NOY A; 1EIX A; 3G1Y A; 2NWR A; 4S2B A; 4IP7 A; 3MV1 1; 3BG3 A; 4G2D A; 4C65 A; 1EOK A; 1GJP A; 2VVS A; 1FHL A; 3HGJ A; 4D6I A; 1H0G A; 3PF3 A; 1QC7 A; 1TV7 A; 1CXK A; 3H54 A; 2R94 A; 3EMC A; 3TQV A; 1W9X A; 4AYS A; 2PWD A; 2VJX A; 4WOQ A; 1QBA A; 5AHI A; 3E0I A; 3MP4 A; 3C2R A; 1FKX A; 4EAN A; 2DT3 A; 1Q6F A; 3RU6 A; 4G7E A; 2VZS A; 4AIO A; 2X2G A; 1XM3 A; 5HR7 A; 5H8P A; 4QE5 A; 3KS6 A; 1BGA A; 1Q45 A; 2GVJ A; 3I6T A; 4XV0 A; 1BFN A; 1HJS A; 2XIM A; 3UGV A; 4C1N B; 3CV7 A; 3E3H A; 4W7V A; 1TPU A; 1WXZ A; 3OHI A; 3DCP A; 1SJD A; 4MDU A; 3M0X A; 3W87 A; 1J0I A; 3B4U A; 3QR1 A; 3DEC A; 1SFT A; 3H4X A; 3URA A; 3AJ7 A; 1MYR A; 3P62 A; 4GIA A; 3GNX A; 3TP4 A; 3VOI A; 3NYZ A; 1B4E A; 2QII A; 3W27 A; 4JQ4 A; 5E0C A; 2Z2B A; 3F2D A; 4DZ5 A; 3FY1 A; 4WQ6 A; 4P8V A; 2DVX A; 2K6X A; 4XKY A; 1PWL A; 3PZO A; 1GOM A; 4MSS A; 2ZDS A; 2B31 A; 1J9Y A; 5D30 A; 1TV8 A; 3W1T A; 2VWT A; 2ATM A; 1FP8 A; 1VEN A; 4F66 A; 1UCW A; 1CGX A; 2YZR A; 4C6E A; 1X8C A; 3PFM A; 1GVO A; 4BWL A; 1WZL A; 3WCZ A; 1FWT A; 3PS7 A; 4MNM A; 4UTT C; 2TRS A; 3WLL A; 4UQ9 A; 4W5Z A; 3KOZ A; 2O55 A; 2NX1 A; 1T8X A; 4LTS A; 4XO7 A; 1A5O C; 4CNU A; 4U3C A; 3TQP A; 4C6O A; 2DY3 A; 1XKY A; 2CLE A; 1TZ9 A; 1C9D A; 4CD4 A; 1K9E A; 4XAF A; 4B2D A; 3UG5 A; 1UWU A; 3PWG A; 3BE7 A; 4M29 A; 1TRD A; 5DF0 A; 3H23 A; 2CBI A; 3LHW A; 4GGH A; 3VIH A; 2ZUT A; 4FR1 A; 2J4G A; 4O1E A; 2QKF A; 1ADS A; 1AG1 O; 1K9T A; 3KVL A; 3HMC A; 4Z2K A; 2R1L A; 3W6M A; 3MDU A; 1FWC C; 2PDH A; 4KIR A; 2J7B A; 4B2D D; 3P7Y A; 1HVD A; 1KW1 A; 1KR0 A; 8XIA A; 1J5S A; 4MLR A; 1NQK A; 1RPX A; 5AYX A; 4MDV A; 2VC7 A; 5I79 A; 3TN6 A; 3SGG A; 4H2C A; 1WUF A; 1X38 A; 3DIP A; 2DSW A; 3NQA A; 2FJK A; 3LU2 A; 3ART A; 1P48 A; 1K8Y A; 2QPS A; 2J13 A; 4O4V A; 4GFI A; 1L2Q A; 3TR2 A; 3LHV A; 4OUI A; 4CD6 A; 3VIL A; 4ZOA A; 3PAN A; 2IKG A; 2G37 A; 2R1M A; 4M51 A; 3H2O A; 1LUC B; 1KQY A; 1Q86 V; 1OT1 A; 4HCL A; 4A4A A; 5KZE A; 2WJF A; 3F5K A; 2OEJ A; 1BG4 A; 4L1W A; 4L64 A; 3PAO A; 5CG0 A; 5MKG A; 3WH8 A; 4FNT A; 4CVU A; 4NJI A; 4JQA A; 5HUO A; 2HIS A; 1F5Z A; 2R14 A; 4OFC A; 2XGI A; 3ECQ A; 3EX3 A; 2PDK A; 3BSM A; 4TTG A; 3S4T A; 2D20 A; 1JJD A; 4U4M A; 7ENL A; 2ODO A; 4QG6 A; 4PSR A; 3VYL A; 1A3X A; 2O7S A; 1MUW A; 2VTF A; 3PJX A; 3H53 A; 4E8C A; 2XIQ A; 3AUO A; 4MY1 A; 3SCT A; 4NYY A; 1H49 A; 3CPW T; 2F2K A; 2VCB A; 4F79 A; 3LRL A; 2DZW A; 1X39 A; 1XLH A; 3K8K A; 3MUI A; 3H12 A; 3SCW A; 4BR1 A; 3ZO9 A; 3R12 A; 1RQB A; 1YEY A; 1L6W A; 5FD0 A; 1JJK A; 5DCD A; 3VKA A; 4R88 A; 4HJF A; 4AEO A; 2EQD A; 1OJX A; 3S1W A; 1C3F A; 5LGZ A; 2ZA2 A; 1WX0 A; 4WVX A; 2EG6 A; 1G67 A; 2WSY A; 2OWW A; 4R85 A; 4WF7 A; 4K36 A; 2JF6 A; 1QI0 A; 4UIO A; 2YCI X; 3MCO A; 2GK1 A; 3S1G A; 3VD3 A; 2VCA A; 1FKW A; 2V82 A; 4KS0 A; 2O9P A; 4PL1 A; 4W86 A; 4Q8N A; 1TX2 A; 4PMY A; 1XD0 A; 1PL9 A; 4GI9 A; 5DLC A; 1ISV A; 1GPW A; 1KQS T; 2NV2 A; 2CXG A; 2P76 A; 2CXE A; 1S46 A; 2IKH A; 4E0C A; 3PWA A; 4MOZ A; 3BOL A; 5I3K A; 3AOF A; 5BOE A; 3UWW A; 3CWO X; 4A3U A; 4EXB A; 5A6B D; 4O11 A; 2VZT A; 4K1G A; 3QEZ A; 3W1N A; 2CHN A; 1D8W A; 4C6F A; 4AD0 A; 2C79 A; 4XBS A; 3UF9 A; 1L8P A; 5DXY A; 2GDV A; 4AWS A; 3NFU A; 3COT A; 4KMQ A; 2W91 A; 1MFV A; 3CQK A; 3ONB A; 2EEX A; 2Q8Z A; 1Q81 V; 4FP1 A; 1J5T A; 1ODU A; 4NRS A; 3KL3 A; 5A6A A; 3M46 A; 4GGB A; 2BFG A; 3WBE A; 3HV8 A; 4FB7 A; 2Z6J A; 3H2C A; 3HGR A; 4IP5 A; 2RFG A; 2A2I A; 4BZY A; 2X0H A; 3P67 A; 2FPY A; 1SJC A; 2PGE A; 4GJI A; 2P0I A; 1LYX A; 1JUK A; 4JKK A; 4PBF A; 1OFE A; 1B3O A; 3R2G A; 4IUO A; 2WMI A; 2CIP A; 3H2E A; 1NSJ A; 4O16 A; 5C9W A; 4EVZ A; 4EKJ A; 3DH7 A; 4O0Q A; 1ZBK A; 2AQV A; 1J6O A; 1Z42 A; 2HSA A; 3EXR A; 5JBD A; 4MF4 A; 4UTH A; 3UPW A; 2EF9 A; 3B0U X; 2WLY A; 3LHU A; 4FX6 M; 3UER A; 2A7P A; 3MCM A; 3MMD A; 1TCD A; 1CEC A; 3DHU A; 5ABF B; 1G9H A; 1Q6L A; 1G7U A; 3B0P A; 4DF1 A; 5AYY A; 4UR9 A; 3SUW A; 4GBU A; 3LER A; 2Y89 A; 3W84 A; 1DID A; 3BW2 A; 2Z1K A; 1J4E A; 1TQX A; 3NR0 A; 4XD5 A; 1TV4 A; 5EL1 A; 3LTL A; 3MI6 A; 3JW7 A; 3HN3 A; 1OD8 A; 4JS3 A; 1KGW A; 2QA4 U; 3SCV A; 2WM0 A; 3HQP A; 1KFB A; 3G6E U; 1H61 A; 2Z29 A; 1FWJ C; 2VDI A; 5HDR A; 3UY7 A; 3AIQ A; 1D7K A; 1OF6 A; 3NM1 A; 2VX5 A; 3NO3 A; 3O6A A; 4PB5 A; 4UFK A; 1VQ9 U; 1KM4 A; 2TSY A; 4QG8 A; 4GC3 A; 3AIC A; 2NW0 A; 3TTY A; 1QRQ A; 2ZE0 A; 3NQF A; 4A8I A; 3QMR A; 4YVV A; 1U5H A; 4C6K A; 3VNJ A; 1TUF A; 1Q85 A; 3I3B A; 4V1X A; 1K7F A; 4QAI A; 1EX1 A; 1KQZ A; 4PH6 A; 1XIJ A; 1KBL A; 1EA9 C; 2EGZ A; 4J9J A; 3M9Y A; 4H8U A; 1WKF A; 3KCJ A; 3LZ5 A; 2V5J A; 3Q2D A; 2VDH A; 4J7R A; 1T8Q A; 3G1H A; 2Z2U A; 4AZC B; 3HQ1 A; 1REQ A; 1KA9 F; 4S23 A; 1CEN A; 4MAV A; 4XIS A; 5BOO A; 3F1Q A; 4S3A A; 4XAG A; 3OZY A; 3S5Z A; 4B5T A; 1TQJ A; 2AAM A; 3W7Q A; 4GK8 A; 4WHB A; 5HNN A; 3S3G A; 5B7Z A; 4UQA A; 3IUH A; 3ITG A; 4GY7 A; 2BS9 A; 4H6R A; 4GG9 A; 4QCE A; 5M17 A; 4AHO A; 3ZR4 A; 4QIU A; 3R4I A; 3L4W A; 3Q58 A; 1J96 A; 2WNW A; 3AHZ A; 4OBS A; 4HYR A; 2HWG A; 5ENL A; 2CLO A; 1R30 A; 1UUO A; 3LCL A; 1D3H A; 1R47 A; 3AYR A; 2ATS A; 4WLC A; 2FGB A; 4JZ5 A; 4FHR B; 3FW6 A; 2QUL A; 4L9Z A; 1OYC A; 2XM2 A; 2Z7G A; 4YHE A; 3RUB L; 1DQW A; 2D2J A; 4AD3 A; 1WXJ A; 1YDV A; 1XRT A; 4NZ4 A; 2KX9 A; 3QC3 A; 4XIA A; 4H00 A; 3I55 U; 4C60 A; 3GFY A; 4POD A; 3ARU A; 3LCW A; 3PPH A; 1W17 A; 4AF0 A; 2D1Z A; 3KP1 A; 1K03 A; 3PUG A; 4O12 A; 2OZT A; 3FYP A; 3LBC A; 1ESW A; 2A6L A; 2DZA A; 5FF7 A; 1IR1 A; 3R7M A; 3WMB A; 4QDM A; 1POJ A; 4EX4 A; 4OJY A; 3MJM A; 3V36 A; 1VQ8 U; 4PUN A; 4PUK A; 1KM5 A; 1FVP A; 2X8R A; 4EE9 A; 1M1B A; 1CGU A; 5FOO A; 5ABG A; 4POC A; 2XPK A; 3T0T A; 1E6Q M; 1F8M A; 4FNP A; 5FJI A; 2FJU B; 2PBG A; 3AQU A; 3V5C A; 1EP2 A; 2E40 A; 2FVL A; 4O57 A; 3DFN A; 1KBK A; 1T40 A; 2DQW A; 3NUR A; 2OPJ A; 1W91 A; 4MIM A; 5A6S A; 1ECQ A; 5DMY A; 2OO0 A; 2CPU A; 3TR9 A; 5FJT A; 1WO2 A; 4F0K A; 3THU A; 2BHU A; 3I65 A; 2PRH A; 4Z1C A; 5DBU A; 4LVF A; 4KWW A; 1B5T A; 1L6G A; 7CGT A; 5CXP A; 2FTW A; 5EMK A; 4CU6 A; 3E38 A; 5D04 C; 2YR1 A; 5DT7 A; 3UWU A; 3T0D A; 3NNT A; 3P74 A; 1PKM A; 1XPY A; 1F73 A; 4GFS A; 4MZ8 A; 1XGD A; 3KW3 A; 1PXG A; 2RDW A; 4DWS C; 1PV8 A; 3UG8 A; 1ANX A; 3EB3 A; 2DIJ A; 3M0H A; 3TUU A; 3UXI A; 3NDO A; 2NXG A; 1PTG A; 1TPF A; 3DXI A; 2I5I A; 2WVU A; 1ZUA X; 4MHB A; 4Q7I A; 2W9M A; 2CKS A; 3W22 A; 4AH7 A; 1OVD A; 4J3W A; 1HVQ A; 1GZ1 A; 4Z2G A; 2XIO A; 2Y70 A; 2XH4 A; 2Q8L A; 2R1N A; 3WJZ A; 4ORI A; 3CUH A; 1SAV A; 4LFY A; 4US6 A; 5LNW A; 4LCR A; 1FDY A; 4XVD A; 5EU9 A; 2E09 A; 4JCL A; 3LLX A; 4NRR A; 4LS2 A; 2AGT A; 1XHG A; 1R3W A; 4ML4 A; 3GVQ A; 5I0D A; 3WH5 A; 1X1O A; 3GVV A; 3AYS A; 2WB5 A; 3UXL A; 1ISW A; 1UAS A; 2WVV B; 1W5O A; 4ACZ A; 3ARR A; 1K02 A; 2NLY A; 1M65 A; 3BVJ A; 5GVJ A; 3TN5 A; 4LUI A; 1UYQ A; 3N2M A; 3SB0 A; 4TUF A; 4HU0 A; 1UKS A; 3MTW A; 3KX6 A; 4F0L A; 4B4F A; 3WBA A; 3GND A; 4UTK A; 4LC6 A; 1EJU C; 4QJ1 A; 3OZP A; 4IFK A; 3I6E A; 1VPX A; 3S9Z A; 2Y85 A; 5LNR A; 1IT8 A; 1E15 A; 1DBT A; 7TIM A; 3ANU A; 3PP7 A; 1SR0 A; 3EWZ A; 3EDD A; 3K4D A; 1K70 A; 4XZM X; 4HA3 A; 3DYM A; 3KOF A; 1F07 A; 3VC6 A; 4GE8 A; 1DE5 A; 3BMX A; 4GI8 A; 5D38 A; 1TVL A; 2EO7 A; 1FH9 A; 3H22 A; 2OSY A; 3D47 A; 4URI A; 3DHP A; 1G4T A; 3EX6 A; 1H0I A; 2BOG X; 2PTD A; 1JDF A; 4Z1A A; 2CHR A; 3QR0 A; 4EWN D; 3N6H A; 4GYP C; 3G1F A; 3W81 A; 4IMA A; 3M0Z A; 3IJ8 A; 3PBM A; 4WKF A; 3JZU A; 3CS2 A; 1I0B A; 1NAR A; 4ATL A; 3WNL A; 2XVN A; 1YJ0 A; 4HN8 A; 3UXK A; 2PWV A; 1VKF A; 1OQF A; 2OQY A; 4TIM A; 3U57 A; 3SN0 A; 2A5H A; 4H01 A; 3C8N A; 3U2C A; 4LAM A; 3KBV A; 4U3A A; 3O05 A; 4W93 A; 3LY0 A; 3TVA A; 1PIG A; 3VGG A; 4A8L A; 1W3K A; 1OFD A; 5HMF A; 4AD1 A; 1OCN A; 3LPG A; 4C6D A; 1EO7 A; 3UWQ A; 1LBM A; 3AU2 A; 3WNK A; 2O9O A; 4N42 A; 3Q65 A; 3CB8 A; 4JTH A; 2BHZ A; 1EEP A; 2ZRZ A; 4MFD A; 4CN4 A; 4X9Y A; 1E6P A; 4TXE A; 4NI3 A; 3GU1 A; 2V81 A; 3BG9 A; 3S6T A; 4K2S A; 3AMG A; 4AUB A; 2YOC A; 1SFL A; 1C1K A; 5J5D A; 2ZBT A; 3EOL A; 3W24 A; 5J04 A; 1K8C A; 4KT2 A; 4O19 A; 4BQ5 A; 1E5J A; 4J27 A; 5FF3 A; 3W73 A; 4Y90 A; 4HMC A; 1JXJ A; 4AWE A; 2RJH A; 2FHB A; 2BOF X; 4O1A A; 3KTS A; 1DOS A; 1XIE A; 5IXE A; 1V77 A; 2VD8 A; 2YJQ A; 2V69 A; 3GDK A; 4TO8 A; 2VQT A; 3F7J A; 1SZR A; 2Z1S A; 1EXP A; 2R1K A; 4G7E B; 4HSO A; 2QCH A; 2Q71 A; 5TF0 A; 5A6K A; 2Y7F A; 4F3H A; 1TPD A; 1M9H