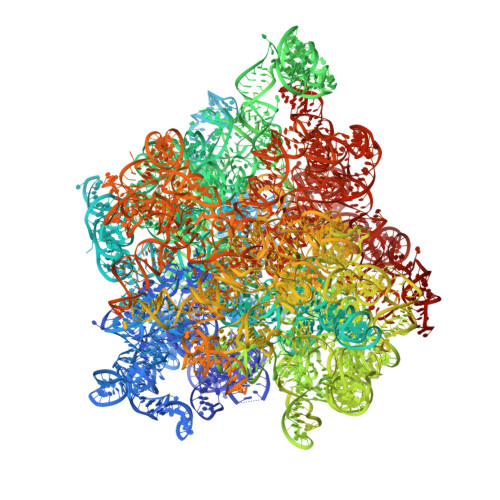
The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
42
|
sequence length |
237
|
structure length |
237
|
Chain Sequence |
GRRIQGQRRGRGTSTFRAPSHRYKADLEHRKVEDGDVIAGTVVDIEHDPARSAPVAAVEFEDGDRRLILAPEGVGVGDELQVGVSAEIAPGNTLPLAEIPEGVPVCNVESSPGDGGKFARASGVNAQLLTHDRNVAVVKLPSGEMKRLDPQCRATIGVVAGGGRTDKPFVKAGNKHHKMKARGTKWPNVRGVAMNAVDHPFGGGGRQHPGKPKSISRNAPPGRKVGDIASKRTGRGG
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA.
pubmed doi rcsb |
| molecule keywords |
23S ribosomal rna
|
| molecule tags |
Ribosome
|
| total genus |
42
|
| structure length |
237
|
| sequence length |
237
|
| ec nomenclature | |
| pdb deposition date | 2004-12-16 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00181 | Ribosomal_L2 | Ribosomal Proteins L2, RNA binding domain |
| A | PF03947 | Ribosomal_L2_C | Ribosomal Proteins L2, C-terminal domain |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Beta | Roll | SH3 type barrels. | SH3 type barrels. | ||
| Mainly Beta | Beta Barrel | OB fold (Dihydrolipoamide Acetyltransferase, E2P) | Nucleic acid-binding proteins | ||
| Few Secondary Structures | Irregular | Ribosomal protein L2; domain 3 | Ribosomal protein L2, domain 3 |
#chains in the Genus database with same CATH superfamily 4IFD I; 3LH0 A; 1I3Q H; 4DV7 Q; 4A3F H; 4GKK L; 3OW2 S; 1G51 A; 3GTK H; 1Q82 U; 1A0I A; 2LCL A; 1BVS A; 3L0O A; 3FKI H; 1PYS B; 1L1O A; 1VQ6 T; 1K83 H; 3PUW A; 2KDS A; 3D31 A; 4GNX B; 1SLJ A; 2MI6 A; 4R4O A; 1QUQ B; 1Q81 U; 1B7Y B; 3STB C; 2WFW A; 3ER0 A; 4JBK A; 3HKZ G; 1M1K C; 4ZN1 A; 1T9H A; 3CCJ T; 4OX9 L; 2PI2 E; 4LFC L; 4GLA C; 3W5O A; 3KBG A; 4B3S Q; 2R7Z G; 3CC4 A; 1SRO A; 2UU9 L; 2JEA I; 3CCE A; 1G29 1; 5LL6 S; 4LUZ A; 3CF5 A; 4JI7 Q; 4DV3 Q; 4JI0 L; 1K73 C; 4IFD G; 1VQM T; 1JJ2 A; 4NXN L; 4DR4 L; 1YHQ T; 4JI1 L; 1IBM L; 3S1M H; 1WCM G; 3UE0 A; 1TWC H; 4A4I A; 1PH8 A; 3UDF A; 4QQD A; 1VQ8 T; 1L2F A; 4GN4 B; 2R92 G; 1EQR A; 3S2H H; 4RG1 A; 1YHB A; 1AE2 A; 2RHQ B; 1Y14 B; 1AH9 A; 4DUZ L; 2OQ0 A; 4LF8 Q; 1EUJ A; 2JNG A; 4IFD J; 1WCM H; 3AQQ A; 2IX0 A; 3DM3 A; 3S15 H; 3HKS A; 2A1A A; 4JCV E; 3IRB A; 1VQ5 T; 1HZC A; 1PH5 A; 3RLF A; 2GVB A; 1RL2 A; 2R92 H; 2E70 A; 3S1Q H; 1KC8 U; 3CCV T; 1HJP A; 3E9H A; 2XTI A; 1HZA A; 3BJU A; 3K8A A; 3I4N G; 3CCM A; 4QIW E; 4PZ6 A; 2VL6 A; 3CD6 A; 3G4S T; 3I56 T; 4A3L G; 2G3R A; 4LF6 L; 2VQF Q; 4A3G G; 1NJI U; 3AYH B; 1IXR A; 3J81 c; 1W2B A; 2JA9 A; 4JOI C; 1HR0 Q; 1VQ4 A; 3I2Z A; 3KF6 A; 3I4N H; 5IYD H; 5E7N A; 5ELO A; 4UY8 C; 3KJP A; 3HOV G; 2QA4 A; 4A3L H; 1VQK T; 2LXK A; 2JE6 I; 4B3S L; 2UXB Q; 3TRE A; 4L5T A; 4A3G H; 1LUZ A; 3K80 C; 1YJN A; 3G6E A; 5DLQ E; 2OTL A; 5M5W H; 1KIX A; 4BA2 I; 4BY7 H; 1CKO A; 5C3E G; 2F4V Q; 2LQ8 A; 3JAM i; 1YIJ A; 1VQB A; 2B29 A; 2I5H A; 5JEA J; 4HIK A; 4JBM A; 1UE1 A; 2AWN A; 2K75 A; 2ZTE A; 4OWT B; 1VQ7 T; 4YCV A; 4POF A; 1O7I A; 2ZTC A; 4DV4 Q; 1SR3 A; 4HJ7 A; 1JMC A; 2JA7 G; 1QZG A; 1HNZ L; 1TWH H; 4OWW B; 1X3F A; 2R7D A; 2NZH A; 2JVV A; 1YI2 A; 4LFA Q; 4BY1 G; 5C3E H; 5M3F G; 1XPO A; 4LF8 L; 1VQJ A; 2HT1 A; 1K8A C; 1E3P A; 4A3M H; 4JYA Q; 2VY3 A; 1IBL L; 3PSI A; 1VQ9 A; 1GO3 E; 1X55 A; 3CPW S; 5FLM G; 3CCQ T; 4B3M Q; 2I5L X; 3H3V I; 3NEN A; 1PH1 A; 4LUV A; 4R4Q A; 1J5E Q; 2ALY B; 3TQY A; 2NVQ H; 3ICE A; 1GKH A; 4JG2 A; 4MZ9 A; 1EQQ A; 5FLX E; 3RR5 A; 3I7F A; 4DV6 Q; 5M3F H; 1E7Z A; 3KF8 B; 5DM7 R; 3GTO H; 4BY1 H; 4DR1 Q; 3S2D H; 4JI5 L; 3CCE T; 1N36 L; 4LW1 A; 2MNA A; 4JV5 Q; 1VQD A; 3RNU A; 2KVQ G; 3J81 E; 3K7A H; 3M4P A; 2MWP A; 4OX9 Q; 3A5Z B; 3U4Z A; 4DV0 L; 4LFC Q; 1VQP A; 4YHH L; 1CSP A; 1U0L A; 3KDF B; 2VQE L; 1HR0 L; 2UU9 Q; 4XIG S; 1N8R C; 3TS0 A; 3B6Y A; 4PG3 A; 2E8G A; 5GAG C; 2VS1 A; 4DV1 L; 4C2M H; 2YU9 H; 2BA0 A; 4DK3 C; 2E6Z A; 3UDG A; 1LYL A; 3HKZ E; 4JOI A; 2E2I H; 4LUO A; 4NXN Q; 3JC6 6; 4JI1 Q; 3AFP A; 2UXB L; 5BR8 Q; 1A63 A; 1YIT T; 1GD7 A; 1M1G A; 2IY5 B; 3CPW A; 4X64 Q; 4X66 L; 4A93 G; 2CKK A; 2LVM A; 2EQS A; 2WB1 G; 2JQO A; 1VQI A; 2F4V L; 2IHE A; 3M7N A; 3H3V H; 3HOV H; 1UEB A; 1BBW A; 5IE8 A; 4L5R C; 2JUF A; 1NBW A; 1Q12 A; 1HZB A; 1E22 A; 3KYH C; 4DV4 L; 4AQY L; 4A3J G; 3HOY G; 3CCM T; 3PCO B; 5IT9 c; 4QGU A; 1VS0 A; 2K52 A; 4JI3 Q; 4LF9 L; 4A93 H; 3CD6 T; 1L0W A; 1LTL A; 4X6A H; 1KXL A; 4A3C G; 3PUZ A; 2N49 A; 3WWV A; 5M3M G; 3L2P A; 4L5Q A; 3J80 c; 3NEL A; 2QEX A; 1M90 U; 2Q2U A; 4LFA L; 2EIF A; 2PQA A; 3BZC A; 1N33 Q; 4JYA L; 1XMO L; 1Z9F A; 1V1Q A; 1HR0 W; 5HY6 A; 1VQ4 T; 4B3M L; 1Q1B A; 1PH2 A; 4BXX G; 2JEB I; 5GRO A; 1J5E L; 2C35 B; 3HOY H; 1VQL A; 5ELN A; 1I94 L; 1I50 H; 2QA4 T; 1OXV A; 2YV5 A; 4LF6 Q; 4A3C H; 5M3M H; 2GN5 A; 4DV6 L; 2UXD L; 4RMF A; 1R9T H; 1YBY A; 4DR1 L; 3FP9 A; 3RZD H; 4JV5 L; 1YEZ A; 3K81 C; 4TQU S; 1YJN T; 3DLL A; 2PNH A; 3G6E T; 2R7F A; 1Y1V G; 1PFS A; 1OXU A; 2OTL T; 3I4O A; 4JI6 L; 2IX1 A; 4BXX H; 2K5V A; 4DKA C; 3D0F A; 2WAQ G; 4BA1 I; 3CC2 A; 5M5X H; 4KHP Q; 4AYB E; 1HH2 P; 1TWA H; 3PUX A; 1Q7Y U; 1Q86 U; 1YIJ T; 4IOC A; 1D7Q A; 4B3T L; 2RF4 A; 4IO9 A; 4DNI A; 1K9M C; 4GKJ L; 4WJ4 A; 1X56 A; 5BR8 L; 2UUC Q; 1YI2 T; 3PO3 G; 4IPH A; 4X64 L; 2R7Z H; 5GQO A; 3S24 A; 1W2B S; 1KQS A; 1B70 B; 1VQO A; 1VQ9 T; 2ZJR A; 1FGU A; 1Z47 A; 4P74 A; 2LSS A; 1HNZ Q; 2UXC L; 1S3O A; 2CFM A; 2EXD A; 3CCR T; 3U58 A; 1FL0 A; 3EBE A; 4HIM A; 3KF6 B; 5IT9 E; 4A3E G; 1NY4 A; 1NPP A; 4YWL A; 3UDI A; 3PO3 H; 1YJ9 A; 1IBL Q; 1VQH A; 4QL5 A; 4JI3 L; 3J80 E; 3ULJ A; 2L3R A; 1FVI A; 1WYD A; 1K8G A; 5M5Y H; 2CCZ A; 3CW2 C; 1N33 L; 1G6P A; 3M4O H; 1M90 C; 4O2D A; 4A3E H; 4JI5 Q; 3UE1 A; 1N36 Q; 3J7Z C; 1B8A A; 4AYB G; 2LXJ A; 1PH4 A; 2CKZ B; 4DV0 Q; 1YJW A; 2OQK A; 4YHH Q; 1BBU A; 4MNO A; 1Q82 C; 1X3G A; 3OTO L; 2KCM A; 1N32 L; 1WI5 A; 2VQE Q; 2VNU D; 1KAW A; 3CME T; 4FDB A; 2HIV A; 2E5L Q; 3H0G G; 2JJQ A; 4JBW A; 5EAY A; 4O0A A; 2K5N A; 1Q81 C; 4DV1 Q; 3S14 H; 4TVA B; 3I4M G; 4KHP L; 1XMQ Q; 1X9N A; 4RG2 A; 2YTY A; 2AWO A; 2VW9 A; 1JE5 A; 1LM0 A; 4X66 Q; 1S72 A; 4ZN3 A; 4MTN A; 3J81 i; 1Q7Y C; 4QY7 A; 4LWC A; 1XNR Q; 2CWA A; 2RCN A; 2CQO A; 4NPS A; 4A75 A; 3H0G H; 2NN6 I; 1VQN T; 2IT1 A; 4YWK A; 5HGQ A; 1KL9 A; 2UUC L; 1XPU A; 5LMN L; 1JJG A; 2QEX T; 3I4M H; 3GD7 A; 3GTP H; 4LF4 Q; 4APV A; 4AQY Q; 3HOW G; 2ZJQ A; 1JJC B; 2MO1 A; 4OWX B; 1KHI A; 3MXN B; 4LF9 Q; 3I55 A; 3CCS A; 1SN8 A; 4HIO A; 1V43 A; 4HJA A; 1MJE A; 1IZ6 A; 1VQL T; 1XNI A; 3KOJ A; 2WB1 E; 4DUY Q; 4A3I G; 4IOA A; 4LFB L; 2D0P A; 1PVO A; 1XMO Q; 1KD1 U; 3J7A F; 3CMA A; 1K0R A; 3KLW A; 2ASB A; 3TRZ A; 1QZH A; 3HOW H; 2R93 G; 2ID0 A; 2JA7 H; 1I94 Q; 3CCJ A; 4C3H H; 1HNX Q; 1Q8K A; 2MFI A; 2DO3 A; 2NN6 G; 1KC8 C; 2QGQ A; 2JA5 G; 4A3I H; 4JI4 L; 2UXD Q; 3CC4 T; 3UDX A; 2ZM6 L; 1JB7 A; 4IJH A; 3CQZ H; 2BH8 A; 5C4J H; 2Q2T A; 2A8V A; 1RIP A; 5FLM H; 3HOX G; 3S1R H; 4JI6 Q; 1X6O A; 5J26 A; 4X62 L; 2R93 H; 3CCL A; 2ZTD A; 1A8V A; 1NZ9 A; 3A0J A; 2HIX A; 3DB4 A; 3Q8D A; 4R1J A; 1P16 A; 1S40 A; 5LMN W; 3OYY A; 1VQE A; 3TEH B; 2JA5 H; 1NJI C; 2E5L L; 4B3T Q; 2Y0S G; 1SE8 A; 1V9P A; 1NTG A; 3T1Y Q; 1VQO T; 3P8B B; 3PO2 G; 2HHH L; 4YY3 L; 3HOX H; 1EFW A; 2PQA B; 1X65 A; 4GKJ Q; 1XMQ L; 2Z0S A; 1UE6 A; 3S17 H; 4DR5 L; 2NZO A; 2WAQ E; 3DB3 A; 3GDE A; 4BBS G; 1VCI A; 1XJV A; 1YJ9 T; 4EX5 A; 1XNR L; 1IBK Q; 3F1Z A; 1NMG A; 1VQ5 A; 3SGI A; 3PO2 H; 4DAY B; 2IHF A; 3RZO H; 2UXC Q; 2AKW B; 4LF7 Q; 5JJI A; 4LF4 L; 1YVC A; 2K4K A; 4K0K Q; 3NEM A; 4TVR A; 1YZ6 A; 3K7U C; 3T1H Q; 2UUB L; 4KHZ A; 1K73 U; 1GVP A; 3G4S A; 4DUY L; 3LGL A; 3TS2 A; 4GNX C; 1XTD A; 1YJW T; 3PIP A; 2OCE A; 1W3S A; 2JOY A; 5K36 G; 1HNW L; 2A19 A; 2KBN A; 4WJ3 M; 1OTC A; 1KD1 C; 4A3J H; 4LF5 L; 3PUY A; 4JI8 L; 2D62 A; 1KQS S; 4IPC A; 5H1S E; 3CC7 A; 1DGS A; 2RHS B; 4DV2 Q; 1PH3 A; 1KRS A; 1HNX L; 1SMX A; 1EIY B; 2B3G A; 1VQK A; 3OTO Q; 5GAD C; 1N32 Q; 4JBJ A; 2KCT A; 1H95 A; 3J80 i; 1J6Q A; 1S72 T; 5IP7 G; 2I0Q A; 4YCW A; 3F2B A; 2D0O A; 3GTM H; 4R4C A; 2OWO A; 2JA6 G; 4EQ5 A; 1EIF A; 2ES2 A; 4HJ5 A; 1C7Y A; 3E0E A; 3LGJ A; 3T1Y L; 4YTL A; 2F3I A; 1N9W A; 5IP7 H; 2PI2 A; 3G71 A; 1Y1V H; 3I55 T; 3CCS T; 2Q2H A; 4OXP A; 2FXQ A; 4C3J H; 5LMN Q; 2H5X A; 2JA6 H; 4B3R Q; 2CWP A; 3EN2 A; 3CMA T; 1IBK L; 3A74 A; 3M4Q A; 3CCU A; 1VQF A; 3S16 H; 3CCQ A; 4ME3 A; 4OO1 I; 4DR7 Q; 1CUK A; 2ATW A; 2GVA A; 1CSQ A; 2AMC B; 4DK6 C; 4L5S A; 4LF7 L; 4JI2 L; 3EIV A; 4LFB Q; 2KEN A; 3W1B A; 1VQC A; 1E1T A; 2K5H A; 4A3D G; 4K0K L; 3PF4 A; 3LGF A; 1UWV A; 1K3R A; 3T1H L; 3GO5 A; 4HJ8 A; 4DR2 Q; 5LMU Q; 3CXC A; 3F2C A; 4GOP C; 3CCL T; 3VDY A; 1VQP T; 1MIU A; 2BA1 A; 5C44 G; 1PHJ A; 4JI4 Q; 4KLK A; 4NB3 A; 1PH7 A; 1FJG Q; 4X65 Q; 2ZM6 Q; 3AFQ A; 1I6H H; 2Q2I A; 2KHJ A; 4A3D H; 3PUV A; 3ZJY C; 4GNX A; 3OW2 A; 4R4T A; 4DV2 L; 1ASZ A; 4X62 Q; 1EOV A; 3GTL H; 1QUQ A; 5C4X G; 2V1C C; 4DV5 Q; 5IY0 A; 1XNQ Q; 4KI0 A; 3M85 A; 3CAM A; 4OO1 G; 1K8A U; 3RLO A; 1YIT A; 5C44 H; 4IPD A; 1EYG A; 2NVT H; 1AE3 A; 3PGZ A; 2MWO A; 3AEV A; 2DGY A; 5C0X I; 3ERS X; 2Z6K C; 2HHH Q; 5JJK A; 4YY3 Q; 5GAE C; 1N34 Q; 2IG0 A; 1WOC A; 3U4V A; 4NXM Q; 2OTJ A; 4UPA A; 3ULP A; 3RN2 A; 1MJC A; 4DR5 Q; 2NVX H; 5C4X H; 3PF5 A; 3S1N H; 1TWF H; 5FJ8 G; 1PA6 A; 1KRT A; 3GTQ H; 1E24 A; 1Q86 C; 5IYD G; 3BZK A; 4GN5 A; 5FLX c; 3U50 C; 3CPF A; 2Y0S E; 3MEF A; 2E2H H; 4DV7 L; 3A5U A; 4IPG A; 2N78 A; 2UUA Q; 1QVF S; 4POG A; 4AIM A; 3HOU G; 3NBH B; 3GTJ H; 4B3R L; 1YHA A; 2MQH A; 4GOP B; 1N8R U; 4A76 A; 5FJ8 H; 1EWI A; 4DAM A; 2UUB Q; 4X34 A; 4DR7 L; 3J7A 5; 4HJ9 A; 4BY7 G; 3CC7 T; 1C9O A; 3JAM c; 5C0X G; 3GTG H; 4QQB X; 5JEA G; 4A3B G; 5GAH C; 4YWM A; 1OXS C; 1A62 A; 3HOU H; 1VQ6 A; 1BKB A; 1WFQ A; 1HNW Q; 2N3S A; 4LF5 Q; 4DR2 L; 4JI8 Q; 5LMU L; 4JI7 L; 3L7Z C; 4DV3 L; 2AHO B; 4DR3 Q; 2VZA A; 3KFU A; 3CC2 T; 2PMZ E; 5AJ3 Q; 1E1O A; 3J81 j; 3H15 A; 4DR6 Q; 4A3B H; 1FJG L; 1Y1W G; 4X65 L; 1OXT A; 2YYZ A; 5C0X J; 4A3M G; 2JA8 G; 4GLX A; 4GOP A; 1UE7 A; 3M9B A; 1PH9 A; 5IYB G; 4HID A; 5IWA Q; 2E2J H; 2B63 G; 1QVF A; 4P72 A; 3G71 T; 3KJO A; 1OXX K; 5IAY A; 1Q46 A; 1U5K A; 1VQM A; 3E9I A; 4DV5 L; 1XNQ L; 1YHQ A; 3PV0 A; 4OON A; 4DPG A; 4XTC S; 1IL2 A; 2DUD A; 2HQL A; 1MKH A; 3K0X A; 1Y1W H; 1VQ8 A; 4PZ7 A; 2JA8 H; 1L1O C; 1X3E A; 3CCU T; 3RN5 A; 5IYB H; 4AH6 A; 1N34 L; 1C0A A; 4CRI A; 3HFZ B; 1JT8 A; 4NXM L; 2B63 H; 3HOZ G; 4GKK Q; 1X54 A; 1PH6 A; 3CCR A; 4IJL A; 5IYC G; 2ZJP A; 2I5M X; 5IP9 G; 4LNQ A; 2Z6K A; 2YTV A; 1QVG S; 1M1K U; 1IYJ B; 1QVC A; 3M3Y H; 4BBR G; 1SRU A; 2WP8 J; 2XHC A; 1TWG H; 3RLN A; 3PIO A; 3CP0 A; 4ALP A; 1GM5 A; 2UUA L; 3STB D; 5A2Q E; 3CCV A; 4Y52 H; 2L15 A; 4JC0 A; 4JI2 Q; 2VQF L; 3HOZ H; 2N8N A; 4J15 A; 4QJF A; 1NPR A; 1CKN B; 1NMF A; 1TXY A; 5IYC H; 4C3I H; 1ASY A; 5IP9 H; 2KHI A; 2R6G A; 4GN3 B; 4BBS H; 3JAM E; 1WJJ A; 1P8L A; 4H02 A; 2YTX A; 4GS3 A; 1K9M U; 1NE3 A; 3I56 A; 1SFO H; 1VQG A; 4BBR H; 4JI0 Q; 2MO0 A; 4UP8 A; 2BH2 A; 4PMW A; 2VUM G; 4DR4 Q; 4A3K G; 2F52 A; 1IBM Q; 4P73 A; 4R4I A; 2K14 A; 3CME A; 1SSF A; 1Q1E A; 3BU2 A; 1VQA A; 1CKM A; 1D8L A; 1YNX A; 2K50 A; 4Y7N H; 4DR3 L; 4UP7 A; 5AJ3 L; 3F2D A; 2OTJ T; 1L1O B; 4P71 A; 5J3B A; 4YCU A; 4GLV B; 1XPR A; 1QVG A; 4UP9 A; 4DUZ Q; 4DR6 L; 2VUM H; 4A3K H; 3ULL A; 3W1G A; 1CKN A; 3F8T A; 3G48 A; 3KDF A; 5IWA L; 1HZ9 A; 4A3F G; 5JJL A; 3FHW A; 1VQN A; 4PZ8 A; 3CXC S; 2JK8 A; 3FKI G; 2XGT A; 1PYB A; 1JJ2 S; 1PV4 A; 2NVY H; 1UE5 A; 4P75 A; 1I5F A; 1VQ7 A; #chains in the Genus database with same CATH topology 3QJ6 A; 1GCQ C; 1MV3 A; 5KNB A; 4IFD I; 2OAU A; 4A3M B; 3GTK H; 1G51 A; 2BE9 B; 2CYZ B; 1HPC A; 1OEB A; 1W6J A; 3NBI A; 2M00 A; 3I5R A; 4RCN A; 1RAF B; 2BIV A; 2D5D A; 2DE0 X; 2MI6 A; 4JJB A; 1QUQ B; 2F03 A; 5HGT A; 4LUD A; 1SNC A; 3ER0 A; 1EZZ B; 5C4Z A; 3GTO B; 4ZN1 A; 2YRD A; 3S2D B; 2DF6 A; 1VQP Q; 4HSP A; 2CH4 W; 3C1E A; 4B3S Q; 2UU9 L; 2JEA I; 1G29 1; 3CF5 A; 3GTP B; 1YNZ A; 4AFU C; 2IL8 A; 1B9N A; 4IFD G; 4N9P A; 4OP0 A; 1QIL A; 3BS1 A; 3RMH A; 2HZY A; 2HDM A; 3H6M A; 3S1M H; 2Z4H A; 1LTR D; 3UA6 A; 3PVQ A; 3TU4 K; 4BDZ A; 3RNU A; 2HL5 C; 1TS9 A; 3HPW A; 2R92 G; 1EQR A; 1STG A; 4O5V A; 2LFN A; 1EY5 A; 1AH9 A; 3RIR A; 2X9H A; 3JCM W; 4Y7N B; 5NUC A; 2M1H A; 3DHQ A; 4IUT A; 3VYG A; 2ZPB B; 4WCI A; 1HZC A; 3MA2 B; 1OV3 A; 2PUK B; 2M0O A; 1PMR A; 1O7Y A; 2B2Y C; 3S1Q H; 9ATC B; 2XTI A; 3BJU A; 1HQR D; 1QLY A; 2P0P A; 4XTU A; 1GUW A; 2RDF A; 4PZ6 A; 4M7D A; 1HA5 A; 1I6V C; 4DA2 A; 5IGD A; 4F04 B; 2FMM A; 3H09 A; 2LT4 A; 4A3L G; 3BDL A; 1QWE A; 1XQH A; 2DBK A; 2EYQ A; 4GBQ A; 4HVV A; 4JOI C; 1CKB A; 3GTK B; 4M75 C; 2QXF A; 1VQ4 A; 2HCC A; 4QF4 A; 4Z8A A; 1I5O B; 3EGX B; 2F3V A; 3DWQ A; 1JOR A; 5E7N A; 4XQ3 A; 3R2I A; 5ELO A; 1ZBJ A; 5LJ5 h; 3K5M A; 2RDH A; 2QA4 A; 4LD9 K; 3BY7 A; 2B2V C; 4HCB A; 2HD3 A; 3VYR A; 4L5T A; 2JMA A; 2JS0 A; 2E2D C; 4A3G H; 2DL5 A; 2XMF A; 3CBO A; 5ISR A; 1YJN A; 4II1 A; 3G6E A; 2QIL A; 1JU5 C; 1JWM D; 2LQK A; 2K2V A; 2F4V Q; 4KH0 B; 3RES A; 3RNJ A; 2E1H A; 4UC4 A; 1YIJ A; 2B29 A; 4HIK A; 1WHM A; 5JEA J; 4OWT B; 3M0T A; 2AWN A; 2K75 A; 2EYL A; 2J06 A; 1JMC A; 1O7I A; 2ZTC A; 5IXB A; 5JPM C; 2A69 C; 1BCP F; 4K11 A; 1MGS A; 4LFA Q; 4Z89 A; 2JVV A; 3OYF A; 4M7D D; 1N8R R; 3LWI A; 1YJ9 Q; 5M3F G; 4LF8 L; 3J7A V; 2HT1 A; 1ZX6 A; 2VY3 A; 3A8I E; 5KND A; 1VQ9 A; 3H3V I; 1PKS A; 3CPW S; 1TQO A; 3H0F A; 4RMO A; 3OS1 A; 1I4R A; 3NEN A; 4M75 B; 3TQY A; 1GKH A; 4FXG C; 1EQQ A; 5FLX E; 3R8B A; 2DYB A; 3HFO A; 5GAN 7; 2EZV A; 5IGF A; 1E7Z A; 4HAE A; 3S1Q B; 4LW1 A; 5JVH 3; 3RKY A; 5GAN d; 2DD4 A; 3GPL A; 1QQJ A; 1YJW Q; 1GUG A; 2EY1 A; 4PQX A; 3M4O B; 4DV0 L; 1TSF A; 4YHH L; 4XU0 A; 1U0L A; 2VQE L; 4QB4 A; 2MF2 A; 1N8R C; 2YSQ A; 4XIG S; 3TS0 A; 2CZ1 B; 4BE0 A; 1SYC A; 2E8G A; 4PG3 A; 2DM1 A; 1TH7 A; 3UWN A; 4FT2 A; 1CFM A; 4XQQ A; 2EGC A; 3R6O A; 1ZSZ C; 3CQT A; 2Q8T A; 3HL8 A; 5I2M A; 3HKZ E; 4RKB A; 4Q5V A; 2UXB L; 3HTR A; 2YLC A; 5BR8 Q; 2FZK B; 1S72 Q; 4OL7 A; 4DK0 A; 3NQI A; 1M2V A; 3REA B; 1QWY A; 3BDO A; 2WB1 G; 3V05 A; 2ENM A; 2RBM A; 4A3G B; 