A; 5CJM A; 2CBU A; 1Z8A A; 5I7G A; 1LLZ A; 2ZX7 A; 4TOQ A; 3SM0 A; 2O4Q A; 1JGI A; 1MUM A; 1VBG A; 3RPT A; 3KRZ A; 1XJL A; 3WDI A; 3QFW A; 2YKK A; 1MW0 A; 1FWF C; 4Q4O A; 1EDT A; 4L9X A; 3CC1 A; 3ZS4 A; 2P0O A; 2O56 A; 2BB0 A; 1FHV A; 2V3G A; 4GRS A; 1FWN A; 3QTG A; 3IV3 A; 1D7F A; 2A7R A; 3MYP A; 3E0W A; 1VAH A; 5AZ1 A; 4N9C A; 1NOF A; 2Z25 A; 3DI1 A; 1XLL A; 2AGK A; 2XIJ A; 4M19 A; 3GD6 A; 1TV5 A; 3ZJK A; 4Q8O A; 3ZYZ A; 2VNC A; 1WE5 A; 1SFS A; 1LRN A; 2ANU A; 2VC9 A; 2ZOX A; 2I16 A; 1EQC A; 4QFU A; 1QAT A; 1QPK A; 4XKY B; 3GRI A; 5A5W A; 2OFJ A; 3GH5 A; 3W83 A; 4E2V A; 5KTL A; 1PSC A; 1KD0 A; 4ALD A; 1HX0 A; 5E93 A; 3ARY A; 4X2R A; 1XC6 A; 1TJ0 A; 6A3H A; 2Y8K A; 4H9V A; 3KTX A; 3EB4 A; 2BV9 A; 3KOY A; 2GQ8 A; 1J10 A; 3V0S A; 3QV9 A; 1XLJ A; 3CQH A; 3LXA A; 3T0B A; 5KSW A; 2DSZ A; 4FMV A; 4RV3 A; 2J24 A; 3LTS A; 3M5X A; 1PTA A; 4IMC A; 4MWA E; 4N0R A; 5JMU A; 5I07 A; 4YLW A; 3IJQ A; 4IQL A; 2BY1 A; 3L4X A; 3SUR A; 3W1Q A; 1GZG A; 1NQ6 A; 4E8D A; 4IIF A; 1YAD A; 2VRK A; 2C1H A; 3MZN A; 1C7S A; 2ESC A; 1WB0 A; 1DJZ A; 3CC7 U; 3SCQ A; 5LIW X; 3MA8 A; 3R8R A; 4DI9 A; 4YWI A; 3BW3 A; 1WDS A; 4Y8E A; 1BQG A; 1C8Q A; 2V5B A; 2Y7D A; 4WDU A; 1VFU A; 1CIU A; 4KFN A; 2VX6 A; 1LBL A; 5F2I A; 4BFA A; 4MVA A; 4UWM A; 3EGJ A; 1DQU A; 3DZ1 A; 1NOU A; 5DEL A; 1R85 A; 4DXV A; 4IXH A; 3TYU A; 3E0V A; 2D3L A; 3IGX A; 4D8L A; 3PFR A; 4CVW A; 3AIW A; 6XIA A; 3H6O A; 5EMY A; 3LLF A; 3CHD A; 3QH2 A; 1YTK A; 1JZ6 A; 1W54 A; 3QYS A; 1H1Z A; 4C1K A; 3M0Y A; 1O7A A; 2FGL A; 4C7P A; 2OTJ U; 1B9Z A; 3TJI A; 3APT A; 2ZID A; 5IFT A; 4TR9 A; 3QN3 A; 3R1Z A; 3GYE A; 3DU0 A; 2XWE A; 3S46 A; 3VDH A; 1TPE A; 1XV8 A; 3GC2 A; 2OWC A; 1LKV X; 5KEZ A; 5H1W A; 2ABB A; 3FA4 A; 1DYS A; 1OIN A; 2A1Y A; 2DSU A; 1JCT A; 2Z7K A; 2X1U A; 4Z08 A; 5HA7 A; 3WUB A; 2X09 A; 4H4I A; 1Q6E A; 4DO5 A; 1V6T A; 3CU2 A; 2HXT A; 4M6U A; 3BV7 A; 4DXK A; 3TSD A; 5MKG B; 4DZH A; 3LEP A; 3EA2 A; 4GO8 A; 5A6J A; 2ACR A; 1MW2 A; 4WJE A; 1HL9 A; 2JBM A; 1FXP A; 3H2A A; 2H0K A; 4WR3 A; 3D46 A; 4ZQR A; 1HVF A; 3ANV A; 4GJ1 A; 1XSZ A; 4NAS A; 1P43 A; 3TYB A; 5HAO A; 3CEU A; 2BY3 A; 4EXA A; 3RX2 A; 4C6I A; 1EZW A; 1LLO A; 3O3R A; 2ZTK A; 1PKY A; 4GUF A; 3BUR A; 4J3U A; 1XLC A; 3UJS A; 5J5R A; 2UY5 A; 3S1U A; 1VQ7 U; 2Z24 A; 3ZXW A; 2W4X A; 2G4F A; 3W70 A; 1FCQ A; 3UT0 A; 3G18 A; 2QCN A; 3RX3 A; 3GXP A; 3VII A; 1GTE A; 3TW6 A; 3SY5 A; 3CCE U; 3II1 A; 3MKV A; 3UG3 A; 3TH6 A; 2GK1 B; 2X0Y A; 1J2B A; 3TJ4 A; 3HTW A; 1QPO A; 4Y9M A; 3JRK A; 4P7L A; 2J47 A; 3L0N A; 4HHH A; 4XD4 A; 4OIY A; 1VEO A; 4B91 A; 1PKN A; 3KBM A; 3UQN A; 4JHM A; 1BO9 A; 1GNX A; 2XTK A; 4LY4 A; 4I90 A; 2NX3 A; 1UWS A; 1JZ5 A; 1PE1 A; 1TFV A; 4PCN A; 1ME8 A; 5DMM A; 3W9Z A; 1NP2 A; 3BAK A; 3VGF A; 3WEO A; 2OOG A; 3PZQ A; 1UKQ A; 1VIZ A; 4EF8 A; 2GSE A; 1XS2 A; 2Y88 A; 4FSA A; 3MYO A; 1AL7 A; 3CEP A; 3TC3 A; 1XQK A; 4MPQ A; 3WQO A; 4ATW A; 3PZM A; 4Q4S A; 4TMB A; 4BF7 A; 4A3H A; 3RM9 A; 4PB4 A; 3A23 A; 3L5A A; 1V0N A; 4G56 A; 5C9Z A; 2AQW A; 3MKC A; 4I5R A; 2EPL X; 2GQ3 A; 1VFL A; 4HNO A; 3XIS A; 3ZT5 A; 5K2Z A; 2BGN E; 3FFS A; 4LYK A; 1VQP U; 3WUF A; 4K8E A; 1VBR A; 2VDG A; 3ZJ6 A; 1W5Q A; 4XZ9 A; 2WWD A; 4P7R A; 1DPM A; 1QT1 A; 4DF0 A; 4O18 A; 2GJX B; 4IFR A; 5LXX A; 3GZA A; 2QS8 A; 1GOW A; 3WQ8 A; 4CN6 A; 3FHQ A; 1VBH A; 1BCW A; 3G1V A; 1TYG A; 2DVU A; 3V16 A; 1JYW A; 3BZG A; 3NL5 A; 5AC7 A; 4H6Q A; 5EY5 A; 3AIV A; 3NG3 A; 3QPE A; 4KIE A; 1RXO B; 1J2W A; 1M41 A; 3VKJ A; 3HJZ A; 3H7V A; 5H8U A; 4K6A A; 1OD0 A; 3ZM8 A; 2FH8 A; 1UP0 A; 3T1G A; 4XZL X; 3WJY A; 3K46 A; 1FWA C; 3FGC A; 1PHW A; 4DBE A; 2Z28 A; 1TA3 B; 2V2C A; 4PTW A; 1BG9 A; 4HZ7 A; 1GOX A; 1GUV A; 1A5C A; 3OPS A; 3NQV A; 3UR8 A; 1GVR A; 2W61 A; 4IM4 A; 3TLL A; 1XYF A; 5HQM A; 1KD1 V; 1C29 A; 3B40 A; 1S2A A; 5LA2 A; 2V3E A; 4BKN A; 5HMD A; 1CB2 A; 5EBU A; 1G7V A; 1KBJ A; 1KCZ A; 1LQA A; 1GVY A; 3RO6 A; 5AEC A; 3PUE A; 3ES7 A; 2ZRW A; 1LF1 A; 1UOK A; 3FSU A; 1M04 A; 2BG5 A; 5AY7 A; 1DJQ A; 4Z0S A; 4YVX A; 1J79 A; 2DVT A; 4Y9O A; 4AX7 A; 1KFK A; 1DJW A; 4CQ9 A; 3O6C A; 5E09 A; 3M0M A; 1VFH A; 3S2N A; 3WNN A; 3N04 A; 4V1Y A; 2XO3 A; 2XM1 A; 5CSR A; 4DWS A; 4MLJ A; 1DTU A; 4S3G A; 1CXI A; 1B3Y A; 3NPW A; 3QHN A; 4R7J A; 3CCL U; 1H5Y A; 4JY8 A; 5LIY X; 1F61 A; 4AVO A; 3VIJ A; 3STP A; 1QAS A; 4TVD A; 1IEQ A; 2PDY A; 3WEL A; 1QCW A; 2V8Z A; 3ERP A; 5AOU A; 1GWJ A; 3UND A; 4DUX A; 2RHG A; 2V38 A; 3I7R A; 1KFL A; 1AH0 A; 1E55 A; 2BOD X; 4O14 A; 4Z1D A; 1R6W A; 3ARW A; 1GKR A; 3RJX A; 1PKL A; 3VIN A; 1W1P A; 4DWS D; 5KET A; 1ITX A; 3M43 A; 3EOE A; 4U28 A; 1FUY A; 3A3W A; 5FEP A; 4TVU A; 1BTM A; 2GGE A; 4FEZ A; 3WEM A; 3ZVH B; 3GK0 A; 4IIB A; 4XO6 A; 1DWF M; 2REQ A; 3BRX A; 3G5E A; 1Q7M A; 3L12 A; 2CHO A; 1UML A; 1K6A A; 4O13 A; 4GXW A; 5ABT A; 1WDD A; 3O63 A; 4K7Y A; 4M8U A; 3TZF A; 1F4A A; 3C0Q A; 3G71 U; 3CHE A; 5EPD A; 3R0U A; 3G1X A; 1ZAJ A; 4JTK A; 1Z41 A; 2WMH A; 4UMC A; 4B3K A; 3ABZ A; 2XVL A; 4X9S A; 3A13 A; 4PUD A; 3GS6 A; 4YPJ A; 4ZOC A; 4JAW A; 4A4P A; 1XUZ A; 3VUP A; 3MUA A; 5C9U A; 1YGM A; 4X63 A; 4E5T A; 1XP3 A; 1CCW B; 3R0D A; 5CPO A; 1FH7 A; 2VYO A; 3NCO A; 4LRS A; 3WD0 A; 1U83 A; 3TW9 A; 4M1R A; 2E6D A; 4AZG A; 3QKE A; 3WLI A; 4QTG A; 2ZZ6 A; 1ZAI A; 2PC8 A; 4AF0 B; 3IAN A; 4I7U A; 2J74 A; 1M6J A; 3VO0 A; 8RUC A; 1IXO A; 3GQY A; 4MTV A; 3QMS A; 3IUD A; 1XC4 A; 3B05 A; 4S3F A; 1IEV A; 2YW3 A; 1HKJ A; 1G5N A; 1YJN U; 2QJJ A; 3W4R A; 4JFW A; 1W31 A; 4LJ3 A; 4MKS A; 4ACY A; 3I68 A; 2H3D A; 4P8W A; 4W87 A; 2C13 A; 4HPG A; 1EWE A; 4UFH A; 2Q09 A; 4GI6 A; 3GHT A; 3KE0 A; 3AML A; 1GQI A; 4UOQ A; 1N7K A; 2ALD A; 2WMK A; 1M7O A; 2DW6 A; 2QDH A; 3AJC A; 3E0F A; 3OO2 A; 2GJN A; 4GH1 A; 1NEL A; 2XVK A; 1EJ7 L; 2ICS A; 5D9O A; 4E4U A; 5E1Q A; 5JIP A; 4EVF A; 4RR4 A; 3CME U; 3LX9 A; 3G1D A; 3RF9 A; 3UD6 A; 3YPI A; 4KCU A; 1P0D A; 1E4M M; 3NUK A; 1AZ2 A; 2D24 A; 1GHS A; 4UNL A; 4MJ2 A; 2WZT A; 1HVE A; 4D6H A; 5C7H A; 4Q7B A; 2OZ3 A; 1VPY A; 5B7Y A; 2YPI A; 1UA7 A; 1YHT A; 2VUY A; 5D4Y A; 4EAM A; 3CPG A; 2QLY A; 4DJF A; 3EWC A; 4HHM A; 3GNO A; 4KES A; 1B54 A; 4E8G A; 1A5S A; 1XXX A; 4UMA A; 3E02 A; 4C6L A; 3WD4 A; 1ITU A; 1FCU A; 2XH7 A; 1TRE A; 1FFQ A; 4UNK A; 1TV3 A; 4ZMG A; 1NY1 A; 5LGX A; 1YTE A; 4A29 A; 2GUU A; 2DZT A; 5I77 A; 2ISD A; 3H2F A; 1IT7 A; 5HPC A; 1K8X A; 5LA1 A; 5HJX A; 1DP0 A; 4KEU A; 1HW6 A; 3LI1 A; 3DFH A; 3SFW A; 1E6X M; 4B5V A; 3LS9 A; 1C8Y A; 3AXX A; 5DML A; 3QLL A; 4H3D A; 4BI6 A; 1MXS A; 4LAK A; 1EO5 A; 1CGW A; 1W9D M; 3CD6 U; 3E2S A; 2DQX A; 2VD9 A; 5HJY A; 1JZ3 A; 2EPN A; 4WJ8 A; 3GIY A; 3WUE A; 5EGR A; 1W9B M; 3WNP A; 5CPN A; 1K9M V; 1E56 A; 2ZZ4 A; 2X1S A; 2B4G A; 4ACZ B; 4UR7 A; 5G1T A; 1UD4 A; 6ENL A; 2I5Q A; 3QYQ A; 3WH6 A; 2VR4 A; 4LXF A; 3ME3 A; 1XIH A; 4L6H A; 1Q4N X; 1UP3 A; 4QS5 A; 4ADU A; 4F0R A; 2OX1 A; 3B97 A; 4OEC A; 2CBJ A; 3IEB A; 4IFK B; 1VCG A; 1Y5X A; 3VU2 A; 4C5Y A; 2YIH A; 3VDG A; 3W72 A; 3VOJ A; 1D9E A; 2XFY A; 2XWD A; 3D6N A; 5CC5 A; 1R87 A; 2VHL A; 1NF7 A; 2DJL A; 1JZ8 A; 1PDY A; 2YL8 A; 3MU7 A; 4CNO A; 1LOQ A; 2GUB A; 2JIW A; 4IHC A; 1A5L C; 4IG2 B; 1P0E A; 3GG8 A; 4KER A; 3EDK A; 1K4G A; 3USY A; 4GH3 A; 3THC A; 3TC7 A; 1R9Y A; 4MNJ A; 2E5C A; 1B31 A; 2FMN A; 2XN0 A; 4QNE A; 3OTR A; 3IIV A; 1JIB A; 1WYI A; 1Z6K A; 3DFY A; 4ZOE A; 1FCV A; 1EQP A; 1TPH 1; 2XVP A; 3K8L A; 1XDM A; 4B9Y A; 1KWG A; 2EPO A; 2FZB A; 1C90 A; 2V3F A; 3M64 A; 1NWS A; 2OZ0 A; 1K9F A; 3GLC A; 5D9N A; 4QQ3 A; 3DGR A; 4F2B A; 4PCS A; 1KV5 A; 1EIB A; 1S38 A; 4OWG A; 2IQD A; 4HCD A; 4YNG A; 2VFF A; 1NVM A; 1IE7 C; 1QVG T; 3W71 A; 5A95 A; 4BF5 A; 3DFS A; 5CCZ A; 2NUX A; 1F3T A; 3TE9 A; 1PTD A; 2VEN A; 1UPM B; 3TNB A; 2G8Z A; 5GJP A; 3CUG A; 2ZZ3 A; 3G22 A; 2IXU A; 5M1T A; 2OYK A; 3U4A A; 3VK5 A; 3P3B A; 4BF9 A; 1V6U A; 4GIS A; 3FXY A; 2OSX A; 3ZWS A; 2ZRU A; 3MMW A; 5AHL A; 1MQR A; 1GPF A; 2DUA A; 3CWN A; 1F3E A; 2WMF A; 2YXO A; 3PUL A; 3CB3 A; 1UH3 A; 3TXZ A; 1XI3 A; 4L4M A; 1B57 A; 1KLY A; 1HF6 A; 5F2L A; 2ZVR A; 2DT2 A; 4U31 A; 3OGS A; 2FZD A; 1J0Z A; 1HUV A; 4DBS A; 2ZC8 A; 3T9P A; 1W1B 1; 4DIA A; 1FTX A; 3HOJ A; 3OOQ A; 4GPN A; 2VFD A; 1CGT A; 5GJM A; 2R91 A; 2G96 A; 1UR8 A; 4AXK A; 2FH6 A; 2R9R A; 3BQ6 A; 4L80 A; 2F6U A; 1IZC A; 1ZQ5 A; 3A3X A; 2ONE A; 1M68 A; 3LCF A; 1AK5 A; 1KXH A; 