2DPP B; 1VQI A; 2EJG C; 1R8O B; 4L5R C; 4ORZ A; 5KIX A; 1M4V A; 1E22 A; 4A3J G; 2BZY A; 3HOY G; 1F2M A; 4LF9 L; 4A93 H; 1LTL A; 3VS7 A; 4A3C G; 2RHU A; 1DXM A; 1LAC A; 2H3E B; 1WTH A; 3J80 c; 1TKW B; 3L2P A; 1M90 U; 2Q2U A; 2Z0W A; 3BZC A; 2ED0 A; 1N33 Q; 4JYA L; 1XMO L; 5AYF A; 5KYW B; 3OW2 P; 5HY6 A; 1VQ4 T; 2JEB I; 3JCM g; 3M5A A; 4CI0 C; 1QVP A; 3FIF A; 4N0A C; 2QA4 T; 2YV5 A; 3MHX A; 3V8K A; 4DAY A; 2XDP A; 1ZSZ B; 2D0N A; 2JA7 B; 1PNJ A; 3RZD H; 2L3S A; 1YEZ A; 4JV5 L; 1SG5 A; 3DLL A; 1YJN T; 3G6E T; 3CCL Q; 1BDO A; 1K28 A; 1Y1V G; 2PNH A; 1IYU A; 4C0D C; 1BCP C; 4M78 G; 2K5V A; 3JB9 H; 5A35 A; 1WTP A; 3EGD B; 3CC2 A; 3NWS A; 4KHP Q; 1HH2 P; 3MVV A; 4C8Q F; 2ED1 A; 5KEE A; 3J7Y W; 4KH1 B; 1TS3 A; 1YIJ T; 4IOC A; 1D7Q A; 2RI3 A; 2RF4 A; 2I0N A; 4J6Y A; 2RQU A; 2MJ8 A; 5A0L A; 2JA5 B; 1BXT A; 5L23 A; 2JP1 A; 3OYB A; 4E7I A; 1Y69 U; 2V8I A; 3HGB A; 4IPH A; 4X64 L; 1WGS A; 1U3O A; 1BNZ A; 2RHZ A; 4J7I A; 1B70 B; 1WHJ A; 1VQO A; 2F0T A; 1VQ9 T; 1ILQ A; 3BZD B; 1FGU A; 4P74 A; 1OUL A; 3VR3 A; 1B3Q A; 1X3Q A; 4K5V A; 1R4P B; 2EXD A; 3U58 A; 1UGS B; 2I2L A; 2B2T A; 5FOB B; 4A3E G; 3PO3 H; 5DLO A; 2FMC A; 4DAV A; 3I72 A; 1VQH A; 4DXF A; 1VUB A; 3J80 E; 4JJC A; 2L3R A; 4OMF B; 2X3W D; 1FVI A; 2DEU A; 1T5E A; 5M5Y H; 2HBP A; 2ZPE B; 1PRM C; 1BCP B; 4JK1 C; 4R56 A; 4HXJ A; 3UE1 A; 4FYY B; 2Y90 A; 2LP5 A; 2ESW A; 1P92 A; 4DV0 Q; 4YHH Q; 2IPO B; 3JCM X; 1BBU A; 1X3G A; 3OTO L; 1M3C A; 1PCX A; 3X28 B; 5FRN A; 4B08 A; 2VQE Q; 5IN1 A; 3CME T; 2VNU D; 5T1I A; 4KGX B; 2JJQ A; 1Y7X A; 2K5N A; 4DOW A; 3QWX X; 1AEX A; 4MN3 A; 2C06 A; 1EFN A; 1X9N A; 2YTY A; 3ZFJ A; 4F7U G; 2VW9 A; 1KQ1 A; 1JE5 A; 2WV6 D; 3BVZ A; 1VIF A; 3INZ A; 2HQX A; 3CBP A; 4ZN3 A; 3G71 Q; 1RAD B; 3U9S A; 3QZ5 B; 2NN6 I; 1G83 A; 2IT1 A; 4GZY C; 5LJ3 f; 2LJ0 A; 3OYD A; 3RD4 A; 4YWK A; 2UUC L; 3OIQ A; 5EBD A; 3D4W A; 2SEM A; 3FDR A; 2CP3 A; 5FU6 C; 3EG2 A; 4K5X A; 4C3J B; 2ZJQ A; 1F1B B; 1KHI A; 3WVD B; 4LF9 Q; 5G3L D; 3I55 A; 3CCS A; 1V1O A; 2AK5 A; 2KBT A; 5AFW A; 5CV8 A; 1IZ6 A; 2HKQ B; 2RMO A; 4PMB A; 3SK8 A; 4C8Q C; 2EYJ A; 3E6Z X; 3G6J B; 4A3I G; 1K8O A; 2KXD A; 2C0T A; 4X3U A; 4HT8 A; 1TY0 A; 3M57 A; 2OXP A; 3CMA A; 5KYU A; 2P4R A; 4TKO B; 1XMO Q; 1UGV A; 2X3X D; 1I4X A; 3EG9 A; 1BK2 A; 1QVG P; 1B1Z A; 2QDB A; 3CCJ A; 1HNX Q; 1HHV A; 3PMT A; 4DEX A; 1U4L A; 3MHB A; 2WSC E; 1CZG A; 3TP6 A; 2LNH B; 2MYS A; 2A36 A; 2R3Z A; 5ECA A; 1V1P B; 2BH8 A; 2L3Q A; 2A8V A; 3D6W A; 5M5Y B; 1RIP A; 2KHS A; 1X6O A; 4Y28 E; 1BI1 A; 3V7R A; 4HCZ A; 2HKN A; 2F4I A; 1RDP D; 3TEH B; 5A0D A; 5C4H A; 5I6W A; 1GHJ A; 2NWM A; 1KNE A; 2EYF A; 3OYN A; 1VQO T; 3CRL C; 4YY3 L; 2VV5 A; 2NVT B; 5FU6 B; 1XMQ L; 1OPK A; 3OYC A; 2D2C C; 3WXM B; 1KD1 R; 2RSO A; 4RFB A; 5CV9 A; 3WOE A; 1XJV A; 2FPF A; 1PTO F; 4EX5 A; 1X01 A; 2F0Q A; 1XNR L; 3JCM d; 5EPK A; 3QDR B; 4C5E A; 2IHF A; 4PK4 A; 1IRX A; 2WKC A; 5CR3 A; 1YZ6 A; 2EY5 A; 3K7U C; 2GF7 A; 4RGM A; 3T1H Q; 2UUB L; 1Z6H A; 5K5P A; 1K73 U; 1K9A A; 3H0H A; 1U1S A; 1F2Y A; 1XTD A; 5LJ3 e; 4GNX C; 5KE2 A; 4JJD A; 2OCE A; 2BUD A; 4LN2 A; 1PFM A; 3D4D A; 4OML A; 3A8M B; 4CKE A; 1GUS A; 2E65 A; 3G7L A; 1GCP A; 2O4X B; 1UHC A; 1KD1 C; 1K0S A; 2EJF C; 4A3J H; 1OTC A; 4KTA A; 1PH3 A; 1XX8 A; 2RHS B; 4IUV A; 4X3K A; 1T3S A; 5CC4 A; 2FYK A; 2DET A; 3OTO Q; 4DNT B; 4K14 A; 5GMK l; 5CR2 A; 3LWE A; 2JM8 A; 5K36 H; 1I4K 1; 2XK0 A; 3EGU A; 4IUP A; 1ZA1 B; 3M56 A; 3S15 B; 4RHS A; 4A3B B; 1WPY A; 1NLP C; 4C1N I; 1LV9 A; 3EA6 A; 3KLS X; 2JA6 G; 2EW3 A; 1QCF A; 4DEY A; 2KK4 A; 4HJ5 A; 2C0I A; 1NCV A; 3E0E A; 1MV0 B; 3U23 A; 2DJG C; 3MFG A; 2GA4 B; 3C12 A; 4OGQ C; 4Z9C B; 4C56 C; 4CJ2 C; 1DCZ A; 1Y1V H; 3I55 T; 3CCS T; 2NUT B; 4OXP A; 1Y1W B; 1EIG A; 2TMP A; 1MI2 A; 5GAM f; 2MP1 A; 1PTO H; 3OYM A; 2NYS A; 3CMA T; 4IZ8 A; 2K03 A; 4E7J A; 2QKI C; 1A2U A; 2ZPG B; 5GAO m; 5C0M A; 1FNV A; 2GVA A; 1IGQ A; 1KLU D; 4DK6 C; 4L5S A; 2DIG A; 3C94 A; 3W1B A; 2K5H A; 4K0K L; 3TP7 A; 1K3R A; 3G4S Q; 1JOK A; 1Q3L A; 4OHJ A; 3GO5 A; 3MAQ A; 3WOF A; 3EHQ A; 4BI8 B; 2IVW A; 3F2C A; 4BBS B; 2ZGW A; 1II3 A; 2BA1 A; 2EJM A; 3CME Q; 1PTO C; 4NB3 A; 1IRE B; 1I6H H; 2DL7 A; 5IHK A; 5IOD A; 2LJ1 A; 1CHQ D; 3GTL H; 3QII A; 3IKJ A; 5CYU A; 3MEV A; 3SB2 A; 4ZKC B; 2IPK D; 3I9Q A; 5COY A; 5IY0 A; 3ZBD A; 4OO1 G; 2EYM A; 3I70 A; 4XTV A; 3BFM A; 1K8A U; 5KCO A; 1YIT A; 5KRU A; 1NR4 A; 2JT4 A; 4LNP A; 2RI5 A; 4KRT A; 1PXU A; 1STB A; 1MND A; 5JJK A; 1F9Q A; 2JMC A; 3UT1 A; 4YY3 Q; 1X27 A; 2IG0 A; 1WOC A; 3QIA A; 1EY4 A; 1JB3 A; 5GJV B; 1PLF A; 4FL6 A; 4UPA A; 4EVO A; 4NXM Q; 3RN2 A; 5C4X H; 1PA6 A; 5FJ8 G; 4KKU A; 3ZYY X; 5FO7 B; 