1QOQ A; 2XGB A; 2OYM A; 4HIB A; 3NVT A; 4HMN A; 3FX4 A; 2WBK A; 1YDO A; 3HQN A; 1GG0 A; 4LEF A; 3BLI A; 2WLZ A; 1TPW A; 2FPT A; 4R7O A; 2YB1 A; 2GLK A; 4N9D A; 1QNQ A; 3WZ0 E; 4DB7 A; 1DJX A; 3PK7 A; 4AC1 X; 1E4N A; 1ONW A; 4QLJ A; 1GCY A; 3UVI A; 4PRT A; 3ENL A; 2DH2 A; 3RPD A; 3CAV A; 1RAK A; 4V27 A; 3CZG A; 2XJ7 A; 4JQ3 A; 1F9C A; 1QFE A; 3BAW A; 1R46 A; 3M6Y A; 3MO7 A; 2XH0 A; 4DJD C; 1K77 A; 4KFO A; 3FDG A; 1R9X A; 3DAQ A; 4KCT A; 4ILS A; 5L6U A; 4ADS A; 1M53 A; 4HB7 A; 3QQ1 A; 1XIN A; 4XSM A; 2LLE A; 2NZL A; 5PTD A; 1GOK A; 3BJT A; 3ANY A; 2YCL A; 3S9Y A; 4WIW A; 3BGJ A; 1R0M A; 3HM7 A; 1ZB5 A; 4BEU A; 2QCD B; 5F7U A; 3QGK C; 3I7S A; 4ZQC A; 4RE4 A; 2HXU A; 4QDW A; 3WQC A; 4HNT A; 3TK7 A; 3QBU A; 1YNP A; 3KDN A; 3FKK A; 5FKY A; 1PCK A; 4DBD A; 3WLR A; 3C3U A; 2J7F A; 5I2U A; 2VFH A; 3TV8 A; 1QUM A; 2DZP A; 2OEK A; 2YLA A; 4OOZ A; 3V75 A; 1XGZ A; 3DC8 A; 3L5M A; 4E38 A; 1PJ9 A; 9RUB A; 2CWN A; 1MI3 A; 1HFB A; 3OG3 A; 3AWO A; 3MUZ 1; 3IJ7 A; 4XAP A; 2ZUM A; 2OT1 A; 4M5P A; 1YI2 U; 2W79 A; 4M3P A; 2PDC A; 4S2C A; 4KCW A; 4V15 A; 1ADD A; 1IXP A; 1K3U A; 3M1Z A; 4QP0 A; 1KLZ A; 4N6E A; 4ERA A; 3A24 A; 3C52 A; 1EUN A; 2Q3O A; 4JTA A; 4NF7 A; 1F6Y A; 3OLG A; 1MXG A; 9CGT A; 1UWQ A; 3EWU A; 4B90 A; 5A8P A; 3QM3 A; 2UZ9 A; 2XG9 A; 1ZY9 A; 1TWI A; 3UCQ A; 3E5B A; 4P61 A; 3SBF A; 4FQ0 C; 1HQT A; 3NA8 A; 1XCF A; 4O4W A; 1EGV A; 1T6M A; 1XBZ A; 3VOH A; 1NU5 A; 4ZFM A; 4JYD A; 4HPN A; 4MZY A; 4WGH A; 1DKW A; 1X1Z A; 4X0N A; 1XLG A; 3TA6 A; 1IZJ A; 5I24 A; 3TSM A; 2X2Y A; 4CEX C; 3GNP A; 1QK2 A; 1W3W A; 4KF1 A; 2GFT A; 1JAK A; 4L56 A; 3UPM A; 3TA9 A; 2GYI A; 3QV7 A; 4V33 A; 5BOF A; 4MB1 A; 2PCQ A; 1JGM A; 1RK5 A; 1B9B A; 1GTH A; 1A3H A; 3KCL A; 1V3I A; 3MP3 A; 4EPD C; 1JZ4 A; 7REQ A; 4D1I A; 4N6F A; 2E7F A; 4FNU A; 1QI5 A; 1HJV A; 4AZH A; 1CB7 B; 3WLS A; 4DXX A; 2VGF A; 3WFA A; 2QCL A; 4F2U A; 3Q67 A; 2VWS A; 5C9X A; 2P3Z A; 3IIO A; 3MPG A; 1OFQ A; 3IJ6 A; 4Q8T A; 1Z7A A; 1ELS A; 1M7P A; 3OIX A; 4AWT A; 3UYI A; 4MJZ A; 3W1R A; 2GVL A; 2NQ9 A; 1Q8J A; 1NFB A; 5HUP A; 2OR2 A; 5C71 A; 5FL0 A; 3VTR A; 3MBF A; 4BWL C; 1XL9 A; 4LS1 A; 1C8V A; 4GVG A; 4JTL A; 4RW3 A; 7TAA A; 4JIE A; 1J0K A; 1VFO A; 3GAY A; 3AZT A; 3UWE A; 4A6G A; 3QOM A; 4W5U A; 3PVF A; 4IGM A; 4FNS A; 4JXH A; 3RU8 X; 5BYW A; 3VA8 A; 2XN2 A; 1UZ1 A; 2WC7 A; 5FJO A; 5B80 A; 3D3F A; 2YXX A; 1K8Z A; 1K7X A; 3WLO A; 3GU2 A; 3UXA A; 1Q7Y V; 2XSB A; 3A21 A; 1UMY A; 1HVG A; 4JTG A; 5D2T A; 5A2A A; 3URQ A; 2I1O A; 1B3V A; 3K8M A; 4R9O A; 3E5P A; 4E4F A; 2IKJ A; 2GOK A; 4JN8 A; 3ZWT A; 1WXY A; 3GG1 A; 3P80 A; 3QY7 A; 1W8G A; 5AVN A; 4PUE A; 3U5Y A; 1WZK A; 4PUJ A; 1KTB A; 1Q3N A; 4HNV A; 2J62 A; 1UBS A; 3MC5 A; 4S3J A; 2DJX A; 3VM5 A; 4KN4 X; 1LBF A; 1XGE A; 3VPL A; 3W2M A; 3BG5 A; 3VAB A; 3N9R A; 3CC2 U; 1I1X A; 2BVW A; 5DY2 A; 3S2J A; 1U2Y A; 6PTD A; 3KM8 A; 3NZG A; 4ZE5 A; 2ZRV A; 1MEW A; 1R2S A; 5JH1 A; 4JTT A; 5M2F X; 2QUV A; 2G3N A; 3N2X A; 4N02 A; 3LTP A; 1E0W A; 2WNQ A; 4WSH A; 1QW7 A; 5G2H A; 2O7Q A; 4NJK A; 4X9P A; 1S5W A; 3IRS A; 3KOW A; 1EPV A; 1NP0 A; 5KZ6 A; 5D33 A; 4GQ0 A; 2ZXB A; 1S5M A; 1OZQ A; 2JIE A; 1UWA A; 1ZCC A; 4MMW A; 3EX2 A; 1EC7 A; 1XCW A; 3ZZ1 A; 3U7B A; 3IGH X; 1Y8B A; 4H2H A; 5EMU A; 4YYF A; 4HME A; 1KFJ A; 1R8S E; 4K37 A; 1PX8 A; 4TX8 A; 4XSL A; 3PPG A; 1KNW A; 2YDR A; 1Q63 A; 3CCJ U; 2PWG A; 4I6V A; 3IJ9 A; 1LCO A; 1JXK A; 2OT0 A; 2A3H A; 1DIK A; 4CQD A; 2J8G A; 1GGO A; 2QW5 A; 3MO4 A; 2UY2 A; 3QGA C; 4GVH A; 2ZUS A; 1NFG A; 4N4P A; 3USW A; 4IP4 A; 1RCX B; 3U5U A; 4ICN A; 1IIG A; 4Z42 C; 2PLM A; 4EPK A; 4DUO A; 3VAV A; 3E6E A; 4OBT A; 4HOX A; 4IQD A; 3EYP A; 3O0K A; 5HWN A; 3RLU A; 3ITC A; 3FGC B; 4HZM A; 3ATY A; 3KRS A; 4DVO A; 4I6K A; 3QQW A; 4IPP A; 3EWD A; 5LKY A; 2ZIC A; 1ZNN A; 1OEP A; 1UD3 A; 4BEQ A; 1F3X A; 5I5R A; 4JKM A; 5AZ0 A; 1EA0 A; 4QG9 A; 4QEE A; 1AJZ A; 3QMT A; 5H2Z A; 4AAJ A; 3REQ A; 4F48 A; 5AYZ A; 1XSI A; 1FHU A; 1YQ2 A; 2QVN A; 2V5L A; 3CQJ A; 4L1X A; 4R34 A; 3VE9 A; 2PGR A; 1NWT A; 3QC0 A; 1M5W A; 5AHE A; 2YA0 A; 1BXB A; 1YLV A; 3C61 A; 4L7Z A; 1WDW A; 2OQH A; 1BCZ A; 2AKM A; 1HJU A; 1ZP3 A; 3P5Y A; 5CPQ A; 4XIM A; 3ITX A; 4C6N A; 1YIT U; 3KS5 A; 1WNO A; 1GQK A; 5E9A A; 3S1X A; 4AZ5 A; 1FY6 A; 1K7E A; 5AYB A; 3PBG A; 3SZ8 A; 1WKD A; 4O28 A; 4Z0J A; 4DWS B; 1TPV A; 3NZQ A; 1FOB A; 5JUV A; 1I1W A; 4UCG A; 1JCX A; 4HT3 A; 5BRO A; 4XBK A; 2SFP A; 3GTN A; 1BKS A; 1OGG A; 2CFF A; 5F2M A; 2E9B A; 1DTN A; 3MI2 A; 3AAL A; 3GY1 A; 3FA5 A; 4XZW A; 1B30 A; 4WY0 A; 2JF7 A; 3GDL A; 1T99 A; 4Q8M A; 4M82 A; 2GSJ A; 2R24 A; 2GDU A; 4LPC A; 3LGD A; 5AHN A; 3OOD A; 1A53 A; 4PYS A; 4LUJ A; 5CC6 A; 3L65 A; 3EDF A; 2Y4S A; 3VGH A; 2JAL A; 4LUY A; 5FEW A; 4N2Y A; 3OVG A; 4ICC X; 4U5I A; 1XKJ A; 4IX2 A; 1I2O A; 4N9B A; 3CIW A; 3HO8 A; 2QCF A; 1P5B A; 2AOS A; 2V5C A; 4D7R A; 2QAF A; 3MY9 A; 5C7V A; 1OAB A; 3CL6 A; 3CQ0 A; 3C2V A; 1FWS A; 3PTQ A; 3SY8 A; 1ZBC A; 3QHO A; 5CZK A; 2PDI A; 4MJ4 A; 4UMB A; 1VCF A; 1V7Y A; 2NX9 A; 4LBS A; 6ALD A; 4I9M A; 3A4X A; 4NAE A; 2BHY A; 2NLI A; 3F2C A; 3PHA A; 4CNS A; 1F4H A; 1JR1 A; 3FVM A; 3HGS A; 2X30 A; 4J4K A; 3DFO A; 2DIE A; 4CQC A; 2OLA A; 4AIB A; 1JUB A; 4DNM A; 2VC6 A; 1EJX C; 3NEH A; 3B06 A; 3N2B A; 4NNC A; 1UKO A; 1HKI A; 3QSR A; 2E68 A; 4MM1 A; 5MF5 A; 1A5U A; 4RE3 A; 2IXV A; 4W9T A; 4IIG A; 5DO8 A; 5AHM A; 5FF2 A; 7A3H A; 3L2L A; 1DJH A; 1OIM A; 3LPO A; 4Y9P A; 4H9U A; 3P93 A; 1TCM A; 1G5A A; 1UR9 A; 4CD5 A; 3O0F A; 3A4A A; 2QGY A; 2R8W A; 4ZB5 A; 3NYD A; 1H2J A; 1OWQ A; 4GJJ A; 3W86 A; 1UP2 A; 1W3I A; 2X0S A; 3BGA A; 4L61 A; 4QJ3 B; 2FZM A; 4E2D A; 5HUL A; 3E74 A; 4FPS A; 4J2M A; 3N2O A; 4J3T A; 1YJW U; 3LLD A; 1IEW A; 4A22 A; 4EIV A; 2WKJ A; 3O9N A; 1ALA A; 1WZM A; 4J28 A; 1BF6 A; 3R8G A; 4NQ1 A; 3OEX A; 2PZN A; 1K4H A; 2JQX A; 4XK2 A; 1E0V A; 4N7Q A; 2J7C A; 4ZO7 A; 2VGB A; 1ONX A; 3L66 A; 1N82 A; 4Y7E A; 4MUZ A; 5BXR A; 2V2D A; 3VFC A; 3TX9 A; 4FAM A; 2HZG A; 1XYZ A; 3G3M A; 5CBJ A; 3VGD A; 4Q3M A; 1K6W A; 1ICP A; 2P8C A; 1UD8 A; 5KIN A; 3A4W A; 1OFO A; 4EPE C; 3URN A; 1F2J A; 5EKY A; 4FOK A; 4AD4 A; 7PTD A; 5MLC Y; 4U2Z A; 1V02 E; 1BTC A; 2QCC A; 2Z0I A; 1UZ4 A; 1PBV A; 3WD2 A; 1QU4 A; 2VXG A; 2QEX U; 2OLS A; 2OSW A; 3LMY A; 4MNK A; 4EF9 A; 2X1I A; 4MKV A; 4C6P A; 2RAN A; 3R8H A; 1JDA A; 5D09 A; 1Q13 A; 4RDW A; 4XB3 A; 1T96 A; 4JYF A; 2DT1 A; 4KRU A; 5IOB A; 2WHK A; 4IFO A; 2R82 A; 3NM3 A; 3L4T A; 1RBA A; 3RPM A; 3MHG A; 1LOR A; 3VKD A; 1N8W A; 3S5O A; 2Y63 A; 4DO7 A; 2EAW A; 1J18 A; 3SCP A; 5K9X A; 4JTQ A; 1Z69 A; 2VUN A; 4QJ4 B; 1XW8 A; 1VH7 A; 3G6L A; 4K8H A; 1RY8 A; 5HWM A; 3TCE A; 2R0D A; 1SUX A; 3BV4 A; 3CK5 A; 3WLT A; 1MZR A; 4H6E A; 1UB3 A; 1N00 A; 5KZD A; 3SCN A; 1KC7 A; 1WDP A; 4FLR A; 3RIT A; 4F4R A; 3FND A; 2VR5 A; 1DWH M; 2PWU A; 5AC8 A; 3M7W A; 4H7Z A; 1WAW A; 1CW2 A; 1TTQ A; 1LM1 A; 4LF2 A; 3O07 A; 1GQJ A; 1FWW A; 3BJF A; 2W0U A; 3PB0 A; 5BU6 A; 3SUS A; 1O68 A; 4HNC A; 3EMQ A; 3A22 A; 3INP A; 4UTW C; 3QGV A; 3UZX A; 2PLC A; 3PTD A; 6CGT A; 5GJN A; 4JQ1 A; 3ZST A; 3LGE A; 1Q6R A; 3W7R A; 4CEU C; 3VOC A; 3UPB A; 3PPF A; 4B15 A; 4ICM A; 4ZVJ A; 2OEM A; 3A64 A; 4CN1 A; 3D3A A; 5A8O A; 3NVD A; 2PWE A; 4FF7 A; 1CGY A; 4DYK A; 3GVW A; 2JKP A; 1O5Q A; 2NQH A; 1R3T A; 2FPV A; 3BMW A; 2E6A A; 4HP5 A; 2OKO A; 4WKH A; 2C16 A; 1OX4 A; 1UD2 A; 4P7Q A; 1EGZ A; 4Q7N A; 2VMF A; 1E71 M; 1BYC A; 1QBB A; 2EPH A; 4J1O A; 5HBF A; 5D05 A; 5FSD C; 3LYE A; 4M24 A; 2X16 A; 3OGR A; 2WV8 A; 3TWA A; 1J01 A; 4NVT A; 1ONR A; 3DFP A; 4O15 A; 2HVN A; 2Q8X A; 3C8F A; 4PRR A; 1AQM A; 3H2N A; 3KHD A; 3UGR A; 1KXV A; 2GVY A; 2RAG A; 3GHR A; 2UV8 G; 3KLL A; 5TBO A; 1WA3 A; 3CCU U; 1LDC A; 3CCQ U; 3TC6 A; 1KFW A; 5A69 A; 3P5Z A; 3MUX A; 2C14 A; 3CIX A; 2Y6Z A; 3T8Q A; 3T07 A; 3UJ2 A; 1DXE A; 3K30 A; 3HPZ A; 2YBT A; 3VGB A; 2TYS A; 5K9D A; 2O9T A; 3AHY A; 4D6E A; 1XLE A; 1OX5 A; 4LGX A; 3KBS A; 2DP3 A; 4JIC A; 4H7V A; 2BLE A; 4AXN A; 4BY3 A; 3VNY A; 5DN1 A; 1P6B A; 1MMF A; 3CV6 A; 4G5D A; 2AAA A; 3GXT A; 1AVR A; 1KTC A; 1FWR A; 2L5H A; 2XSX A; 3ALF A; 3E49 A; 4NXK A; 4ZOD A; 1H7O A; 1IWA A; 3GE7 A; 4TX6 A; 5EYW A; 2QDD A; 4XD6 A; 5E70 A; 3CBW A; 4PCT A; 1ZJB A; 3TTO A; 5I78 A; 1Y5V A; 2GDQ A; 2XFR A; 2ZE3 A; 4A35 A; 1O1Z A; 1YHQ U; 1RVK A; 4YJ5 A; 4DJE A; 4ROQ A; 2CLK A; 1VD6 A; 3T40 A; 4IJB A; 4J3V A; 3HPA A; 4MUP A; 4ZXL A; 3F2B A; 3MWC A; 1RLD A; 3QFE A; 2ON3 A; 2E4T A; 4QXI A; 2WW5 A; 1F76 A; 2Z26 A; 2OVL A; 2PTW A; 4D6J A; 5DEZ A; 2GUY A; 2OMA A; 1P6W A; 2WWC A; 4DJE D; 1D2K A; 3H24 A; 2ISS A; 3GG0 A; 3VCC A; 1VRX A; 4LAO A; 3W3O A; 3DC0 A; 1KC8 V; 2J77 A; 3RIL A; 3C3N A; 1HJQ A; 3OJG A; 4LBR A; 1E6S M; 4O10 A; 4US5 A; 3BSH A; 3MQT A; 3MWA A; 3GNR A; 1BHG A; 1EJT C; 1EJR C; 5BRQ A; 4JMO A; 2X1T A; 1PAM A; 2W3Z A; 1VLI A; 3UCD A; 1YNQ A; 3CC4 U; 3SY1 A; 1XX1 A; 3LI0 A; 4NAF A; 4WUI A; 3FJ4 A; 3SSJ A; 1NM9 A; 3M6D A; 3LRM A; 1Q2S A; 1XG4 A; 1KO0 A; 2DOR A; 4US5 D; 1L0P A; 4RXG A; 3EKG A; 3HPF A; 2C1I A; 2QZR A; 5AC5 A; 4S3B A; 1XPG A; 2J7D A; 2HCV A; 2OO6 A; 1XIC A; 2XVG A; 2PDP A; 4GYF A; 4F0M A; 4UOZ A; 1G94 A; 3WQ4 A; 3E2R A; 4FIQ A; 3TCS A; 1AJ0 A; 1VFS A; 4WJX A; 4C5Z A; 3OIH A; 4I7V A; 3WML A; 1L6S A; 4FA3 A; 5CJ5 A; 3B8T A; 4LVG A; 1GYN A; 1W3T A; 3GVR A; 1QI3 A; 1H1N A; 2WHM A; 4EPB C; 1XUU A; 5DMN A; 1E73 M; 3V4B A; 1C7T A; 4D6D A; 3LQL A; 1BXN A; 2BDQ A; 4OU1 A; 3BLE A; 3ZT7 A; 5LNU A; 3VIP A; 3RME A; 3S9I A; 4FK9 A; 3CIV A; 1XFB A; 4FX7 A; 4UC5 A; 3GHS A; 3GZ3 A; 4O53 A; 4PBG A; 4CD7 A; 1MWO A; 1MEH A; 1L2U A; 4R9X A; 1PZ0 A; 3VX1 A; 4J2P A; 5JU6 A; 1BYA A; 1TRI A; 3Q37 A; 4RE2 A; 3WQ5 A; 4F9J A; 4RUB A; 5JQ9 A; 1QHZ A; 2WQD A; 3CZ8 A; 3GU9 A; 4OTK A; 1RBO B; 1E1F A; 1H7N A; 3QW4 B; 1DE6 A; 2HDJ A; 1UJQ A; 3DHD A; 2ZRX A; 1BC0 A; 4M7S A; 3VZY A; 4LWW A; 3KAO A; 5M5D A; 7REQ B; 2ZWY A; 3DER A; 1ADO A; 4RPP A; 1QWG A; 5D2V A; 3RFA A; 4C6Q A; 2BY2 A; 1AH4 A; 4KN7 X; 2Z4G A; 2HYW A; 3ABX A; 1AEI A; 3UWV A; 2PDW A; 3UYC A; 3M5Y A; 3EX1 A; 2ABA A; 4L4L A; 1U1H A; 2ISW A; 3AZR A; 3DBN A; 1W5P A; 4LT9 A; 4DLF A; 2J8T A; 1EDG A; 1EEX A; 2OYL A; 2E77 A; 1V03 A; 3ITO A; 4S3K A; 3AAM A; 3SO7 A; 5F2G A; 1V08 A; 4WRH A; 1JZ7 A; 4JDY A; 3OVQ A; 4JL2 A; 3EMZ A; 2CES A; 1KRC C; 1MZH A; 3PUD A; 5IHR A; 5LSM A; 1KRA C; 3QZE A; 4QGC A; 2NVC A; 3KFT A; 4S3E A; 1TE1 A; 1YMY A; 1TB3 A; 1TG7 A; 2O0T A; 3OQ8 A; 3NTY A; 5KIT A; 2E1W A; 5E0K A; 4M0X A; 3ABR A; 1MRQ A; 2YL6 A; 3FKR A; 2Z1B A; 1SYT A; 3GFZ A; 3ONC A; 4GI4 A; 4QNW A; 3VIO A; 4NI8 A; 1AOD A; 1VB9 A; 3TYE A; 2WCG A; 1E51 A; 1BD0 A; 2I17 A; 1Q6O A; 3H8A A; 3WK2 A; 1DOR A; 2Q6Z A; 5AN7 A; 2VGG A; 5I03 A; 3TZN A; 4E6M A; 1RA0 A; 4PMZ A; 1UKP A; 4ZR8 A; 5ABH A; 4LS0 A; 3RAO A; 1LUC A; 2OXN A; 3GHU A; 3NQE A; 3QOK A; 2Y2W A; 1TWD A; 4W8A A; 1ZCO A; 4IF2 A; 2OJP A; 1BRL B; 3PY2 A; 1A49 A; 1PUD A; 3TYZ A; 1D3G A; 4H19 A; 4F2T A; 2EG8 A; 4HA0 A; 3Q4D A; 2W66 A; 1OT2 A; 4IGS A; 1W1I E; 4RJE A; 3H55 A; 1MRA A; 2VRQ A; 4DLM A; 3W2L A; 1VJS A; 2X40 A; 4PEK A; 1LRT A; 4ROP A; 1QW8 A; 4M6Q A; 1ZJA A; 2PDM A; 1AVC A; 5AVH A; 2B81 A; 3WQP A; 2H9A A; 1HZY A; 3TJL A; 1LOL A; 2JE8 A; 2ZRY A; 3CH9 A; 1ISX A; 3MZ2 A; 3QQ0 A; 4M81 A; 3U3H A; 4HCH A; 3MHU A; 3FV9 A; 3AI0 A; 1U5J A; 1Z44 A; 8CGT A; 5CKS A; 1TZ7 A; 3FHA A; 3SQM A; 4BQ4 A; 1QHO A; 4XZH A; 4DZI A; 1GG1 A; 2VOT A; 3MSG A; 4GVI A; 3JU2 A; 4FOJ A; 3W7K A; 3TFX A; 3GFX A; 3J7Z X; 1O5X A; 3NQ7 A; 4QS6 A; 1KM1 A; 1JNE A; 3KOX A; 2E8Y A; 3SGZ A; 3SN1 A; 3A5V A; 1QXL A; 2PDU A; 4Q8P A; 3KP0 A; 1YIJ U; 3FAX A; 3WDP P; 1I2N A; 1SJB A; 3NXL A; 4LQ1 A; 2IPF A; 3NOE A; 3HA1 A; 3VIG A; 3GDR A; 1AJ2 A; 3WJX A; 5IRP A; 4AWU A; 4RE0 A; 3O6Y X; 3O15 A; 5HK4 A; 2H0M A; 1JEZ A; 1RA5 A; 1W37 A; 3FAW A; 2E0P A; 3TAO A; 4GUG A; 1YE4 A; 3A3H A; 1VYP X; 5CJN A; 2HE5 A; 4MDP A; 1XG3 A; 4QLK A; 1OK4 A; 1XCX A; 3CO4 A; 4IL0 A; 1X6U A; 3REQ B; 4DBW A; 2FMO A; 2XUC A; 1J8V A; 2Y5J A; 1O60 A; 3W01 A; 2GL5 A; 3TAK A; 3DYO A; 3T44 A; 3WDJ A; 4REQ A; 3FOK A; 3DGB A; 4CKQ A; 1US0 A; 2XSA A; 5LIK X; 4DYE A; 1SW3 A; 4WK9 A; 2PAJ A; 1JVN A; 2CCR A; 2VZV A; 4J5N A; 1JUE A; 2H6R A; 1NH6 A; 1GOI A; 1B90 A; 4P7N A; 3ZJ8 A; 2D0G A; 4ZST A; 1KGX A; 4R53 A; 3W7J A; 4IMF A; 2H8Z A; 1RP9 A; 3HB9 A; 3RX4 A; 2A7N A; 5K4Z A; 2HRW A; 5EA9 A; 4ZQP A; 1R86 A; 1E1C A; 3N6Q A; 5TIM A; 2QUN A; 2QF8 A; 4WY0 B; 4WKA A; 1UIP A; 1SPQ A; 3PJW A; 4B5X A; 2FHF A; 1I0D A; 3B9E A; 2DH3 A; 5HQE A; 3MMU A; 1WDR A; 3TU9 A; 3N19 B; 3W00 A; 4H04 A; 4JDB A; 4PRW A; 4D2J A; 3HJE A; 4TV5 A; 1BXC A; 3BAI A; 5HPO A; 4S3D A; 4UTU A; 2OGY A; 4LU0 A; 3AHT A; 1TZZ A; 2E5D A; 2C0H A; 3BOX A; 1SSD A; 4EK7 A; 3CZE A; 3GWQ A; 3QTP A; 3AYT A; 1GON A; 1KCK A; 1E5N A; 1PYF A; 1AD4 A; 1V0L A; 3KBN A; 4HSH A; 1EUA A; 1ZJI A; 3R79 A; 2FFC A; 2XHY A; 3VM7 A; 1R38 A; 4G7F A; 5G4H C; 3OBA A; 4PMQ A; 1Q82 V; 1EZ2 A; 3N2C A; 3MAN A; 4WCJ A; 5D37 A; 1TMH A; 5DT5 A; 4ZBC A; 3VVG A; 2X7W A; 2D69 A; 2HYU A; 4LCS A; 3WN6 A; 4LB4 A; 4NAE B; 1KXT A; 3KHJ A; 3PTM A; 5JH2 A; 3N17 A; 4C7F A; 2GGI A; 3HZ3 A; 4DO4 A; 1ZBV A; 4F9D A; 5FF4 A; 2H90 A; 1K9D A; 4QRN A; 3SQS A; 1WKE A; 3H4G A; 1A4L A; 3FIG A; 5C70 A; 2IMR A; 4RV8 A; 1QI4 A; 4IZG A; 4IIC A; 4C6J A; 1LT8 A; 4X22 A; 2J25 A; 1TEL A; 3PG8 A; 2YDS A; 1XII A; 3LCG A; 4ZA0 A; 3W37 A; 3WKZ A; 1S3H A; 1OAD A; 4RDY A; 1KKR A; 4UTW A; 5A6L A; 1NDW A; 4GOA A; 3TLQ A; 3OBE A; 4LVA A; 2CZ5 A; 3T2O A; 2VO5 A; 3N2T A; 3EOU A; 1UH2 A; 2P10 A; 3G8R A; 1OHL A; 1RVG A; 2VEF A; 3BUV A; 3EOO A; 4BS0 A; 2QIW A; 1R9Z A; 3LEN A; 2DZV A; 5DXX A; 1Q4W A; 3K1D A; 3LGG A; 2CKR A; 1EF2 A; 2PGW A; 1TJ2 A; 2E6Y A; 4IUG A; 4CNN A; 4D7Q A; 1GYM A; 3UZY A; 2BNJ A; 2I9U A; 4RDV A; 4PCF A; 2X41 A; 3B8S A; 1UQY A; 4WEV X; 1U33 A; 2G5W A; 1DHP A; 4A3Q A; 4GYJ A; 5EFF A; 3GB6 A; 3BMV A; 3VMP A; 1RK6 A; 3K13 A; 4Y6G A; 5FES A; 1IXQ A; 2WPG A; 3ARQ A; 1DIO A; 4UQE A; 3VDB A; 4KCV A; 4IOT A; 3VI2 A; 4HN4 A; 1R7A A; 1GW1 A; 1QVB A; 4LYR A; 3BJS A; 4JL1 A; 1ZL1 A; 1XLB A; 1JYN A; 2J9Z A; 1B1Y A; 5GNX A; 5FIP A; 3F03 K; 3H4W A; 4H8N A; 3OW1 A; 2QF7 A; 2BXY A; 4UCF A; 2OOD A; 4LNC A; 5D03 A; 3OU8 A; 4J3S A; 3WL1 A; 1V4Y A; 1ZS2 A; 5DM6 U; 4LV4 A; 2DZB A; 1MCX A; 4GNJ A; 3BGG A; 1N44 A; 2F8Q A; 5G1M A; 3WG6 A; 1TWW A; 2WM1 A; 3WLK X; 1UR1 A; 3NPU A; 2QMK A; 4PUW A; 2YCE A; 4IPN B; 2QPU A; 5A8N A; 4GIN A; 2C19 A; 2PFH A; 4LSB A; 4N4Q A; 5HWJ A; 3ARX A; 3NL3 A; 1SR9 A; 2GQ9 A; 5C0Q A; 4P5U A; 2BOE X; 2GJR A; 2ZX6 A; 2ZXQ A; 5KZM A; 3T2P A; 4BHY A; 2ZZ1 A; 1EH9 A; 1QK0 A; 1UG6 A; 1FDZ A; 3XIM A; 3GKA A; 2GOU A; 1G6C A; 3SJN A; 1TV2 A; 1MXD A; 1BKH A; 5I23 A; 1DWJ M; 2B0M A; 1O1H A; 4D71 A; 3NSN A; 4PMU A; 2PA6 A; 1E43 A; 3GNN A; 4OQV A; 3L0K A; 3WK3 A; 3L0G A; 3WK0 A; 5F1V A; 3PZT A; 3VNI A; 3P2V A; 4O1B A; 3B8W A; 3ARP A; 4AIS A; 2ZWZ A; 4AZ6 A; 5IHS A; 2PC4 A; 1XYC A; 3KZP A; 1IEI A; 4ERG A; 2XO2 A; 3OLD A; 4DZA A; 3VIK A; 2D22 A; 4Q22 A; 1JYX A; 3TKF A; 1M1K V; 4DAI A; 3CCM U; 3UWY A; 2E28 A; 3C0L A; 2J75 A; 4HQV A; 2HNE A; 1XBY A; 3RYS A; 1S5N A; 4YU1 A; 1OG0 A; 4JCM A; 4GUH A; 4OOU A; 3T42 A; 3AO0 A; 3NGF A; 1UPT B; 4DRS A; 1BVN P; 1MQQ A; 3V0U A; 1JPI A; 2OX4 A; 5CPM A; 4DJF C; 1IXN A; 5CPL A; 3JUG A; 3F4V A; 4Z2I A; 3MW7 A; 2WAN A; 2V5K A; 3W75 A; 2A21 A; 3OZM A; 5DX7 A; 1RH9 A; 1ZAL A; 2DZU A; 3L9C A; 4MFE A; 3RCN A; 4GQB A; 2WJZ A; 4V2X A; 4NZJ A; 2F9R A; 1QHP A; 4P7O A; 2WZM A; 2V67 A; 1BSL A; 4P8U A; 3VZX A; 1CXH A; 4Q4M A; 1W5M A; 3GM8 A; 1OFR A; 2IW0 A; 1XQL A; 1NDV A; 1S2V A; 2OEL A; 4XUG A; 1ICS A; 2PLJ A; 2XN1 A; 3GDM A; 2GVE A; 2KZH A; 3LRK A; 4JEQ A; 3W1U A; 2PLK A; 4PUM A; 3PUO A; 4HK7 A; 1AMK A; 3NAV A; 4GYK A; 1M3U A; 3H21 A; 1CT5 A; 1HO1 A; 4DQW A; 4A05 A; 1AOW A; 5D8W A; 5FSE C; 3L4V A; 4NP7 A; 1OCB A; 1QOP A; 3GUG A; 4NX0 A; 4AY8 A; 2DPE A; 4PFH A; 1I45 A; 3D0C A; 3CMJ A; 3CHV A; 3AXK A; 4PB3 A; 3OYX A; 4Z1B A; 3RR1 A; 2Y0F A; 4GUI A; 2C8N A; 3OQE A; 3SRF A; 3ITT A; 3VMO A; 1XFC A; 3IAC A; 3UW6 A; 3G2G A; 3M41 A; 1P6C A; 3IV8 A; 1VP5 A; 1ZZM A; 4OBR A; 1EC9 A; 4HKP A; 1FH8 A; 2HJP A; 1IHI A; 4I7W A; 4Q8W A; 4ZGY A; 4O8R A; 1ML1 A; 3W25 A; 1EKO A; 4ATD A; 2CLH A; 3JS3 A; 1QWK A; 4CCD A; 4Q4Q A; 1OBA A; 3PJU A; 2WC4 A; 1N41 A; 1JYV A; 3VKC A; 5REQ A; 1VQ4 U; 2JGQ A; 4I5U A; 1BVW A; 3HFY A; 3MSD A; 3CUZ A; 3SW6 A; 1CX9 A; 1KR1 A; 3UJR A; 3C2Y A; 2CYG A; 1AII A; 3DHF A; 2VKZ G; 4HD5 A; 1HGW A; 4EAY A; 4IIH A; 4J3X A; 2ZXD A; 1CXF A; 4DX3 A; 2A4A A; 3MHF A; 4BI7 A; 1G4S A; 4M80 A; 1ICQ A; 3RXZ A; 5A2B A; 1Q7Q A; 1VFF A; 1YXC A; 5CSS A; 3R25 A; 1ZLP A; 3KVM A; 3P6L A; 1W45 A; 2GJL A; 4HA4 A; 2PGF A; 3LPP A; 1C8X A; 3QV8 A; 4YTS A; 4KWV A; 3EDJ A; 4H9M A; 1EYE A; 3O7U A; 5FAC A; 3QJA A; 3BSG A; 4MWA A; 3SFK A; 3B0V C; 4IXX A; 1X97 A; 4W84 A; 4ZSU A; 2OGJ A; 7ODC A; 3KBW A; 2DZX A; 4G8T A; 4O17 A; 2CU0 A; 4LBU A; 2IPW A; 3SAZ A; 3HQO A; 5FI8 A; 1ISZ A; 2E67 A; 4QAW A; 2BXV A; 1L6F A; 4XG1 A; 2D2G A; 3KVK A; 4NAF B; 4TQT A; 4LVB A; 4MWA D; 3UWZ A; 2Y5S A; 4GX8 A; 2ACU A; 2POZ A; 2VZU A; 1JFH A; 1DM5 A; 4O8A A; 5XIM A; 1LL4 A; 3MVT A; 2PDL A; 3MDW A; 3VDA A; 3KTC A; 3BAN A; 3EWW A; 1W9V A; 7XIM A; 2YYU A; 5I3H A; 2QV4 A; 4UTL A; 2BVD A; 4K3Z A; 2INE A; 2BAS A; 4ONV A; 2TMD A; 4HMD A; 3KXQ A; 3MI3 A; 3CUI A; 5LIU X; 1R8Q E; 3UJF A; 1L8N A; 3PTK A; 2IE6 A; 3S47 A; 1H50 A; 1ITC A; 1JCL A; 3PZI A; 2DSK A; 3STE A; 3EKZ A; 1LTD A; 4FI4 A; 3ARS A; 2C6Q A; 2QJM A; 4JTJ A; 3UA3 A; 2WOQ A; 3K5X A; 1YPI A; 2VXN A; 5C54 A; 2G50 A; 2PDF A; 4OHQ A; 3RG6 A; 3M44 A; 4XAY A; 2GGJ A; 5JBE A; 1RZM A; 4JTF A; 3QY8 A; 1BAG A; 3S5S A; 3N4A A; 2YLL A; 3DFT A; 3HG2 A; 4GCA A; 2PCE A; 3SCU A; 3S6O A; 1EDQ A; 4Z1Y A; 3NQ6 A; 1SW7 A; 4QEH A; 1MUC A; 3WJS A; 2F6X A; 2G41 A; 3SDO A; 4YS1 A; 4C6M A; 5EFD A; 4L4P A; 2EG7 A; 4DF2 A; 1VQ6 U; 4G6C A; 1MQP A; 1HNY A; 3UCC A; 3NQD A; 1PX4 A; 4H8H A; 3IJI A; 4HNN A; 5AN9 G; 5D2W A; 1VHC A; 1GQN A; 3UZJ A; 3TQK A; 1N8I A; 4ZQM A; 1ZAH A; 2E9M A; 3LM7 A; 4PN2 A; 1F6K A; 1EGM A; 1BSI A; 3DUG A; 4HVX A; 1KGU A; 2O92 A; 1TR9 A; 4DNH A; 5LIX X; 3CZL A; 3CCV U; 1SO5 A; 2WSQ A; 4QR6 A; 2F6K A; 3R5E A; 3G8E A; 3PZV A; 1FWH C; 1B3W A; 2ZZ2 A; 3J7H A; 1Q6Q A; 4GHR A; 3W6L A; 2J27 A; 1R8M E; 1FRB A; 3T55 A; 2WNZ A; 2G95 A; 3EAU A; 5BXT A; 2WKG A; 4PMJ A; 1WOA A; 2XIS A; 2FLI A; 2VP8 A; 2WMG A; 4A3Y A; 3WDR A; 1OG6 A; 5BW6 A; 5BZA A; 2HVM A; 1SSG A; 3IIX A; 4DEL A; 2JEQ A; 2ACQ A; 2QAP A; 4XTI A; 1AXN A; 1CLK A; 2PTY A; 2NUW A; 1IF2 A; 3L68 A; 3N14 A; 3UR2 A; 4RNJ A; 5M03 A; 1QW9 A; 3GBD A; 3LN3 A; 1UHV A; 4UXD A; 4GWE A; 3EEZ A; 5FEX A; 4RXF A; 4ECL A; 4FXS A; 4YTT A; 1RJP A; 2Z27 A; 4JKL A; 5A1A A; 3H2M A; 1QOX A; 2ZXA A; 4KEV A; 2V63 A; 1E0U A; 1KFE A; 2ZYF A; 2QHA A; 4IPI A; 1U5V A; 4PTM A; 5FJU A; 4P2V A; 2WK2 A; 4QE4 A; 3RM4 A; 3VUS A; 3ELF A; 3UZ5 A; 1A0D A; 3P0W A; 4YCP A; 1CZ1 A; 1ANW A; 4AZI A; 2XU2 A; 4GQG A; 4M6P A; 1GJW A; 4UTJ A; 1JPK A; 1S2T A; 3TWB A; 4EWJ A; 3N15 A; 5ABE A; 3FEM A; 4TTU A; 4UZU A; 1XSJ A; 4C7D A; 4DWD A; 5FJJ A; 2CW6 A; 2XFF A; 3W88 A; 2DT0 A; 2BVT A; 3LG3 A; 2PEV A; 3LTY A; 3GAK A; 4UNI A; 3WMS A; 1JQV A; 1QO2 A; 2C0A A; 1EB3 A; 1ZTV A; 4OV4 A; 3G0X A; 3RHG A; 3C5Q A; 3MSY A; 1W3J A; 4JIH A; 2NSX A; 4L1G A; 4C6C A; 2WMI B; 4CGT A; 3PM6 A; 3VD7 A; 2HEJ A; 3KBJ A; 4C91 A; 4IX0 A; 3DG6 A; 4PFM A; 5ECU A; 3QXB A; 1Q2R A; 4B4H A; 3VC5 A; 3IGS A; 5FL1 A; 2JKE A; 1XLD A; 2DUZ A; 3W0K A; 1AL8 A; 5EB8 A; 2WBG A; 3L23 A; 4KGL A; 5HMS A; 3WF1 A; 1PYM A; 2H9A B; 3LD5 A; 4HPH A; 4KRT A; 1BC9 A; 1E9L A; 3AIE A; 1JQX A; 2ISV A; 3WH9 A; 1P1M A; 3GIQ A; 1ITQ A; 2QV5 A; 4C1L A; 4MB5 A; 3V3W A; 2OU4 A; 5BXS A; 3M0L A; 1QDS A; 1WZA A; 1V7A A; 1XLI A; 5A6B A; 1U30 A; 4FXJ A; 3VNK A; 4EVH A; 1EJW C; 5G4W A; 4B5W A; 3GW0 A; 4NJG A; 1QJW A; 1X98 A; 2W5F A; 3W7O A; 4E4P A; 3LQG A; 2NXH A; 1HKV A; 3LNM A; 3VKB A; 4QDP A; 3EBV A; 2HS6 A; 1V3K A; 3HJL A; 2QCG A; 5I00 A; 5DFA A; 1UKT A; 3G0U A; 4DI8 A; 1MEI A; 5GNZ A; 3ZLF A; 4GD0 A; 5D9M A; 1XJB A; 5C2G A; 4NZ3 A; 3WEZ A; 4F40 A; 3BZJ A; 4GIE A; 5E6Z A; 3SCS A; 5LZL A; 3PG9 A; 2ZX5 A; 3U6W A; 3L4Y A; 4LYP A; 3EKL A; 4ZEN A; 2QEZ A; 1TPB 1; 1S2C A; 1TJ1 A; 5FJP A; 3OA3 A; 4GXM A; 1IV8 A; 3GDB A; 4PE8 A; 5LNS A; 1M7J A; 3PZU A; 3OBK A; 2IIC A; 5AYI A; 2A6N A; 1IGS A; 1KTN A; 4REQ B; 4NY2 A; 1XRS A; 3R13 A; 4AIE A; 2E5B A; 2ALR A; 1I60 A; 2A79 A; 5T9C E; 4XBJ A; 2WSR A; 3S42 A; 3VDC A; 1OB0 A; 3P61 A; 1CEO A; 1LG1 A; 4W85 A; 4MAD A; 1REQ B; 4NL1 A; 1QNR A; 5CSY A; 5CZT A; 3T4W A; 5BNK A; 2VFI A; 2ZUW A; 1JD9 A; 4R33 A; 2FIQ A; 1PIF A; 5EMM A; 3QKZ A; 1TIW A; 1DJI A; 2GJX A; 4YTR A; 1CBG A; 5H6H A; 3N6J A; 3MKK A; 3DKJ A; 1YNY A; 3WLN A; 3TYC A; 1E1C B; 3SWV A; 3SAD A; 3W2U A; 1IWB A; 4DUV A; 1B4K A; 3I4E A; 2ZHI A; 3BIC A; 3FDK A; 4NHV A; 4O0Z A; 2QPX A; 5T26 A; 5HAN A; 1XYM A; 3FK4 A; 1DED A; 3RMJ A; 3P84 A; 1WV2 A; 5SWU A; 3CYV A; 4UR8 A; 3AWN A; 1TMQ A; 1UUM A; 5A8Q A; 3AHX A; 2QDE A; 3GX9 A; 3MS8 A; 1H09 A; 5T91 A; 4AG0 A; 5E6Y A; 1WDQ A; 2AMX A; 3VD9 A; 1MDL A; 5I6S A; 3LOT A; 1BEU A; 3KWS A; 3W1P A; 1TA3 A; 6REQ A; 1V6X A; 4XZI A; 1QPR A; 3LHY A; 3OB8 A; 3ZFH A; 3BL3 A; 1VGA A; 2HRO A; 3DOP A; 3T6C A; 2ZOC A; 4JBR A; 4LAZ A; 5K9C A; 3CL8 A; 4DO6 A; 3ARZ A; 2NQZ A; 4FXR A; 2VT0 A; 1KBI A; 3B9O A; 1A8B A; 4D9P A; 3WQE A; 3DDM A; 5E9X A; 4KWO A; 1HO4 A; 4JTS A; 3SQL A; 4D1J A; 3O1N A; 3T2Q A; 3E96 A; 4HPX A; 1GK8 A; 2HV5 A; 1A4M A; 3W38 A; 1U1J A; 1AVA A; 3PZG A; 5BCA A; 3DES A; 1HIZ A; 1EOM A; 4JY9 A; 3M4H A; 1LJY A; 4AVN A; 1W2B T; 5CAK A; 4C6B A; 1S9D E; 4HU3 A; 2HSN A; 4L5Z A; 3MCN A; 5I02 A; 4GQQ A; 2ACS A; 3C86 A; 5L9F A; 3VD4 A; 2QNE A; 3PNZ A; 4ADT A; 4UQB A; 4YHG A; 3DBP A; 2XGZ A; 2P8B A; 1JI1 A; 1UD6 A; 4KWS A; 3NZ1 A; 1RE0 B; 2NV9 A; 1KKO A; 2GZX A; 2NT0 A; 4P8X A; 5GNY A; 5LA0 A; 4INF A; 1LRO A; 4HTY A; 4GUJ A; 4L5M A; 1LLW A; 3W26 A; 4CQ8 A; 4UTI A; 5A3H A; 3CM4 A; 2ZC1 A; 1VPQ A; 1TR1 A; 4PR4 A; 2J66 A; 1XD1 A; 3MUC A; 3WYZ A; 4LB3 A; 5IM5 1; 1S39 A; 4FLQ A; 3GTX A; 2HS8 A; 3CV1 A; 3ZVH A; 5CSU A; 4KMU X; 4ES4 A; 3LA4 A; 1UZD A; 2EJA A; 3SIM A; 1PBG A; 2DGA A; 1L6Y A; 4A3R A; 1ONE A; 1ZFJ A; 3HPX A; 5J8L A; 1MO0 A; 4JXC A; 4IPL A; 3FYY A; 3RCY A; 2NQ5 A; 2ZDR A; 4MDO A; 1CDG A; 4PUU A; 1IIH A; 1JDD A; 1E3Z A; 3F4W A; 2J78 A; 3NDY A; 1VF8 A; 3TXV A; 4QCF A; 1CXE A; 1HXJ A; 4PTD A; 4LUS A; 3K2G A; 4DJF D; 2QUU A; 1BPL B; 1HKM A; 1E0X A; 1QML A; 3ZR5 A; 4C0A A; 5I3D A; 2YB4 A; 3RLH A; 5CQ1 A; 4QE1 A; 3SEP A; 2NQJ A; 4AMA A; 1B3Z A; 4Q4P A; 1XRV A; 2D0H A; 2Y8U A; 2F61 A; 3HFT A; 4NKC A; 2YL9 A; 4Q6X A; 2JKA A; 4GY0 A; 2XIN A; 4BJH A; 1HVX A; 1CI1 A; 2V3D A; 4MKN A; 2PF0 A; 1O94 A; 1FX6 A; 2CNC A; 2HTM A; 4RZ5 A; 3BLP X; 4IJR A; 2WCS A; 1WCG A; 3FEQ A; 1BF2 A; 1H7R A; 2FQI A; 4UZS A; 4AEF A; 1V93 A; 1Q5M A; 4HHP A; 3QPZ A; 1IZK A; 5LXE A; 4U2Y A; 4QM1 A; 4OV9 A; 3IWP A; 1Z89 A; 3UFY A; 4J3Z A; 4W88 A; 3VXG A; 3LNP A; 3CCR U; 3CPU A; 4CU7 A; 3RRA A; 2PRL A; 2CER A; 4Q8Q A; 5LHF A; 2ZA3 A; 3BLK A; 3CUX A; 1N2V A; 5F4X A; 4KRZ A; 2PSN A; 3MV0 1; 1RY0 A; 1JD7 A; 5CZJ A; 4NNB A; 3I56 U; 4S3C A; 3SUT A; 3M0K A; 2PP0 A; 2A85 A; 2DIK A; 3TIM A; 3OYZ A; 5F4S A; 4B16 A; 1O12 A; 3ZSS A; 1RHC A; 5IBX C; 4ZTX A; 5LHE A; 1QNO A; 1LWH A; 1VQN U; 3G3D A; 1ZRQ A; 2HBX A; 3NSX A; 1JUL A; 1UUQ A; 1M9I A; 4PEE A; 4WDT A; 1M03 A; 2BTM A; 5CGQ A; 3UJE A; 2FDM A; 2Y1H A; 1ZGD A; 4JTU A; 4HU4 A; 4HK6 A; 2VEG A; 4DN1 A; 4IGH A; 3W2N A; 1MNS A; 5DY3 A; 2C91 A; 3I7Q A; 5G1Y A; 2Q02 A; 4M1B A; 5A2C A; 4G9P A; 1J0Y A; 2BSL A; 3K1G A; 4R5E A; 2ZUN A; 2NUY A; 2D23 A; 3UR5 A; 3ITL A; 2OLH A; 1E9I A; 2ZUV A; 4OVX A; 5CEW A; 4W89 A; 3IPW A; 2VR2 A; 1HJW A; 3RGT A; 1SQ7 A; 2PE4 A; 3IVS A; 4GNK B; 3LXC A; 2FGZ A; 3O16 A; 2II2 A; 4AIU A; 2CZV A; 3SRH A; 3BPW A; 3WH7 A; 4C90 A; 5D32 A; 3M0V A; 4O1C A; 2EPM X; 2AL2 A; 4GX9 A; 3A9B A; 4LEQ A; 4NJJ A; 3FJL A; 1J0J A; 4XQ6 A; 1T5A A; 2WZI A; 3F4N A; 4PTN A; 1E70 M; 3W82 A; 2EKG A; 4LNY A; 4YN5 A; 2QR6 A; 1KM6 A; 1W5N A; 3EPO A; 5AKQ A; 1QR7 A; 3H7U A; 3C2O A; 2I56 A; 3W85 A; 4ZL1 A; 1QNV A; 1KCX A; 5A6T C; 1TML A; 3AIR A; 1VLW A; 2Y24 A; 2P5N A; 2Y62 A; 3CQI A; 4U10 A; 4ZB0 A; 1UWR A; 3KRU A; 3KRB A; 4B5U A; 1XWY A; 1NTH A; 1OCJ A; 3BAY A; 3N18 A; 3U2Z A; 2G0W A; 3ZOA A; 3DG7 A; 4TMC A; 4V28 A; 1PCW A; 2ASH A; 3WBW A; 4I8Y A; 2QJI A; 5DCE A; 3ABQ A; 3FVD A; 5C9R A; 1L9W A; 4QIT A; 3VOG A; 3UNT A; 3VFL A; 2OKT A; 1LWI A; 4DUW A; 3P81 A; 1TTP A; 3R0K A; 2WVS A; 2G3J A; 1FXQ A; 1GOR A; 1BWK A; 3BCJ A; 2ZZ5 A; 3ABS A; 3UR7 A; 2PU0 A; 3W1A A; 3WF4 A; 4O52 A; 1SZN A; 2V5D A; 4IG2 A; 3CUF A; 2GJP A; 1H7W A; 3OGV A; 5H91 A; 3NEV A; 2YL5 A; 3WUG A; 1QNP A; 4GA8 A; 3PAJ A; 4UTF A; 4GIR A; 1NWU A; 1F6P A; 1LL7 A; 1M01 A; 3MUY 1; 5REQ B; 1UJP A; 1UQZ A; 3WQ6 A; 3B8V A; 4E3T A; 3SUU A; 4BCE A; 4AB4 A; 3B8D A; 5C6M A; 4JQ2 A; 2E25 A; 4CUC A; 3KZS A; 5A94 A; 3NO1 A; 5KMY A; 3A47 A; 1CPU A; 2NQL A; 