2RQW A; 1JQQ A; 1N9S A; 3CPF A; 3GBQ A; 2XWJ B; 1R77 A; 3MXP A; 3U50 C; 2D8H A; 4IPG A; 4DV7 L; 2N78 A; 2JZ2 A; 5ELC A; 3HOU G; 2DVJ A; 2X35 A; 2JSO A; 1YCY A; 4M7A E; 1PRL C; 3EHR A; 3H6Z A; 2UUB Q; 1PTO B; 4DR7 L; 4HJ9 A; 1C9O A; 3GTG H; 3JAM c; 3M58 A; 3CPW P; 5GAH C; 4IQZ A; 2C0O A; 1F2L A; 1X69 A; 4IUR A; 5GAM e; 3OHX C; 5FWB A; 3DWP A; 4DR2 L; 4JI7 L; 5LMU L; 2MK5 A; 1ZEQ X; 4EGV A; 2KMT A; 4DR3 Q; 1UUE A; 1PD0 A; 4X9D A; 1YWP A; 4RGN A; 3L2U A; 5C0X H; 4DR6 Q; 1PRT D; 1FJG L; 5IGC A; 4A3M G; 2A5H A; 2YYZ A; 5C0X J; 4HVW A; 2LQW A; 3M9B A; 2MIO A; 2E2J H; 1DOK A; 1QVF A; 2B63 G; 2FG0 A; 1I6H B; 3FEO A; 1OXX K; 3E9I A; 1VQM A; 4DV5 L; 2EYX A; 5CCA A; 1LO5 D; 4OON A; 3GTL B; 5CQY A; 1I0C A; 2J6F A; 2HQL A; 4XTC S; 2SRC A; 5C3X A; 1RD9 D; 2ZPF B; 1X3E A; 1JT8 A; 4PJN A; 3HFZ B; 2GTO A; 1UDL A; 4J83 A; 3UDC A; 3HOZ G; 3ND9 A; 1FNW A; 2ZJP A; 2L5Q A; 3VRZ A; 3OYA A; 5IP9 G; 3S3O A; 1D1K A; 3R93 A; 1M1F A; 1M1K U; 3CD6 Q; 5C4X B; 3SD4 A; 3SVM A; 1TWG H; 1CKA A; 1GM5 A; 3L2R A; 1ACM B; 4Y52 H; 4DK1 A; 1G2T A; 4ZQ3 A; 2N8N A; 4QJF A; 1NPR A; 5IYC H; 5FKA C; 1V8Q A; 3A98 A; 3C0C A; 3EFR A; 4PJO E; 3JAM E; 1KNA A; 2KHI A; 4H02 A; 4MIU A; 5EPL A; 3GOX A; 1NE3 A; 1NYG A; 2B8G A; 1KTK A; 1GBR A; 4UP8 A; 1D1C A; 4PMW A; 2VUM G; 4DR4 Q; 1KQS P; 2X6G A; 4P73 A; 2EVB A; 1SNM A; 1BBX C; 2B2W A; 1FX7 A; 2E6N A; 5FO8 B; 1VQA A; 1WD0 A; 3O5Z A; 2RQX A; 4UP7 A; 3L42 A; 4YCU A; 1M9S A; 4GLV B; 5C5L A; 3EG1 A; 2RD1 A; 3M0P A; 2J7Z A; 1VOM A; 3W1G A; 4BY7 B; 1W1F A; 3M53 A; 1G3W A; 5GAN h; 2BE5 D; 1X3P A; 3T4A C; 5GMK j; 4PZ8 A; 3JCM P; 4C8Q E; 2P84 A; 2XGT A; 3UA7 A; 4C5I A; 3RDV A; 2AKK A; 3V96 A; 1TU0 B; 1PV4 A; 4P75 A; 4HKE A; 1DD2 A; 4E7K A; 3L2Q A; 3DM1 A; 3LH0 A; 1SDF A; 4E6I A; 1AZG B; 1D09 B; 2CP0 A; 2DNU A; 2HAX A; 1I3Q H; 4IIO A; 4YIJ A; 4DV7 Q; 1LJO A; 3OW2 S; 3NGP A; 1EL0 A; 4E47 A; 2LCL A; 3H3V C; 1PYS B; 3T56 B; 2D0Q B; 2KDS A; 1LOJ A; 2K3X A; 2F0G A; 3D31 A; 5FW9 A; 2AZS A; 1LT3 D; 4J9C A; 1Q81 U; 2WFW A; 2F3W A; 3NE5 B; 3MEH A; 1X2K A; 1ZYR C; 2MUY A; 3T13 A; 3TCJ A; 2NUP B; 1X54 A; 1B3A A; 4GLA C; 1TWG B; 1SRO A; 1H9J A; 1H9S A; 5LL6 S; 4JI7 Q; 3VUZ A; 1AZQ A; 2XG8 D; 3EVQ A; 4JI0 L; 1UGR B; 1VQM T; 4CGY B; 2A37 A; 3RER A; 5D14 A; 1IBM L; 4WTO B; 1TWC H; 1STE A; 3D4R A; 3S3M A; 3UDF A; 3PNW C; 5D6W A; 1D2B A; 1E0B A; 4GN4 B; 3S2H H; 1EYC A; 2E64 A; 3VA5 A; 3OS2 A; 2RHQ B; 4XQJ A; 4DUZ L; 2HD0 A; 2QW7 A; 5GAN 5; 2OQ0 A; 2RN8 A; 1WVL A; 4CBV A; 3DM3 A; 2DK3 A; 8AT1 B; 2V4D A; 3HKS A; 4BB7 A; 1VQN Q; 3IRB A; 3HOV B; 6AT1 B; 5DY9 A; 1WXT A; 1DOL A; 1ZLM A; 5EJW A; 1ZUK A; 3RLF A; 1L0X B; 2K05 A; 3K59 A; 1Y07 A; 1IYV A; 1V76 A; 1AN8 A; 1C4Q A; 1HJP A; 1PZJ D; 2FZG B; 1HZA A; 2K7A A; 3K8A A; 3T16 A; 2R57 A; 3HRZ C; 3CCM A; 2J6O A; 1QB5 D; 4KKT A; 5LL6 X; 3OWE B; 4KHV A; 4A3G G; 4RTZ A; 1NJI U; 3J81 c; 1W2B A; 2JS2 A; 3I2Z A; 1CM9 A; 3I4N H; 4JZ3 A; 2KQW A; 3TQ7 P; 3SK6 A; 1CHP D; 4L6T B; 1VQK T; 3HRU A; 2CP7 A; 4B3S L; 1I1J A; 1H9M A; 1JIC A; 2DVE A; 2QCP X; 1KK7 A; 3KBX A; 1PYW D; 2AKL A; 4WFN 3; 4FXD A; 1KIX A; 2LTO A; 3KFV A; 4JVZ A; 1CKO A; 1KFZ A; 3MET A; 5CO7 A; 1VMC A; 3HKZ B; 1UE1 A; 3VR5 A; 1K0X A; 4POF A; 5KYI A; 4OMM A; 3TEE A; 3TSS A; 8ATC B; 4HJ7 A; 1SR3 A; 1TWC B; 3N52 A; 2JA7 G; 2CVR A; 2XRQ D; 4J9I A; 3DKM A; 1TWH H; 1D3B A; 1RAG B; 5LJ5 j; 4X9C A; 1U8R A; 1SNO A; 2NZH A; 1YI2 A; 4CJ0 B; 2SEB D; 2F0K A; 4A3M H; 3DUZ A; 3TZD A; 1PDQ A; 3PSI A; 4G7Z C; 1LNX A; 1GUT A; 4C92 E; 1X55 A; 1A6X A; 2I5L X; 4LDW A; 3LX7 A; 4DBQ A; 4JG2 A; 1EZ8 A; 4MZ9 A; 3JCM Q; 2PW5 A; 3RR5 A; 4CHT B; 1OX8 A; 1YP5 A; 2GBQ A; 5IXF A; 3GTO H; 3KF8 B; 5DM7 R; 2GQI A; 2EJ9 A; 4L5G A; 4ME5 A; 4UHV A; 1H9R A; 4DR1 Q; 1C8C A; 3S2D H; 3ITP A; 2MNA A; 3A8L B; 1YWO A; 2KVQ G; 3G1J A; 2HSP A; 4M78 A; 2DLP A; 3J81 E; 4K5W A; 4XT1 B; 1WLP B; 4DAP A; 3A5Z B; 4FYX B; 3KLR A; 2CZ6 B; 2M9U A; 4VUB A; 1STA A; 3KDF B; 1W6X A; 4DFA A; 4FU6 A; 4ZGJ B; 1LT6 D; 3FVQ A; 1RF2 D; 5GAG C; 1I4P A; 2VS1 A; 4M77 E; 5EKI A; 2BA0 A; 3MTS A; 3JV3 A; 3R3O A; 1LYL A; 3UDG A; 3I4N B; 3QXE B; 1H3Z A; 3JC6 6; 2P5Z X; 1EY7 A; 3LNN A; 2D8J A; 3L2Z A; 5KYN A; 4A93 G; 4XTX A; 1I5L A; 2B2T C; 4PFZ A; 4FNF A; 2JQO A; 2QGG A; 2K5I A; 2PVO B; 1TT2 A; 1TUG B; 2F4V L; 4EMG A; 4IUN A; 3L2V A; 1Y8O B; 3Q0A A; 2V1Q A; 1BBW A; 2M51 A; 4KMU C; 1QCO A; 3CXC P; 5IOC A; 3CCM T; 4JRI A; 3PCO B; 4FVM A; 4QGU A; 2K52 A; 4JI3 Q; 3MEA A; 1L0W A; 1G3Y A; 2N49 A; 4JW0 A; 4LFA L; 2M02 A; 2QEX A; 1RAI B; 4JLI A; 4CC4 B; 4JS4 A; 2PQA A; 4M78 D; 1TWH B; 2IIM A; 1SMY D; 1PH2 A; 1K9M R; 2XD0 A; 4H7B A; 4ODG A; 3VS4 A; 2ROT A; 3JB9 