1XYA A; 2YA1 A; 4PGL A; 4UTT A; 3WMC A; 1R4A E; 1E9Z B; 3U7V A; 4ZTY A; 3L67 A; 4H9T A; 1SO3 A; 1V5X A; 1E6N A; 3W2J A; 4PMN A; 1Q6D A; 3QLD A; 4JIP A; 3W7M A; 5IDZ A; 5H8M A; 6TIM A; 1HT6 A; 5HIN A; 3NL6 A; 4BI5 A; 1PVN A; 8A3H A; 3F5L A; 3IS4 A; 4WSJ A; 1JJ2 T; 3TMQ A; 3V18 A; 4W4Q A; 1E9Y B; 4JII X; 1GEQ A; 3G1R A; 2PD5 A; 1T41 A; 3LDV A; 4IWW A; 1O5K A; 4TU1 A; 3ICJ A; 3VCR A; 4F0S A; 4F8X A; 1R8L A; 4MWA B; 4LRT A; 1RD5 A; 3QW3 A; 1S2U A; 3WLH A; 1UD5 A; 1YPF A; 4D6C A; 3I3D A; 1KM0 A; 1UC4 A; 4M56 A; 3GUW A; 4S39 A; 1YII A; 3LK6 A; 4NIR A; 3S5N A; 3S83 A; 1URO A; 1GOQ A; 5AB3 A; 5EMJ A; 1QI2 A; 4PUL A; 1TUX A; 3B03 A; 3G24 A; 4O1F A; 4OKD A; 3CNY A; 3RIK A; 4GO9 A; 1PO9 A; 1GOO A; 4ZO8 A; 3APY A; 3OHM B; 4JR5 A; 3CGT A; 4AY7 A; 3W02 A; 1YTD A; 3BLD A; 3DN5 A; 1I75 A; 2WYS A; 1XDJ A; 2MAN A; 4E2O A; 2JFE X; 3L7R A; 4JEJ A; 2ZFA A; 1J11 A; 4YTU A; 3RLV A; 4X60 A; 3KL5 A; 4PTX A; 1VCV A; 3USB A; 3PSV A; 2VCC A; 2WC3 A; 2OY9 A; 1O95 A; 3A6O A; 4UFI A; 3FST A; 2A3C A; 4IMG A; 3OVP A; 3TRO A; 1SU5 A; 3KLK A; 4IPJ A; 1W8S A; 2W67 A; 1V51 A; 3QHM A; 1ECE A; 3ITY A; 1RPK A; 2CZF A; 4NZC A; 4PMO A; 1UOZ A; 3MVI A; 1GVS A; 2ZX8 A; 1RCQ A; 5F4W A; 4ZFC A; 1XYB A; 2FB3 A; 5JIW A; 3UVA A; 1CLX A; 2F84 A; 3E2Q A; 1W1T A; 3IJL A; 3L5L A; 1HN1 A; 4WDW A; 2FVK A; 4S38 A; 5FA5 A; 4HOW A; 1PII A; 1H3G A; 2YA2 A; 1DV7 A; 3ZMR A; 3LCI A; 4HTB A; 2VEP A; 2CLP A; 1HP5 A; 3PPC A; 2ZCG A; 4DJE C; 3DKL A; 1EBG A; 3VCN A; 4PTV A; 4O1D A; 3BOF A; 3HUR A; 4HBK A; 4GLE A; 4Q8V A; 3QR3 A; 1H11 A; 2WAG A; 2P53 A; 3OZO A; 4WOZ A; 2D73 A; 3VGE A; 2WNN A; 3EEG A; 3AXI A; 1A5N C; 2J6V A; 2QXW A; 5D8Z A; 5HNR A; 4UTM A; 1W7B A; 1HJX A; 2QCD A; 3N13 A; 1BVZ A; 4BK9 A; 2UBP C; 1YXY A; 1GVI A; 2OB3 A; 5AC4 A; 1I2Q A; 2HMC A; 9XIA A; 1V3L A; 2V9D A; 1EP3 A; 4NIL A; 4PZV A; 3BH4 A; 3USZ A; 1QTW A; 1W32 A; 1Y65 A; 3L2I A; 1SVD A; 2I57 A; 3LSB A; 4GY1 A; 4H9Y A; 2VEL A; 4XD3 A; 1JND A; 4WZH A; 2NSO A; 1XLF A; 4F0H A; 1JCN A; 2A3E A; 1NWR A; 3UG4 A; 2VRJ A; 2P50 A; 1UIO A; 1RR2 A; 4NYU A; 1H1Y A; 1MW3 A; 2WJD A; 2Y7G A; 4RX0 A; 1JRB A; 1RLC L; 4QJ5 B; 1DHK A; 3EX4 A; 1I6N A; 1XH1 A; 4JTR A; 4H8V A; 3LPF A; 3PBU A; 2B7Q A; 2WHL A; 4GQR A; 1V0M A; 4GME A; 1Q66 A; 1YRR A; 3MMT A; 1VEP A; 4L65 A; 3ZLH A; 5I3G A; 5I3F A; 1Q8D A; 1AQ0 A; 2IUZ A; 2E9L A; 4RZ6 A; 4NT0 A; 5BWA A; 1UW9 A; 4HZ8 A; 2C18 A; 4GKT A; 1MNZ A; 3B04 A; 4LW7 A; 1RUS A; 3W7N A; 1S2W A; 4L9Y A; 3GXD A; 5CG7 A; 5I3I A; 1GEH A; 4I9J A; 1SZF A; 3E9A A; 2HPI A; 4QBX A; 3AJX A; 3FJN A; 2HK0 A; 3BC9 A; 1YDY A; 1Z48 A; 1DJN A; 1HTI A; 1O6I A; 3B9N A; 5TAO A; 2YGY A; 2ZTJ A; 4KPL A; 3ID7 A; 3BDK A; 4B5S A; 1BLI A; 1E4I A; 3ITV A; 2X42 A; 3VMN A; 4A2S A; 1SM9 A; 3AS2 A; 5JXQ A; 2ZX9 A; 1VYR A; 2B7O A; 4LV9 A; 5FAJ A; 5K3V A; 1HP4 A; 3CHF A; 1CYG A; 3W7P A; 4KWU A; 1SO4 A; 2R1P A; 3SRD A; 5HAT A; 5HXM A; 1ANN A; 1POK A; 3ZQ9 A; 1WQ5 A; 3RCM A; 2UY3 A; 3TTE A; 3MB9 A; 1KWK A; 1QPN A; 4RKA A; 2VEM A; 4FX8 A; 4H9Z A; 2FTP A; 4WD0 A; 3FLU A; 1AA1 B; 2Q01 A; 2C7F A; 3IVU A; 2FHC A; 3GVG A; 4NUW A; 1M90 V; 1EP1 A; 4Z2H A; 3SEC A; 3NIY A; 3RM8 A; 4QRO A; 3S0C A; 2RDU A; 2J7E A; 3V1H A; 1TWS A; 3EA1 A; 1VEM A; 5U4N A; 3IAP A; 4LYQ A; 5A8G A; 3AXM A; 3LUT A; 5AFD A; 2HBV A; 2WMJ A; 2EHH A; 4QQU A; 1GYL A; 1NDZ A; 1Z32 X; 5CH9 A; 3ZVI A; 1BC3 A; 3NXF A; 3WO8 A; 2WHJ A; 3A12 A; 1ZU8 A; 2RUS A; 3SCR A; 4NJH A; 2VQU A; 4Y8F A; 1UWT A; 2RDX A; 5DRI A; 2PDX A; 4FLO A; 4W7U A; 1V6V A; 2PUZ A; 1KB3 A; 1GVQ A; 1NOW A; 5BMW A; 3HF3 A; 3GR4 A; 4FS9 A; 3QST A; 3V35 A; 1H51 A; 1P0N A; 2V6A A; 3B9A A; 3NQB A; 1FQ0 A; 5BXP A; 4Y9N A; 1H7X A; 3KEH A; 2H8X A; 3W28 A; 1H5V A; 2DUX A; 3WQD A; 1JDC A; 1W2V A; 2Y5E A; 3ZT6 A; 3W53 A; 3LV6 A; 4EXQ A; 1H60 A; 4WXY A; 1ME7 A; 3STF A; 4AST A; 3CO8 A; 1KRB C; 3M07 A; 4XZN X; 3N1A A; 4KEM A; 1BQC A; 4KW2 A; 3B0Y A; 1WUE A; 1JPM A; 1SGJ A; 1S1R A; 4TVC A; 4XZB A; 4Y2W A; 2Y7E A; 1TJR A; 4HPJ A; 3PWI A; 3THQ A; 1ALD A; 1PZ3 A; 3SUV A; 3SRK A; 1DIE A; 4PCP A; 3BTN A; 4EAC A; 5BU9 A; 4UQC A; 3TEV A; 3DX5 A; 3CTT A; 3IK2 A; 1W1Z A; 3DG3 A; 1YE6 A; 1VHN A; 3N12 A; 1JL8 A; 2D3N A; 4IID A; 2YSW A; 3G6M A; 2E8Z A; 4Q6J A; 4K7V A; 1O4U A; 2YX0 A; 1JF5 A; 1K73 V; 2BGS A; 1E6R A; 2PS2 A; 3VE7 A; 3XIN A; 5G5I A; 2QT3 A; 2R6O A; 1W3N A; 2YBU A; 2ZAD A; 1JI2 A; 3C2E A; 3TTS A; 2V30 A; 3BW4 A; 1VBJ A; 2G3I A; 4PL2 A; 2PDN A; 2DEP A; 3G1S A; 4AYG A; 5I06 A; 1C91 A; 1OIF A; 5TD4 A; 1WBJ A; 3PZN A; 1KFC A; 1EJV C; 1XIG A; 3O6D A; 1SW0 A; 1AH3 A; 4JIR A; 3WY1 A; 2NVD A; 2REQ B; 3R94 A; 3ZR6 A; 1KM2 A; 2JEP A; 3IRD A; 4OUE A; 3STG A; 1E6Z A; 1XYL A; 3U9I A; 4OGZ A; 2FFI A; 1UZH A; 5F1U A; 4CD8 A; 4ZO9 A; 1AQH A; 1EXB A; 3S6D A; 1EYW A; 1Q6C A; 1D8C A; 2CLM A; 4Z21 A; 4YWS A; 4RLI A; 4NSB A; 4B92 A; 1VQK U; 1GW9 A; 1V0K A; 1K8A V; 2BXZ A; 3VOF A; 4JNM A; 4RNH A; 1XR2 A; 2PP3 A; 1PWM A; 4AY1 A; 4LOC A; 1HAK A; 2NT1 A; 3IIZ A; 3VIM A; 4FNQ A; 6REQ B; 3N73 A; 3N11 A; 1UR3 M; 3HG5 A; 5FCZ A; 2QJH A; 5H1S Y; 4CU8 A; 3GTH A; 1VYS X; 4YIW A; 4HNL A; 1X8F A; 5U7S A; 2IKI A; 1D3C A; 2JZ6 A; 1U22 A; 3P14 A; 3CWH A; 2PF8 A; 1MSS A; 4H9X A; 1H7P A; 3PZ9 A; 2CLI A; 4D6G A; 2X1R A; 4FZI A; 3W2K A; 5IBX A; 4RL3 A; 1R3Y A; 3F5J A; 3O8A A; 3V0T A; 1X1N A; 4MB3 A; 1Y7V A; 3TW7 A; 2AKZ A; 1Q8A A; 1JCM P; 3GTI A; 3WNO A; 2ZUU A; 2C71 A; 4UOJ A; 3LSC A; 1EF3 A; 1CNV A; 3WLQ A; 2AQO A; 2BMB A; 2WO5 A; 3BWW A; 1X6N A; 4YL2 A; 3U2O A; 4DEF A; 1GHR A; 1IR2 A; 2D2O A; 2FZ8 A; 2Z00 A; 2I5G A; 5DAN A; 2PDG A; 3OLI A; 5HNU A; 4IH3 A; 2VX4 A; 3DYP A; 1I4A A; 3WLM A; 1G4E A; 3P82 A; 4CQA A; 4ZE4 A; 3TFC A; 3SI9 A; 1RBL A; 1A3W A; 4MG4 A; 4B9Z A; 1GT8 A; 1QNS A; 3Q45 A; 2PTX A; 4RK8 A; 4O0K A; 3I6R A; 1EHN A; 2FDS A; 4U5K A; 2ADA A; 2QGH A; 1XBV A; 3CAW A; 4A8R A; 2TAA A; 3R58 A; 1V02 A; 4GZI A; 4CCZ A; 2VX7 A; 3TQ0 A; 5AC6 A; 1S5V A; 4AVF A; 1H4P A; 2FZN A; 1XLM A; 2PDJ A; 4W8L A; 1VFM A; 3TDF A; 4ZO6 A; 4FNR A; 4MGG A; 4YVP A; 4KQN A; 4HGX A; 5CGT A; 2XDF A; 2CLL A; 3WBX A; 3L4Z A; 2CWX A; 4Y9A A; 1ZP4 A; 2ZQ0 A; 2IS7 A; 1VQL U; 4DY1 A; 1BWV A; 4GNV A; 1NJI V; 3WX7 A; 1S5T A; 4X61 A; 1HM6 A; 4UQD A; 2FB2 A; 2JK2 A; 4LUS B; 1W9U A; 1XSK A; 4TV6 A; 2PWF A; 4X0V A; 4KKX A; 1T7L A; 2OQL A; 4MAZ A; 4ZB2 A; 4Q2S A; 3T0C A; 1K1D A; 5AHF A; 1W1A 1; 4NG3 A; 4G1N A; 4X2P A; 3H26 A; 1HGY A; 1BCY A; 1XIS A; 4AD5 A; 3L21 A; 2EKC A; 4QIS A; 3AS3 A; 3T0A A; 1VFT A; 1W56 A; 1RJQ A; 2YCK X; 1OYA A; 3TW5 A; 3BQ5 A; 3T08 A; 1F3W A; 5A8M A; 3VNM A; 4K39 A; 1HL8 A; 3W7D A; 1SJA A; 1VS1 A; 3M47 A; 5DCB A; 1HL2 A; 4FF5 A; 4HHL A; 3M5Z A; 4JNY A; 4DPQ A; 4JIQ A; 4NZF A; 4RNI A; 1B2Y A; 3IVT A; 4JTC A; 3CXC T; 1P7T A; 3BCD A; 5K7J A; 2BX7 A; 1FWB C; 4GYP A; 3CT7 A; 4AX6 A; 3RO8 A; 3E1F 1; 3K4A A; 3VX0 A; 3CMG A; 1DWA M; 4JX6 A; 1YW1 A; 2UWF A; 1GZJ A; 4WX2 A; 1B3X A; 1US2 A; 2EXO A; 3KCO A; 4HSN A; 3G0S A; 1U1U A; 3SCO A; 1OZM A; 2WZE A; 4IL2 A; 3O06 A; 1EJS C; 4LY8 A; 1CTN A; 1Z3N A; 4CCE A; 2OZ8 A; 3UZW A; 1DK5 A; #chains in the Genus database with same CATH homology 3A21 A; 3B0V C; 4IXX A; 4JTG A; 2DZX A; 1K7F A; 4O17 A; 2CU0 A; 4QAI A; 5D2T A; 4PH6 A; 5I3J A; 2I1O A; 2J6X A; 4B5N A; 5FI8 A; 2QUT A; 3ZWT A; 2EGZ A; 5G4E A; 3LBM A; 4J9J A; 3M9Y A; 3P80 A; 2BXV A; 1LOS A; 1YYA A; 3KVK A; 3Q2D A; 1KTB A; 1Q3N A; 4LVB A; 4HNV A; 1UBS A; 3UWZ A; 4MYA A; 3MJY A; 4JGD A; 2DJX A; 3G1H A; 2Z2U A; 3HQ1 A; 1LBF A; 1KA9 F; 3W2M A; 3BG5 A; 5BOO A; 3N9R A; 3F1Q A; 3EWW A; 5DY2 A; 2YYU A; 5I3H A; 4UTL A; 4OE7 A; 4ONV A; 2TMD A; 3S5Z A; 3KXQ A; 3MI3 A; 3STC A; 2ZRV A; 1TQJ A; 2AAM A; 1MEW A; 1R2S A; 3W7Q A; 4JTT A; 2QUV A; 2GVG A; 8TIM A; 3N2X A; 4N02 A; 3LTP A; 1H50 A; 2WNQ A; 1JCL A; 3STE A; 3EKZ A; 1LTD A; 5G2H A; 2C6Q A; 2O7Q A; 4NJK A; 4AHO A; 3ZR4 A; 4QIU A; 4JTJ A; 5MHB A; 2WOQ A; 1S5W A; 3Q58 A; 1YPI A; 3W7G A; 2VXN A; 1XBX A; 5C54 A; 4OHQ A; 1FDJ A; 3M44 A; 5D33 A; 2CLO A; 1OFP A; 3SGU A; 3LCL A; 1R30 A; 1UUO A; 2A0N A; 1RZM A; 4JN6 A; 4JTF A; 1D3H A; 1R47 A; 3EX2 A; 2ATS A; 2DU2 A; 2VOM A; 3DFT A; 3HG2 A; 1OYC A; 5EMU A; 1DQW A; 1WXJ A; 3NQ6 A; 1SW7 A; 1KFJ A; 1YDV A; 4MZ1 A; 3DFQ A; 4K37 A; 3WJS A; 3QC3 A; 4WXZ A; 4POD A; 3LCW A; 4AF0 A; 4DF2 A; 1K03 A; 4O12 A; 3LBC A; 3FYP A; 4RZ4 A; 4A2R A; 4H6K A; 3IIP A; 2A6L A; 3NQD A; 2YYT A; 1LCO A; 5FF7 A; 3W07 A; 4XTD A; 2OT0 A; 4IMD A; 4HNN A; 5D2W A; 5G2W A; 1VHC A; 4S1F A; 1GQN A; 3UZJ A; 3TQK A; 3NQ8 A; 4N4P A; 4ZQM A; 1ZAH A; 3LM7 A; 1KM5 A; 3HGO A; 1F6K A; 4ICN A; 2CZD A; 1IIG A; 4POC A; 3NPV A; 4FNP A; 1SO5 A; 2WSQ A; 3C56 A; 1EP2 A; 3R5E A; 4O57 A; 1OYB A; 3G8E A; 1NEY A; 3DFN A; 4OBT A; 4E2B A; 4MIM A; 2ZZ2 A; 5HWN A; 3RLU A; 1Q6Q A; 1PS9 A; 2J27 A; 3ATY A; 3KRS A; 5LKY A; 5K4X A; 3T55 A; 1ZNN A; 2WNZ A; 2G95 A; 3GKF A; 3I65 A; 2PRH A; 4Z1C A; 5DBU A; 4Y96 A; 4LVF A; 1EA0 A; 1EPX A; 4KWW A; 1WOA A; 1H63 A; 3QMT A; 2FLI A; 5H2Z A; 4AAJ A; 5D04 C; 2YR1 A; 3UWU A; 2H3B A; 4AHP A; 5AYZ A; 4UFM A; 2V5L A; 3NNT A; 3P74 A; 5BW6 A; 2J9X A; 1SSG A; 1F73 A; 3IIX A; 4GFS A; 4MZ8 A; 4DEL A; 1A5A A; 4R34 A; 3VE9 A; 2QAP A; 2RDW A; 1PV8 A; 4XTI A; 1M5W A; 1SZE A; 2IQT A; 4LC8 A; 5AHE A; 3TUU A; 1YLV A; 2NUW A; 3NDO A; 1IF2 A; 2NXG A; 3C61 A; 3L68 A; 1TPF A; 3W7E A; 3DXI A; 1WDW A; 3N14 A; 3P5Y A; 3W22 A; 4AH7 A; 4UXD A; 1OVD A; 4GWE A; 5FEX A; 4RXF A; 4M6V A; 1VRD A; 4FXS A; 3S1X A; 1FY6 A; 1K7E A; 3SZ8 A; 2Y70 A; 4O28 A; 4Z0J A; 3PSW A; 3LB0 A; 3E12 A; 1TPV A; 2Q8L A; 3WJZ A; 4ORI A; 1KFE A; 4UCG A; 1FBA A; 2ZYF A; 1JCX A; 4HT3 A; 4IPI A; 4XBK A; 1BKS A; 1R2T A; 2CFF A; 5F2M A; 4P2V A; 3MI2 A; 5LNW A; 1FDY A; 3FA5 A; 3ELF A; 4WY0 A; 3UZ5 A; 2E09 A; 3GDL A; 1T99 A; 4YCP A; 4PC8 A; 4M6P A; 4LS2 A; 4UTJ A; 3W6Y A; 5AHN A; 1A53 A; 4LUJ A; 3FEM A; 3L65 A; 3R89 A; 3WK1 A; 1X1O A; 3HG4 A; 5FEW A; 4N2Y A; 3C6C A; 3NL2 A; 2CW6 A; 3W88 A; 1UAS A; 1W5O A; 3NQ2 A; 4IX2 A; 3EWY A; 1I2O A; 4N9B A; 1K02 A; 3CIW A; 3HO8 A; 3LTY A; 3GAK A; 2QCF A; 1P5B A; 4UUI A; 4MJM A; 3BVJ A; 2QAF A; 1JQV A; 5GVJ A; 1QO2 A; 1OAB A; 3CQ0 A; 2C0A A; 4LUI A; 3C2V A; 1EB3 A; 3N2M A; 1QAP A; 5HR6 A; 1FWS A; 1X7F A; 4OV4 A; 3G0X A; 3KX6 A; 1I2P A; 3GND A; 1WAU A; 4LC6 A; 4UTK A; 4QJ1 A; 4UMB A; 3LHZ A; 1VCF A; 1V7Y A; 1VPX A; 2Y85 A; 2NX9 A; 3PM6 A; 5LNR A; 6ALD A; 1DBT A; 3PC0 A; 4IX0 A; 4PFM A; 7TIM A; 2RDT A; 1I4N A; 2NLI A; 3EWX A; 3EWZ A; 3MP5 A; 3VND A; 1G4P A; 3IGS A; 1JR1 A; 2JKE A; 2QDG A; 1AL8 A; 3HGS A; 2X30 A; 3KOF A; 5HMS A; 4GE8 A; 3DFO A; 1QO5 A; 5D38 A; 5D2X A; 3TSB A; 1GVF A; 2Z6I A; 4Q4K A; 3W1M A; 1JUB A; 1G4T A; 3EX6 A; 2VC6 A; 1DL3 A; 3P8J A; 1JQX A; 3B06 A; 4NNC A; 3QSR A; 2ISV A; 2E68 A; 4C1L A; 4Z1A A; 4EWN D; 1LRQ A; 4PA8 A; 3G1F A; 3M0Z A; 4W9T A; 2OCZ A; 5AHM A; 5FF2 A; 4JYE A; 1QDS A; 4X3Z A; 3QF0 A; 2R8W A; 4UFL A; 3W86 A; 4TIM A; 5G4W A; 1W3I A; 2BWG A; 2A5H A; 4NJG A; 4E2D A; 5HUL A; 3W7O A; 2NXH A; 4DPP A; 2VEK A; 3LLD A; 3PBW A; 4A22 A; 4EIV A; 2YFN A; 2WKJ A; 3RK8 A; 3O05 A; 3QRH A; 1AW5 A; 2E6F A; 2HS6 A; 3GXN A; 1YXD A; 2QCG A; 3G0U A; 3W76 A; 3T78 A; 1OFD A; 1MEI A; 3S8H A; 1OF8 A; 4NQ1 A; 3OEX A; 3UWQ A; 1LBM A; 3N9S A; 5LZL A; 3PG9 A; 3U6W A; 4NU7 A; 3CB8 A; 1TTJ A; 4JTH A; 2W6R A; 1EEP A; 2ZRZ A; 3EKL A; 3L66 A; 4MFD A; 2GPT A; 2QEZ A; 4MUZ A; 2V2D A; 5LNV A; 1TPB 1; 3TX9 A; 2V81 A; 3BG9 A; 3OA3 A; 4GXM A; 3G3M A; 1A5B A; 5LNS A; 1SFL A; 3OBK A; 4EOU A; 1DVJ A; 5J5D A; 2ZBT A; 2A6N A; 1ICP A; 1IGS A; 1FCB A; 4N9E A; 5KIN A; 1OFO A; 4O19 A; 1KTN A; 4ZZ9 A; 4JD4 A; 5FF3 A; 3R13 A; 3W73 A; 2E5B A; 2XFW A; 1P1X A; 1F2J A; 4Y90 A; 2C3Z A; 5EKY A; 2WSR A; 3S42 A; 4A21 A; 3P61 A; 3DEN A; 4CNP A; 4Q32 A; 4O1A A; 3U8G A; 1DOS A; 1YDN A; 2QCC A; 3TML A; 5CZT A; 5BNK A; 2Z0I A; 2VFI A; 3GDK A; 4TO8 A; 4R33 A; 2FIQ A; 3TNO A; 1BWL A; 1OFA A; 2C15 A; 4HSO A; 4EF9 A; 2QCH A; 3BLF A; 1A50 A; 5D09 A; 2Y7F A; 3DKJ A; 2JGY A; 1T96 A; 4JYF A; 1TPD A; 2PRM A; 1LLZ A; 1H62 A; 4ZQN A; 3W2U A; 4AHQ A; 1B4K A; 3NM3 A; 4O0Z A; 5T26 A; 3MHG A; 3JR2 A; 1LOR A; 3RMJ A; 3S5O A; 3P84 A; 2Y63 A; 2EAW A; 1WV2 A; 3KRZ A; 5SWU A; 4UR8 A; 2ZA1 A; 1UUM A; 5K9X A; 3GX9 A; 3RND A; 3MBD A; 1VH7 A; 3CC1 A; 3ZS4 A; 2P0O A; 2NWS A; 4K8H A; 3LOT A; 4MYX A; 1BEU A; 3W1P A; 4GRS A; 1FWN A; 3QPY A; 5HWM A; 2YXG A; 3IV3 A; 3TCE A; 2A7R A; 3MYP A; 1SUX A; 1NAL 1; 3BV4 A; 1QPR A; 3LHY A; 3ZFH A; 1VGA A; 4N9C A; 5K9C A; 4HNU A; 4DO6 A; 1UB3 A; 3DI1 A; 4IME A; 5KZD A; 2AGK A; 4M19 A; 1TV5 A; 4FXR A; 1VZW A; 1KBI A; 3LXB A; 4CLM A; 5AC8 A; 3M7W A; 4ORM A; 1T0O A; 1VQT A; 1LRN A; 1HO4 A; 1CW2 A; 1TTQ A; 1WOB A; 1LM1 A; 3O07 A; 4JTS A; 3NO5 A; 4ZQO A; 4XKY B; 5A5W A; 1FWW A; 3O1N A; 3E96 A; 4HPX A; 3W7H A; 2W0U A; 3PB0 A; 3W83 A; 5KTL A; 4ALD A; 3A22 A; 3INP A; 5E93 A; 4UTW C; 3HG3 A; 4X2R A; 1RQE A; 3W23 A; 3LGE A; 1Q6R A; 3W7R A; 3LV5 A; 1TTI A; 4JY9 A; 2GQ8 A; 4FO4 A; 3V1P A; 3UPB A; 1W0M A; 4ZVJ A; 5DBT A; 3LXA A; 5KSW A; 4MY8 A; 2J24 A; 4FF7 A; 3LTS A; 3M5X A; 1RQH A; 4IMC A; 3HIJ A; 5L9F A; 2JKP A; 1NF0 A; 4ADT A; 2FPV A; 4YLW A; 2E6A A; 4IQL A; 3DBP A; 2C16 A; 3NZ1 A; 1OX4 A; 3W1Q A; 1GZG A; 1LRO A; 1YAD A; 4GUJ A; 2C1H A; 3ATZ A; 3NQM A; 1LLW A; 1SZG A; 2PUR A; 3GDT A; 4CQ8 A; 2EPH A; 4UTI A; 5D05 A; 1OX6 A; 1EWD A; 3CM4 A; 4JTI A; 2X16 A; 3R8R A; 2WV8 A; 4YWI A; 3BW3 A; 4NVT A; 2VEI A; 1ONR A; 5IM5 1; 3SR7 A; 3DFP A; 4O15 A; 3C8F A; 2V5B A; 2HS8 A; 2Y7D A; 4FHA A; 1AW1 A; 2UV8 G; 4KFN A; 3FS2 A; 5TBO A; 1LBL A; 1WA3 A; 3N34 A; 4JX5 A; 2Y61 A; 5F2I A; 4BFA A; 4MVA A; 1LDC A; 1L6Y A; 1ZHA A; 2I14 A; 5FEZ A; 3TC6 A; 1ZFJ A; 3DZ1 A; 2E1D A; 5DEL A; 3HPX A; 4DXV A; 1MO0 A; 3P5Z A; 4IXH A; 4JXC A; 3MUX A; 2C14 A; 3CIX A; 3LHT A; 2Y6Z A; 2Q3R A; 4CCC A; 2ZDR A; 3IGX A; 1IIH A; 3CTL A; 3K30 A; 3HPZ A; 1HG3 A; 3F4W A; 3LLF A; 2TYS A; 5K9D A; 3QH2 A; 1PHQ A; 3TXV A; 2B7P A; 1YTK A; 1OX5 A; 1TSI A; 1W54 A; 1H1Z A; 4NX5 A; 4C1K A; 3GR7 A; 2QUU A; 2DP3 A; 1TJP A; 3NQG A; 4JIC A; 3PBV A; 2BLE A; 4GC4 A; 1QML A; 5DN1 A; 4TR9 A; 3GR8 A; 3GXT A; 3GYE A; 3DU0 A; 4AMA A; 1TPE A; 2I9E A; 1KTC A; 1FWR A; 3SIZ A; 2JKA A; 3E49 A; 3HBL A; 4NXK A; 1H7O A; 4OT7 A; 1CI1 A; 2ABB A; 2A1Y A; 4MKN A; 5EYW A; 5C55 A; 1O94 A; 3CPR A; 3FJ6 A; 2X1U A; 1FX6 A; 2HTM A; 4Z08 A; 4RZ5 A; 3HPS A; 4H4I A; 4DO5 A; 3CU2 A; 1H7R A; 1Y0E A; 3TSD A; 1QPQ A; 2FQI A; 4HHP A; 3QPZ A; 3T7V A; 3Q94 A; 1VR6 A; 4QM1 A; 4OV9 A; 5FF0 A; 4WJE A; 1LZO A; 2CLK A; 1FXP A; 2JBM A; 3T40 A; 2VFE A; 4IJB A; 4ZQR A; 2PRL A; 5LHF A; 2ZA3 A; 4GJ1 A; 1LP6 A; 5F4X A; 1ZEN A; 3QFE A; 3BAR A; 4JX4 A; 4O54 A; 5CZJ A; 4NNB A; 3CEU A; 1F76 A; 2QJG A; 2OMA A; 2A85 A; 3W1X A; 3TIM A; 2ZTK A; 5F4S A; 4XWU A; 2ISS A; 4GUF A; 3UY8 A; 5IBX C; 5LHE A; 3SJ3 A; 3W3O A; 5J5R A; 2B7N A; 3G3D A; 1JUL A; 2DZS A; 3C3N A; 3S1U A; 2V2H A; 4O10 A; 3M5V A; 2BTM A; 5CGQ A; 3FYO A; 3MWA A; 3W70 A; 4JTU A; 1FCQ A; 2X1T A; 3G18 A; 2QCN A; 1F7B A; 3W7L A; 3GXP A; 1GTE A; 2QCE A; 1VLI A; 3TW6 A; 3SY5 A; 4IGH A; 3W2N A; 1THF D; 5DY3 A; 3LI0 A; 4WUI A; 3SSJ A; 3I7Q A; 5G1Y A; 3TH6 A; 2P1F A; 1SO6 A; 4YMZ A; 3LCX A; 2GQA A; 3LRM A; 2BSL A; 3C4U A; 2NUY A; 2WPB A; 3W1L A; 1QPO A; 2DOR A; 3JRK A; 2YDA A; 3L0N A; 4RXG A; 2QCM A; 3UQN A; 1SQ7 A; 2PE4 A; 3IVS A; 3LXC A; 3O16 A; 1ME9 A; 2NX3 A; 3BPW A; 1PE1 A; 5D32 A; 2WQP A; 4FIQ A; 4O1C A; 1ME8 A; 3A5F A; 4NJJ A; 3FJL A; 3W9Z A; 4XQ6 A; 4I7V A; 1L6S A; 3F4N A; 1N8F A; 3ABO A; 4LVG A; 4PTN A; 4LNY A; 4EF8 A; 2QR6 A; 1GYN A; 1KM6 A; 2Y88 A; 3MYO A; 1W3T A; 1W5N A; 1AL7 A; 3CEP A; 4YCO A; 1QR7 A; 3C2O A; 3W85 A; 4LVD A; 4ZL1 A; 5D2Y A; 1QNV A; 4MPQ A; 1XUU A; 1TO3 A; 5BMX A; 1VLW A; 2Y62 A; 4TMB A; 3KRU A; 3A23 A; 3L5A A; 3S5Y A; 2AQW A; 4K38 A; 4OU1 A; 3TJX A; 3BLE A; 5LNU A; 5K2Z A; 4TMC A; 1PCW A; 3FFS A; 1XFB A; 4FX7 A; 4UC5 A; 4K8E A; 1VJI A; 3OVR A; 3GZ3 A; 2QJI A; 5DCE A; 1W5Q A; 4O53 A; 4XZ9 A; 3ABQ A; 1MEH A; 1L2U A; 4DF0 A; 4O18 A; 1L9W A; 4QIT A; 1TRI A; 3Q37 A; 3G1V A; 3VFL A; 4MY9 A; 1TYG A; 4O50 A; 3NL5 A; 3P81 A; 4KFP A; 5AC7 A; 3NZR A; 1H7N A; 3QW4 B; 3NGJ A; 5CZS A; 1TTP A; 5EY5 A; 5TQL A; 3DHD A; 3PBY A; 3C0J A; 2ZRX A; 4M7S A; 3NG3 A; 1J2W A; 4LWW A; 3KAO A; 1FXQ A; 1BWK A; 2ZZ5 A; 3ABS A; 3VKJ A; 3W1A A; 3HJZ A; 1ADO A; 4O52 A; 2NXI A; 3WJW A; 1SZN A; 4K6A A; 3THA A; 1QWG A; 1H7W A; 5D2V A; 5H91 A; 3RFA A; 3NEV A; 3WJY A; 3HMJ G; 3UWV A; 3UYC A; 1N55 A; 1PHW A; 4DBE A; 3M5Y A; 3PAJ A; 3EX1 A; 3P8I A; 2ABA A; 1DQX A; 3EXS A; 4L4L A; 2ISW A; 2V2C A; 1GOX A; 1W5P A; 1F6P A; 1A5C A; 3NQV A; 4LT9 A; 1GVR A; 1UJP A; 1C29 A; 2E77 A; 4AB4 A; 3B8D A; 4NXS A; 5C6M A; 3M16 A; 1P4C A; 5KMY A; 5EBU A; 5F2G A; 