G; 2PVD B; 3MCA B; 1VQL A; 5FRO A; 3DXJ C; 1OXV A; 4A4K A; 2X51 A; 1HFG A; 2F0O A; 1ZQ1 A; 2MPX C; 1B4O A; 2M2L A; 1WQW A; 1QJO A; 2PYK A; 2O5I C; 2DJZ A; 2KEC A; 4ATO A; 3FP9 A; 2A6E D; 4C3H B; 2EPB A; 3JCM h; 1PM3 A; 4HA8 A; 4L59 A; 4NL3 A; 1J9O A; 1PFS A; 3NQT A; 1WTR A; 4BXX H; 1HQM C; 3D0F A; 4BDY A; 3CQZ B; 4BA1 I; 4C3J G; 1K3B C; 4F7U A; 1WFW A; 4KJN A; 4CDD C; 3PUX A; 1FWZ A; 1Q7Y U; 1Y8P B; 1MD2 D; 4J8Q A; 2R5A A; 3A8H B; 4RGO S; 4LUE A; 1MMD A; 1QOH A; 5D3I B; 2F0P A; 4DNI A; 1K9M C; 1V29 B; 1D0X A; 3BDC A; 5EQ0 A; 4WJ4 A; 3C4S A; 5BR8 L; 5JPN C; 1YI2 T; 3PO3 G; 4QMG A; 2ECZ A; 2ZCF B; 5GQO A; 4ELY C; 2CS7 A; 2JXM B; 1O7Z A; 5D3D A; 1HUM A; 1F2Z A; 2A68 C; 2KYM A; 3UPU A; 3T51 B; 1E7O A; 2UXC L; 1S3O A; 2OJ2 A; 2F2V A; 2GFA A; 2KY9 A; 4FT4 A; 1Q82 R; 1KAB A; 3EBE A; 2P0Q A; 3KF6 B; 4HIM A; 4DF7 A; 2L4N A; 2K5F A; 1NY4 A; 5I21 A; 3JCM b; 1HD3 A; 3KUP A; 4YWL A; 2PQW A; 3QWN A; 2NUC A; 4AFQ C; 1EOT A; 3I91 A; 2P0K A; 4QL5 A; 1KAA A; 2KXC A; 1I94 L; 4OEL B; 5JOB A; 2CCZ A; 1N33 L; 1YN8 A; 2Q8I B; 3M4O H; 1UHF A; 4TSS A; 1M90 C; 1AHJ B; 5GAO p; 1SYD A; 4KUI A; 5COR A; 1XJW B; 4ESR A; 1B8A A; 2QDL A; 1VQK Q; 3E19 A; 4AYB G; 4RG8 A; 4XK8 E; 1PH4 A; 1R4Q B; 2HQH A; 1BF4 A; 4FSS A; 4LD6 A; 1Q82 C; 2XDB A; 4MNO A; 1QNK A; 1WI5 A; 1N32 L; 2QTX A; 1Q90 A; 3OIP A; 1NBE B; 5LJ3 h; 2A08 A; 1AT1 B; 1WTW A; 3ONA B; 2HDA A; 1WX6 A; 3S14 H; 4XYO A; 2MOX A; 4BI3 A; 4M7D E; 4KHP L; 3N5B B; 3EH2 A; 4RG2 A; 2LX9 A; 5CK9 A; 2KJX A; 3GTM B; 2DJQ A; 1LM0 A; 4HR7 B; 1B34 B; 5A2Q L; 1BCO A; 5CV5 A; 1TUD A; 4MTN A; 1E6H A; 2E74 C; 4LDV A; 2CWA A; 1GUO A; 2RCN A; 4A75 A; 4J9F A; 4F17 A; 4IIM A; 4G57 A; 1DON A; 1GXD C; 4LDX A; 1KC8 R; 2G5D A; 2AQ2 B; 2YHT A; 1JJG A; 2QEX T; 2E5K A; 5FWC A; 1HYO A; 4LF4 Q; 1LVK A; 1AD5 A; 4M7A F; 4PLL A; 1JJC B; 2Z6B A; 2PZT A; 4X3S A; 4HIO A; 2AT1 B; 1MJE A; 1V43 A; 5GAN 8; 1VQL T; 1PVO A; 1H3H A; 1EWC A; 4A63 B; 3E1S A; 3S16 B; 4F8M A; 4BY6 C; 2SOB A; 1A3V A; 1QZH A; 4KUD K; 1RAH B; 2LNV A; 2D1X A; 1SSO A; 4FSX A; 1AWX A; 3TSZ A; 4HT9 A; 1I94 Q; 2JA7 H; 2NB0 A; 1Z9Q A; 1EIH A; 3URG A; 3NMZ C; 2QGQ A; 1H64 1; 1R0B G; 1KC8 C; 5I2Y A; 1NG2 A; 3NXW A; 1RAA B; 3UDX A; 1O7L A; 2FRW A; 1YNJ D; 2QQS A; 4CKC A; 3VKI A; 2JN4 A; 2ZM6 L; 2KQ3 A; 3S1R H; 3QKJ A; 5KYY A; 2QDY B; 5D7U A; 3MXN A; 2NTS A; 3Q8D A; 1NZ9 A; 3AB9 A; 4R1J A; 1ABO A; 4ZYS A; 1OYX A; 1A2T A; 4DU9 A; 5LMN W; 2JA5 H; 1JWS D; 1H8K A; 1SE8 A; 1V9P A; 1XWE A; 2RA4 A; 2HHH L; 5C44 B; 1WHH A; 4NW2 A; 3HOX H; 2F5K A; 2L9H A; 1TR5 A; 4I65 A; 2XKO C; 4DR5 L; 2NZO A; 2F5T X; 3S14 B; 3H9N A; 2FRY A; 2WAQ E; 3GDE A; 4N9T A; 4BBS G; 2RNA A; 1CT1 D; 2FIN B; 2A73 B; 4PNY A; 3F1Z A; 1VQ5 A; 1TWF B; 3PO2 H; 2UXC Q; 3HOZ H; 1PFN A; 2AKW B; 5JJI A; 5GMK i; 3EQL C; 1Y96 B; 1KDC A; 2E2H B; 3P8H A; 2PVG B; 3P8D A; 4OGE A; 3AHU A; 2HEQ A; 1BYM A; 5AT1 B; 2H8H A; 4PJO F; 1ET9 A; 5GAN 4; 3I35 A; 3G4S A; 2FZC B; 3EJI A; 3VU3 C; 3HFN A; 2AUK A; 3L1A A; 3VR6 A; 2LCC A; 3MEW A; 5K36 G; 3HHT B; 3OY9 A; 4XU1 A; 4WJ3 M; 1MHW C; 3PUY A; 2QEJ C; 2D62 A; 1KQS S; 4IPC A; 5H1S E; 3CC7 A; 4QT7 A; 4DV2 Q; 1HNX L; 1KRS A; 3I4L A; 2CQA A; 3C2P A; 1N32 Q; 5GAD C; 1GOZ A; 4JBJ A; 5A0G A; 1EY0 A; 3KK7 A; 2CSI A; 1H95 A; 4M7A C; 1T0H A; 1G2S A; 1G5V A; 3F2B A; 4OEM B; 2I46 A; 4R4C A; 2OWO A; 5KCH A; 2KCN A; 4EQ5 A; 3PT9 A; 3JB9 J; 1VYU A; 1DZ1 A; 3IQL A; 1KDB A; 2F2X A; 1JBM A; 1H92 A; 1W70 A; 1C7Y A; 1K4U S; 3LGJ A; 4YTL A; 1QCB D; 3SHL A; 2O88 A; 2PI2 A; 5IP7 H; 1N6A A; 4C3J H; 4ZTJ A; 1CA6 A; 2CW0 C; 2RHY A; 3HOX B; 4IUQ A; 2E10 A; 3WVE B; 2X4Y A; 2CWP A; 4H44 C; 1PKT A; 3CCU A; 3OMG A; 1VQF A; 2CP2 A; 3CCQ A; 4OO1 I; 2ATW A; 2VKN A; 2H3J A; 1IB8 A; 4JI2 L; 3M3Y B; 5C3W A; 3O8V A; 3PO2 B; 1EFI D; 3PVL A; 1E1T A; 1VQC A; 2EYZ A; 1DOM A; 2E12 A; 1VYT A; 2J7I A; 4N4G A; 3T1H L; 4LMQ D; 4M7A B; 4F14 A; 5GMK m; 3VDY A; 1PHJ A; 4C0D B; 3T53 B; 1Q86 R; 2D9T A; 1A0N B; 4BBR B; 2B8A A; 4X65 Q; 4OGC A; 5LJ3 g; 2O9S A; 4U67 3; 2Q2I A; 2ZM6 Q; 3PUV A; 2AIR B; 3V8L A; 4QKU A; 1AWJ A; 2CSQ A; 1QUQ A; 4PMC A; 1ESF A; 2EQK A; 2R61 A; 5FO9 B; 5C4X G; 4KI0 A; 3M55 A; 2H1E A; 4OIJ A; 3M85 A; 3PMI A; 3RLO A; 2NVT H; 4PJO C; 1AE3 A; 5FLX L; 2BE5 C; 3GP7 A; 3PIO 3; 3S1N B; 2L8D A; 1F9R A; 3ERS X; 2HHH Q; 2CP5 A; 1U4P A; 5GAE C; 2WII B; 4A3K B; 4E7H A; 3U4V A; 2OTJ A; 5F2D A; 1MMA A; 1X43 A; 1MJC A; 1X6B A; 5CBE E; 4DR5 Q; 3PF5 A; 3V8J A; 1E24 A; 3QB3 A; 1Q86 C; 4BS9 A; 3BZK A; 3NSW A; 4GN5 A; 1SAP A; 2ZXR A; 2KVM A; 2HCI A; 2Y0S E; 2YUN A; 1MSH A; 2UUA Q; 1QVF S; 4OB1 B; 4AIM A; 4POG A; 3NBH B; 3TN2 A; 4LE9 A; 1N8R U; 1PG5 B; 1IKL A; 3VYT