1G7V A; 1KBJ A; 4ML9 A; 4JDY A; 2RH9 A; 3OVQ A; 4UTT A; 5C5Y A; 2G97 A; 3PUE A; 2CLF A; 4I9A A; 3L67 A; 2ZRW A; 1SO3 A; 1V5X A; 1MZH A; 3PUD A; 3W2J A; 1DJQ A; 1F74 A; 4JIP A; 4Z0S A; 3W7M A; 5LSM A; 3QZE A; 6TIM A; 5HIN A; 3NL6 A; 4BI5 A; 1PVN A; 1AW2 A; 4FIR A; 3KFT A; 3TMQ A; 1KFK A; 1TB3 A; 4CQ9 A; 1GEQ A; 3O6C A; 3CLM A; 3LDV A; 4IWW A; 1O5K A; 4TU1 A; 3VCR A; 5KIT A; 3PB2 A; 4LRT A; 5E0K A; 4G1K A; 1RD5 A; 3QW3 A; 1I8J A; 3ABR A; 1YPF A; 1SFJ A; 2BZN A; 5CSR A; 3FKR A; 1KM0 A; 4MLJ A; 2Z1B A; 1VC4 A; 4TX9 A; 3BO9 A; 3S5N A; 3NPW A; 5AB3 A; 3NQC A; 4R7J A; 3UXD A; 3B03 A; 3C2F A; 3G24 A; 3T4C A; 4QNW A; 4JY8 A; 1H5Y A; 5CZ0 A; 2ZZ7 A; 1E51 A; 1OX4 B; 1Q6O A; 4JR5 A; 1QCW A; 2V8Z A; 3WK2 A; 1YTD A; 5AOU A; 1GWJ A; 3UND A; 3BK0 A; 2RHG A; 1DOR A; 3I7R A; 1KFL A; 5AN7 A; 4O14 A; 4JTE A; 4Z1D A; 5D02 A; 1OFB A; 4RCX A; 2ZFA A; 4LS0 A; 2RKX A; 3RLV A; 3M43 A; 3NQE A; 1VCV A; 1WBH A; 3USB A; 4U28 A; 1FUY A; 1RV8 A; 3PSV A; 5FEP A; 1BTM A; 4FEZ A; 1OK6 A; 1O95 A; 4UFI A; 3GK0 A; 1ZCO A; 4IMG A; 2OJP A; 3OVP A; 3TRO A; 1SU5 A; 5BRB A; 3PY2 A; 4IPJ A; 4O13 A; 1W8S A; 5ABT A; 3NPX A; 2CZF A; 1JRC A; 3O63 A; 1GVS A; 4K7Y A; 1D3G A; 3S1V A; 5F4W A; 2FB3 A; 2J9Y A; 4UFJ A; 5EPD A; 3G1X A; 1JCJ A; 1ZAJ A; 4JTK A; 4Q33 A; 2F84 A; 4RJE A; 1Z41 A; 3L5L A; 3G1A A; 3H55 A; 4UMC A; 3W2L A; 4PEK A; 1LRT A; 3EWB X; 4X9S A; 1PII A; 1DV7 A; 4M6Q A; 3KVJ A; 2F7F A; 1XUZ A; 3LCI A; 3A9I A; 2VEP A; 2ZCG A; 3TJL A; 1LOL A; 5CPO A; 1F05 A; 3DKL A; 2ZRY A; 4LRS A; 3QQ0 A; 1U83 A; 1JCY A; 4O1D A; 5G2I A; 2E6D A; 3EXT A; 3MHU A; 2ZZ6 A; 1ZAI A; 4AF0 B; 1U5J A; 1Z44 A; 5CKS A; 4I7U A; 1TIM A; 4YXG A; 1M6J A; 4WOZ A; 2D73 A; 3PR2 A; 1G69 A; 1GG1 A; 2WNN A; 1IXO A; 3EEG A; 4QSH A; 2TPS A; 3QMS A; 5HNR A; 2J88 A; 3IGU A; 1TPC 1; 4UTM A; 1XC4 A; 3W7K A; 4NNA A; 3B05 A; 3TFX A; 1O5X A; 2QCD A; 3LCH A; 3NQ7 A; 2YW3 A; 3W7C A; 1KM1 A; 4BK9 A; 1YXY A; 2YW4 A; 2VFG A; 3SGZ A; 1I2Q A; 2HMC A; 1R2R A; 3A5V A; 1W31 A; 2NV1 A; 2CZE A; 3W7I A; 2V9D A; 4WY0 F; 1EP3 A; 2YFO A; 1I2N A; 3I68 A; 2H3D A; 3NOE A; 3GDR A; 1KV8 A; 3L2I A; 2C13 A; 1EWE A; 3WJX A; 4UFH A; 2VEL A; 4AWU A; 3EB2 A; 4M7T A; 4WZH A; 3O6Y X; 3O15 A; 1JCN A; 5C2X A; 1N7K A; 1W37 A; 2ALD A; 1M7O A; 2QDH A; 3DI0 A; 3TAO A; 4GUG A; 1RR2 A; 1H1Y A; 1VYP X; 2V0T A; 1I2R A; 2GJN A; 2Y7G A; 1JRB A; 4RX0 A; 3EX4 A; 3N3M A; 1EX5 A; 1OK4 A; 3W74 A; 5E1Q A; 1X6U A; 3PBU A; 2B7Q A; 4RR4 A; 1KM3 A; 3P60 A; 3LX9 A; 3MMT A; 3G1D A; 3RF9 A; 5I3G A; 1P0K A; 1XDL A; 5HJL A; 3UD6 A; 5I3F A; 3YPI A; 1EIX A; 3G1Y A; 2NWR A; 1O60 A; 4S2B A; 4UNL A; 4RZ6 A; 3TAK A; 3BG3 A; 4NT0 A; 3T44 A; 1GJP A; 3HGJ A; 2C18 A; 2YPI A; 3FOK A; 3B04 A; 3PF3 A; 4LW7 A; 3W7N A; 1TV7 A; 5CG7 A; 1SW3 A; 1JVN A; 3H54 A; 5I3I A; 1JUE A; 2H6R A; 2R94 A; 3TQV A; 1A5S A; 1XXX A; 1SZF A; 4UMA A; 3E9A A; 4R53 A; 3W7J A; 4IMF A; 2H8Z A; 3E02 A; 3AJX A; 3HB9 A; 4WOQ A; 5AHI A; 3E0I A; 3MP4 A; 1FCU A; 1Z48 A; 2A7N A; 1DJN A; 1TRE A; 3C2R A; 5K4Z A; 1HTI A; 4UNK A; 3RU6 A; 5EA9 A; 4ZMG A; 5LGX A; 1YTE A; 4A29 A; 2GUU A; 2X2G A; 2DZT A; 4ZQP A; 1K8X A; 2YGY A; 2ZTJ A; 1XM3 A; 5TIM A; 5HR7 A; 3LI1 A; 4WY0 B; 1SPQ A; 4A2S A; 1Q45 A; 2GVJ A; 4H3D A; 1VYR A; 2B7O A; 4LV9 A; 4BI6 A; 5K3V A; 1MXS A; 5HQE A; 3TU9 A; 3N19 B; 3W7P A; 3GIY A; 4JDB A; 4D2J A; 1SO4 A; 5CPN A; 1TPU A; 3OHI A; 2ZZ4 A; 2X1S A; 4UTU A; 4LU0 A; 2B4G A; 3W87 A; 4UR7 A; 1WQ5 A; 5G1T A; 2E5D A; 3QYQ A; 3B4U A; 1SSD A; 1QPN A; 4RKA A; 2VEM A; 4FX8 A; 2FTP A; 4WD0 A; 3P62 A; 3FLU A; 3IVU A; 3NYZ A; 1B4E A; 3GVG A; 4ADU A; 4NUW A; 1EP1 A; 1EUA A; 2OX1 A; 1ZJI A; 3SEC A; 2FFC A; 3IEB A; 3S0C A; 1VCG A; 2RDU A; 4WQ6 A; 5D37 A; 1TMH A; 3W72 A; 5U4N A; 5A8G A; 4XKY A; 1D9E A; 3KHJ A; 5AFD A; 1NF7 A; 2EHH A; 1GYL A; 2DJL A; 4DO4 A; 5D30 A; 1TV8 A; 5FF4 A; 3W1T A; 1LOQ A; 4CNO A; 3NXF A; 2ATM A; 2H90 A; 1UCW A; 4NJH A; 2YZR A; 4Y8F A; 1GVO A; 4BWL A; 3FIG A; 4RV8 A; 1FWT A; 3PS7 A; 1GVQ A; 5BMW A; 4UTT C; 2TRS A; 3HF3 A; 4X22 A; 3QST A; 2NX1 A; 1T8X A; 4LTS A; 3TC7 A; 1H51 A; 2E5C A; 2XN0 A; 1P0N A; 3PG8 A; 4QNE A; 3IIV A; 3LCG A; 1WYI A; 1S3H A; 4UTW A; 1FCV A; 1TPH 1; 1FQ0 A; 1XDM A; 1XKY A; 1H7X A; 2H8X A; 4LVA A; 2CZ5 A; 2CLE A; 1C9D A; 2OZ0 A; 3GLC A; 4QQ3 A; 3DGR A; 3LV6 A; 2P10 A; 3G8R A; 1KV5 A; 1OHL A; 4OWG A; 1TRD A; 1RVG A; 1H60 A; 4WXY A; 1ME7 A; 2VFF A; 3STF A; 1NVM A; 3W71 A; 3DFS A; 2NUX A; 3LHW A; 3TE9 A; 2VEN A; 2DZV A; 5DXX A; 2QKF A; 2Y7E A; 1TJR A; 4HPJ A; 2ZZ3 A; 1AG1 O; 3THQ A; 1ALD A; 3G22 A; 2E6Y A; 4BF9 A; 4CNN A; 3KVL A; 4PCF A; 3ZWS A; 2ZRU A; 3P7Y A; 1W1Z A; 5AHL A; 1KW1 A; 2G5W A; 1VHN A; 1DHP A; 4MLR A; 3CWN A; 1RPX A; 3PUL A; 5AYX A; 3GB6 A; 3TXZ A; 1XI3 A; 2YSW A; 4L4M A; 4Y6G A; 1B57 A; 1KLY A; 4K7V A; 5FES A; 1O4U A; 2YX0 A; 1IXQ A; 5F2L A; 3NQA A; 2FJK A; 3VE7 A; 5G5I A; 4IOT A; 3VI2 A; 1HUV A; 1K8Y A; 1W3N A; 4HN4 A; 3C2E A; 2V30 A; 4O4V A; 3BW4 A; 3HOJ A; 3TR2 A; 3LHV A; 2VFD A; 4PL2 A; 2J9Z A; 2R91 A; 2G96 A; 3F03 K; 4AXK A; 3G1S A; 2QF7 A; 5D03 A; 3LCF A; 1WBJ A; 5KZE A; 1AK5 A; 1KFC A; 1QOQ A; 4HIB A; 3O6D A; 3NVT A; 4LV4 A; 1SW0 A; 4GNJ A; 3BGG A; 1YDO A; 1GG0 A; 1KM2 A; 3BLI A; 3IRD A; 4FNT A; 3NPU A; 1TPW A; 2FPT A; 3STG A; 2YCE A; 4NJI A; 4OGZ A; 4N9D A; 5HUO A; 1F5Z A; 2R14 A; 2C19 A; 5F1U A; 3EX3 A; 3S6D A; 4N4Q A; 2CLM A; 5HWJ A; 3NL3 A; 4RLI A; 1SR9 A; 4U4M A; 2GQ9 A; 2O7S A; 4JNM A; 5KZM A; 3H53 A; 2ZZ1 A; 1QFE A; 4LOC A; 1R46 A; 3M6Y A; 1FDZ A; 3MO7 A; 3GKA A; 4MY1 A; 2GOU A; 3IIZ A; 4FNQ A; 4KFO A; 1G6C A; 3N73 A; 3DAQ A; 2B0M A; 3HG5 A; 2QJH A; 3LRL A; 2DZW A; 5L6U A; 4ADS A; 1VYS X; 3QQ1 A; 3GNN A; 1X8F A; 4BR1 A; 5U7S A; 3R12 A; 4OQV A; 1RQB A; 3L0K A; 2LLE A; 2NZL A; 3WK3 A; 1L6W A; 1JJK A; 3L0G A; 3WK0 A; 3ANY A; 5DCD A; 5F1V A; 1MSS A; 4AEO A; 4O1B A; 1H7P A; 3S9Y A; 1OJX A; 3S1W A; 3BGJ A; 5LGZ A; 2CLI A; 2ZA2 A; 1WX0 A; 2PC4 A; 1G67 A; 2WSY A; 2QCD B; 2X1R A; 4K36 A; 3W2K A; 5IBX A; 4UIO A; 3O8A A; 3I7S A; 3TW7 A; 4ZQC A; 3TKF A; 1JCM P; 2V82 A; 4HNT A; 3UWY A; 4PL1 A; 3TK7 A; 5DLC A; 1PL9 A; 3FKK A; 1GPW A; 2NV2 A; 1PCK A; 4DBD A; 4E0C A; 3PWA A; 4MOZ A; 2VFH A; 2WO5 A; 1XBY A; 5I3K A; 3TV8 A; 3UWW A; 3CWO X; 4A3U A; 4YL2 A; 1OG0 A; 4O11 A; 3U2O A; 2DZP A; 4DEF A; 4GUH A; 3QEZ A; 3W1N A; 3V75 A; 3L5M A; 4E38 A; 3AO0 A; 2CWN A; 1HFB A; 3OG3 A; 5CPM A; 2OT1 A; 1G4E A; 4M5P A; 4XBS A; 3P82 A; 1IXN A; 2W79 A; 5CPL A; 4CQA A; 5DXY A; 3TFC A; 4AWS A; 3MW7 A; 4S2C A; 3SI9 A; 1IXP A; 1K3U A; 3M1Z A; 3W75 A; 2A21 A; 1ZAL A; 2DZU A; 3L9C A; 4MFE A; 1GT8 A; 1KLZ A; 4N6E A; 2Q8Z A; 4RK8 A; 4O0K A; 3I6R A; 2FDS A; 1J5T A; 3A24 A; 3C52 A; 1EUN A; 2Q3O A; 2WJZ A; 1XBV A; 4NZJ A; 3EWU A; 3TQ0 A; 1W5M A; 5AC6 A; 3QM3 A; 1OFR A; 1S5V A; 4AVF A; 1ZY9 A; 4XUG A; 4FB7 A; 1ICS A; 2XN1 A; 2Z6J A; 3GDM A; 3HGR A; 2KZH A; 4P61 A; 2RFG A; 2A2I A; 3NA8 A; 3LRK A; 1XCF A; 4JEQ A; 3P67 A; 4O4W A; 2FPY A; 3TDF A; 3W1U A; 4FNR A; 1XBZ A; 3PUO A; 1LYX A; 1JUK A; 1AMK A; 3NAV A; 4JYD A; 1OFE A; 1B3O A; 2CLL A; 3R2G A; 1HO1 A; 4DQW A; 4Y9A A; 4IUO A; 2ZQ0 A; 4MZY A; 1QOP A; 4NX0 A; 1DKW A; 1X1Z A; 1NSJ A; 3TA6 A; 4O16 A; 3TSM A; 1I45 A; 1S5T A; 3D0C A; 4EVZ A; 3DH7 A; 3CHV A; 2FB2 A; 2JK2 A; 4Z1B A; 4GUI A; 1Z42 A; 2HSA A; 3EXR A; 4UTH A; 3UPW A; 2EF9 A; 2PCQ A; 3B0U X; 1B9B A; 1GTH A; 3M41 A; 3LHU A; 3MP3 A; 4FX6 M; 2A7P A; 4KKX A; 4HKP A; 1TCD A; 4N6F A; 1Q6L A; 1G7U A; 3B0P A; 4I7W A; 4DF1 A; 5AYY A; 4O8R A; 4FNU A; 4GBU A; 5AHF A; 1ML1 A; 3LER A; 2CLH A; 2Y89 A; 3JS3 A; 3W84 A; 4CCD A; 3BW2 A; 3WFA A; 1J4E A; 3L21 A; 2EKC A; 4QIS A; 1TQX A; 3NR0 A; 1W56 A; 5EL1 A; 2QCL A; 1OYA A; 3IIO A; 3MI6 A; 1OFQ A; 4K39 A; 3W7D A; 1VS1 A; 1M7P A; 3M47 A; 3OIX A; 4AWT A; 2JGQ A; 5DCB A; 4MJZ A; 4JS3 A; 1HL2 A; 3W1R A; 2GVL A; 3M5Z A; 1NFB A; 4DPQ A; 5HUP A; 4JIQ A; 4NZF A; 3SW6 A; 1CX9 A; 1KFB A; 3IVT A; 3MBF A; 1H61 A; 3DHF A; 2VKZ G; 5K7J A; 4BWL C; 1XL9 A; 4LS1 A; 2BX7 A; 1C8V A; 3UY7 A; 3CT7 A; 4JTL A; 1OF6 A; 2A4A A; 3GAY A; 3MHF A; 4BI7 A; 3NM1 A; 1G4S A; 1ICQ A; 4JX6 A; 1YXC A; 5CSS A; 3KVM A; 4UFK A; 4WX2 A; 2GJL A; 1KM4 A; 2TSY A; 4HSN A; 3G0S A; 3PVF A; 4FNS A; 3O06 A; 4KWV A; 2XN2 A; 4LY8 A; 3NQF A; 4CCE A; 3QJA A; 3QMR A; 1K8Z A; 1K7X A; 3SFK A; 3UXA A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...