A; 4A76 A; 1OPL A; 1DJR D; 2DKG A; 3JCM V; 2B2W C; 3MXU A; 4BY7 G; 3CC7 T; 5C0X G; 2LMJ A; 5C5M A; 1NEB A; 4RCO A; 4ZUI A; 1VQ6 A; 2R58 A; 3FRP B; 2N3S A; 1C48 A; 1SJE D; 3L7Z C; 4DV3 L; 3LX0 A; 1QNU A; 3W2D A; 4PJO B; 4OB0 B; 3CC2 T; 2RCF A; 2RQT A; 1Z9Z A; 1E1O A; 5AJ3 Q; 1BCM A; 4A3B H; 1XOV A; 3H8Z A; 1IXD A; 5LJ5 f; 1IHW A; 1BOS A; 1PH9 A; 5IWA Q; 2BZ8 A; 4LK1 C; 2XNA C; 1HSQ A; 3MJX A; 1FD7 D; 5IAY A; 3EGX A; 1U5K A; 1ZSG A; 2WSF E; 3M54 A; 1YHQ A; 3PV0 A; 1C1A A; 2EKH A; 2A3L A; 2A0F B; 1Y1W H; 1VQ8 A; 4PZ7 A; 1J8I A; 3CCU T; 4C92 F; 1STY A; 5IYB H; 1N34 L; 1C0A A; 2PW7 A; 3NFC A; 2O2O A; 2QAS A; 4NXM L; 4GKK Q; 1ICW A; 3D7S B; 1QP2 A; 2RF0 A; 1N9R A; 4KBM B; 2YXY A; 3MKD A; 3CCR A; 1LTA D; 1SRL A; 4IJL A; 2I5M X; 3UI2 A; 5IYC G; 4LNQ A; 2A6H D; 4C8Q B; 2YTV A; 1B9M A; 1IYJ B; 4GLM A; 4JS5 A; 4N4I A; 2XHC A; 4C0G A; 3PIO A; 5I9O A; 3MP8 A; 3NWT A; 3CCV A; 3OOC A; 2L15 A; 2X69 A; 4JI2 Q; 2QI2 A; 3OB9 A; 1VQ4 Q; 1TXY A; 4K8I A; 4BBS H; 1I4Q A; 1OU8 A; 2R6G A; 4GN3 B; 4A4F A; 4QQG A; 3HSB A; 2OAW A; 3RKX A; 1D1A A; 3M0U A; 5I11 A; 5KYX B; 3I56 A; 2QA4 Q; 1VQG A; 2DX1 A; 4M77 F; 1LTI D; 2PO4 A; 5CAI A; 1NR2 A; 3AU7 A; 4R4I A; 1YJN Q; 2K14 A; 2XRS D; 1SSF A; 3G6E Q; 1Q1E A; 2XN9 C; 5GAO n; 3BU2 A; 2EY2 A; 2QGF B; 4EMK A; 5GAM g; 4ZAI A; 2MCN A; 1D8L A; 2JXB A; 2K50 A; 2CUD A; 5CIU A; 4RA8 A; 4DR3 L; 4KN7 C; 2OTJ T; 4DR6 L; 5J3B A; 1YIJ Q; 1IW7 D; 1EZ6 A; 5M5W B; 3ULL A; 3NHN A; 2FB7 A; 2RQR A; 1CKN A; 2JTM A; 1WJQ A; 1M1K R; 3F8T A; 1G8X A; 5JJL A; 1VQN A; 3CXC S; 2EXZ A; 2JK8 A; 4XTW A; 5CKD A; 1H9S B; 1ZUU A; 1MBY A; 1VQ9 Q; 1FYC A; 5LJ5 e; 1UE5 A; 3FB9 A; 1UGQ B; 1VQ7 A; 2M0Y A; 5CVI A; 1JO8 A; 3OPO A; 4FF4 A; 2Z8L A; 1A0I A; 3L0O A; 2M9V A; 1L1O A; 1VQ6 T; 1K83 H; 4GNX B; 1B50 A; 3M9Q A; 5LJ3 d; 1SLJ A; 2XVS A; 2EVR A; 1SYF A; 3SWN A; 4HN7 A; 3STB C; 4JBK A; 1KSW A; 4M75 D; 3HKZ G; 1M1K C; 1T9H A; 2X4J A; 4QVC A; 3CCJ T; 5GAN j; 4OX9 L; 1AZP A; 3OSO A; 2KE9 A; 1S5F D; 5IGB A; 3CC4 A; 1HXY D; 3K7A B; 4G4K A; 3CCE A; 4C92 C; 1CA5 A; 1HTP A; 4LUZ A; 2RI2 A; 4DV3 Q; 5KYW A; 2YT6 A; 4RCB A; 1GFD A; 2F0L A; 1JJ2 A; 1WDX A; 1SE4 A; 1KG1 A; 4EQO A; 1YHQ T; 4DR4 L; 4D8D A; 4JI1 L; 2KRO A; 1RLP C; 3MP6 A; 2CH4 A; 4A4I A; 2K1B A; 1PH8 A; 4QQD A; 1VQ8 T; 1D5M C; 2XIW A; 2KGT A; 1L2F A; 1ZSZ A; 4EQN A; 4MMK A; 1ENF A; 1YHB A; 4RG1 A; 4UAI A; 1AE2 A; 1Y14 B; 4MHE A; 2VB2 X; 2W10 A; 4Z02 A; 3IL8 A; 2KEA A; 4F87 A; 1WCM H; 2IX0 A; 2K32 A; 2JJ9 A; 5L37 A; 1PH5 A; 4M77 C; 1RAE B; 3QO3 A; 2ICE C; 4RHR A; 1RL2 A; 1GUN A; 5C6A A; 3CCV T; 2XSC A; 3E9H A; 2IWH A; 1EY6 A; 1RAB B; 1SMY C; 1LTS D; 4J6H A; 3I4N G; 4AFS C; 1BQQ T; 3X25 B; 4QIW E; 3KXT A; 2KYB A; 4I7A A; 3I56 T; 2B86 A; 1G2B A; 4J9E A; 1DJ7 B; 4C92 B; 2G3R A; 1TU2 B; 2HUG A; 2N88 A; 4LF6 L; 1HRJ A; 1IXR A; 2JA9 A; 5ELD A; 1HR0 Q; 4CDE C; 4FF1 A; 4LXC A; 5IGE A; 1K83 B; 4UY8 C; 1D3B B; 3CC2 Q; 1BI3 A; 2LXK A; 4M7D F; 1NM7 A; 1TUC A; 1GFC A; 1AWW A; 3SE0 A; 3M0Q A; 1BIA A; 2F0W A; 4H13 C; 2OTL A; 4BY7 H; 2B8F A; 2LQ8 A; 1OO9 B; 3X3N A; 4M77 B; 3VRY A; 3ND8 A; 2RHX A; 1Y0M A; 3MWY W; 1VQO Q; 1RHP A; 4DV4 Q; 4YCV A; 1VQ7 T; 3EG3 A; 2NYZ D; 2FPD A; 2DER A; 3K57 A; 3OBW A; 1QZG A; 2R7Z B; 1HNZ L; 4WF2 A; 4OWW B; 1X3F A; 2K5D A; 2R7D A; 1SJH D; 5C3E H; 2DXC A; 1E2V A; 1YNN D; 3EFO B; 2DD5 A; 2EYY A; 1IBL L; 3L2W A; 1U06 A; 4B3M Q; 4KGV B; 1LLR D; 1HXD A; 4LUV A; 1WCM B; 1J5E Q; 1RTN A; 1HCZ A; 2ALY B; 4J1M A; 2NVQ H; 2KEE A; 3ICE A; 4CJ1 B; 5CQX A; 4DV6 Q; 4BWG B; 4BY1 H; 4JI5 L; 3CCE T; 4J9H A; 1RLQ C; 4HCE A; 5GAM d; 1GJX A; 1PSF A; 3SR1 A; 4JLG A; 1U5S A; 3NPP A; 4L8J A; 2FPE A; 1QMC A; 3HEJ A; 3PQI A; 3U4Z A; 1VQP A; 4OX9 Q; 1BCP D; 1STH A; 4LDY A; 4DBR A; 2ATC B; 3B6Y A; 5FRM A; 2K9G A; 5CGK A; 3BVM A; 4C2M H; 2YU9 H; 1MNE A; 5HUR A; 2E6Z A; 2E2I H; 1G6Z A; 1JXO A; 4KT0 E; 3DEP A; 4JI1 Q; 3AFP A; 2EFI A; 1YIT T; 2L89 A; 2RVL A; 5DEH A; 2IY5 B; 4NOY A; 3JB9 I; 1IL8 A; 2ZXO A; 3BYT B; 3K5L A; 2LVM A; 2K04 A; 4O42 A; 1GBQ A; 3OQ5 A; 4LJZ C; 3H3V H; 2HMA A; 3HOV H; 3JB9 D; 5IHI A; 2EYW A; 1T0J A; 4OVA A; 4N8X 1; 2JUF A; 4DXG A; 3KYH C; 1EY9 A; 3I55 Q; 3CCS Q; 1SYE A; 5IT9 c; 1VS0 A; 3OWF A; 4M7D C; 1A15 A; 1ICF B; 1EQT A; 1ENA A; 3WWV A; 3SDZ A; 2IX1 A; 2ZPI B; 3NEL A; 4BE2 A; 5TSS A; 3CMA Q; 1F9S A; 3A8G B; 1MSG A; 1SEM A; 1KLG D; 2LZ6 B; 1V1Q A; 1HR0 W; 1Q1B A; 5C3E B; 4BXX G; 3AT1 B; 4D6W A; 2C35 B; 5GRO A; 4DA4 A; 1FMV A; 4PJJ A; 5ELN A; 1I50 H; 5JJZ A; 2D6F A; 4LF6 Q; 5M3M H; 2UXD L; 1O9S A; 1CZW A; 3KLS A; 1YBY A; 1R9T H; 4DR1 L; 2NVQ B; 1WI7 A; 2BDN A; 3K6O A; 2CZ7 B; 2JTE A; 2LKV A; 3K81 C; 3TP8 A; 2L0C A; 2R7F A; 1IO6 A; 3I5S A; 4J6X A; 2OTL T; 1OXU A; 3I4O A; 4BY1 B; 4DKA C; 4K2K A; 1C6V X; 1L0Y B; 1D0Z A; 2PPB C; 4AYB E; 4C5G A; 1TWA H; 2K01 A; 4IO9 A; 2DBM A; 2O4X A; 4GKJ L; 2P4T A; 4O6J A; 1E6G A; 1K8M A; 1X56 A; 2DNC A; 4M75 G; 5CV4 A; 4C2M B; 2YU9 B; 3VS2 A; 4M7D B; 2DRK A; 2E2I B; 3S24 A; 4HCK A; 3LH5 A; 2ZJR A; 4E7S A; 2X4X A; 1HNZ Q; 1M8A A; 2L12 A; 5HK3 A; 1B53 A; 1FL0 A; 1NSN S; 4BE1 A; 5IT9 E; 3RZO B; 1LCK A; 1YJ9 A; 1IBL Q; 5E3F A; 2K2M A; 3GNF B; 2F0H A; 2IW3 A; 3VS5 A; 4JI3 L; 1AVZ C; 3ULJ A; 3QR8 A; 1WYD A; 1A3T A; 4HTH A; 3QZ9 B; 3F70 A; 2RKS A; 4IAL A; 3HS0 C; 3CW2 C; 4RH6 A; 2NUT A; 1Q7Y R; 1QVF P; 5IHN A; 4O2D A; 5CKF A; 3SEM A; 4JI5 Q; 3PE0 A; 2GCX A; 4ZGD B; 1K76 A; 3J7Z C; 1PZI D; 3VS6 A; 2CKZ B; 2VXW A; 1YJW A; 3M4G A; 1CQF A; 3MPU B; 2ICI A; 1KAW A; 1I50 B; 2HIV A; 2DN8 A; 2E5L Q; 1WA7 A; 5M3M B; 5G3L F; 1R9T B; 4JBW A; 2E4H A; 2VGM A; 3A1P A; 3QBY A; 2F0M A; 1UTI A; 2K5Q A; 4O0A A; 5EAY A; 2DXB A; 2AQ1 B; 2AWO A; 2MHV A; 4L6V 5; 3MP1 A; 4TRD A; 1S72 A; 2OEO A; 5CMD A; 4NS5 A; 1Q7Y C; 2A7Y A; 2E3I A; 4QY7 A; 3VS0 A; 2K57 A; 1TWA B; 1CTM A; 2CQO A; 3H0G H; 1ML0 D; 1M3B A; 2RSN A; 5CV7 A; 3JAM L; 1WTQ A; 1XPU A; 5LMN L; 3I4M H; 4AMC A; 2O2L D; 1ZXT A; 1PHT A; 3ZL7 C; 3TME A; 5KYY B; 2MO1 A; 2WAQ B; 1J3T A; 3MXN B; 3TP5 A; 4M1U B; 5LJ3 j; 4HJA A; 1D5Z C; 5T1G A; 4FF2 A; 1OX9 A; 4DUY Q; 1XNI A; 1VF5 C; 2WB1 E; 4IOA A; 2EYO A; 1I8F A; 1AW7 A; 1SND A; 3EAE A; 1UJ0 A; 4AG2 C; 1U4R A; 3J7A F; 3TRZ A; 3LW5 E; 1K0R A; 2F0N A; 3MFY A; 1X6G A; 1PH7 A; 3HOW H; 3M7G A; 1HJD A; 1NNX A; 1EU4 A; 4C3H H; 2NBF A; 3GP8 A; 1Q8K A; 2MFI A; 2FL7 A; 2JA5 G; 2UXD Q; 5J39 A; 2DIQ A; 1LT4 D; 1M2V B; 3FMC A; 4XTY A; 4IJH A; 3CQZ H; 2Q2T A; 3VS1 A; 3OSV A; 3HOX G; 2DNE A; 5J26 A; 4X62 L; 2EGE A; 3CCL A; 2R93 H; 3RMS A; 2ZTD A; 4EDU A; 4JPB W; 1P16 A; 2G9D A; 2QYZ A; 1S40 A; 1VQE A; 2NN6 H; 3SFM A; 1AWO A; 2CDT A; 4KK7 A; 3T1Y Q; 3P8B B; 3PO2 G; 2J05 A; 1B7T A; 1N5Z A; 1I4G A; 1TTH B; 2OX7 A; 3GNG A; 2VQF L; 1EFW A; 1X65 A; 4OMN A; 4GKJ Q; 2Z0S A; 1UE6 A; 4M78 E; 1R0C B; 4M7A G; 1DYQ A; 2MMP A; 1VCI A; 1HUN A; 2JW4 A; 3UF4 A; 1YJ9 T; 2JYO A; 1IBK Q; 1NAP A; 3R2T A; 4KGZ B; 4DAY B; 5I22 A; 1SXT A; 3GTQ B; 3EG0 A; 1PWT A; 2EGA A; 2L1B A; 1UEC A; 2EPD A; 2NVY H; 2RH2 A; 2QQB A; 3H0G B; 2K5L A; 5LHY 1; 4KY5 A; 5J22 A; 4LF4 L; 1ZA2 B; 3NEM A; 4TVR A; 1RKN A; 3SHW A; 3WDN A; 3BDU A; 3I4M B; 3I3C A; 2DEQ A; 1GVP A; 3X26 B; 3GQB A; 2QK0 A; 4HCC A; 2GNC A; 1Y8N B; 1Y71 A; 3QO2 A; 1YJW T; 4ZCK A; 4HED A; 1W3S A; 5CBA E; 2JOY A; 2DRM A; 5GJW B; 3SXH A; 4QQ6 A; 3CRK C; 1JXM A; 3K58 A; 1HNW L; 4DOV A; 2BKD N; 1H9K A; 1PTO D; 2FHD A; 5CV6 A; 2E75 C; 4A4G A; 1EQV A; 2J4X A; 4DN4 M; 1EIY B; 3HOW B; 1TG0 A; 2DL8 A; 1JWU D; 4RTU A; 1Y57 A; 3MC0 B; 3J80 i; 5E1F A; 1J6Q A; 2NZ1 D; 1NLO C; 1S72 T; 5IP7 G; 3RKW A; 4YCW A; 5ELB A; 3P1H A; 2VXF A; 1MHN A; 4ELZ C; 1VQM Q; 3GTM H; 2BZX A; 1QP3 A; 5DAU A; 5ELE A; 1T3L A; 1RAC B; 4QVZ A; 1EIF A; 5EC7 A; 4EPC A; 3S3N A; 5EPJ A; 2DYI A; 2R93 B; 1SPK A; 2F0F A; 2DTI A; 1IGU A; 1GL5 A; 1GHK A; 5FU7 C; 4E7L A; 1KDA A; 2W0Z A; 1N9W A; 1BIB A; 3G71 A; 4KY6 A; 4QUF A; 2Q2H A; 1LTB D; 1W2B P; 3CBM A; 1M90 R; 4ANJ A; 2A6H C; 2H5X A; 1WHG A; 3H94 A; 5GAN f; 4PJO G; 5LMN Q; 4V19 0; 4B3R Q; 3EN2 A; 3R6N A; 2B63 B; 4F7U E; 7AT1 B; 3A74 A; 1ZHI A; 1TWB A; 3M4Q A; 5ELF A; 3A7A B; 3S16 H; 2Y4X A; 2IJ0 A; 4C5H A; 4CC2 A; 1CUK A; 4JZ4 A; 4RCC A; 5GAM j; 2CP6 A; 1CSQ A; 4J8O A; 1PZK D; 2K28 A; 2Y0S B; 3CKI B; 3EIV A; 1WTX A; 2CGH A; 1Q8I A; 2KEN A; 5EKK A; 1PD1 A; 2MAM A; 5EKL A; 1JR0 D; 4V0B A; 3E9F A; 3LLR A; 3RUX A; 3HOZ B; 3BG3 A; 2DIL A; 3LWH A; 1OU9 A; 1JE4 A; 5EBC A; 3CCL T; 1UG1 A; 2O9V A; 4J6W A; 1MIU A; 4RTW A; 5M9O A; 5C6S A; 2V26 A; 2RJC A; 2V8K A; 2VXE A; 4LDU A; 1XYI A; 4A3D H; 1KHC A; 3OW2 A; 4R4T A; 4DV2 L; 4C8Q G; 1G8Z D; 1EOV A; 5FU7 B; 4X62 Q; 1ASZ A; 1IW7 C; 1HFN A; 4FW2 A; 2V1C C; 1XNQ Q; 2BDO A; 3CAM A; 2F0E A; 5C44 H; 1EYG A; 2NUP A; 4IPD A; 4N4H A; 2GTJ A; 3PGZ A; 2MWO A; 3V7S A; 2DGY A; 3DWA A; 4XT3 B; 2KNB B; 1S5C D; 5C0X I; 5EGT A; 2VUM B; 1SEB D; 1WIE A; 4B9W A; 5KYU B; 3S1R B; 2NVX H; 1TWF H; 3J81 L; 5I4D A; 1KRT A; 3EG9 B; 5IYD G; 3FPP A; 2SNS A; 3D6C A; 5FLX c; 3SK4 A; 2E2H H; 2KSS A; 1N27 A; 4RTY A; 2K5W A; 2NVY B; 2PQE A; 3MZ5 A; 3G7Z A; 3M0R A; 1PRT F; 1JC7 A; 5CKE A; 1ABQ A; 3DLM A; 1ENC A; 1EWI A; 4MTK A; 4X34 A; 4E2F B; 2E41 A; 2VVK A; 4EMK C; 4QQB X; 4A3B G; 4YWM A; 3GV6 A; 5GAN e; 1BUV T; 1BKB A; 1WFQ A; 1FGB D; 1PFB A; 2ZT9 C; 1HNW Q; 4KKS A; 2K4Y A; 3M59 A; 1RUW A; 2B39 A; 3URY A; 3OYH A; 2XI9 A; 1UFF A; 2PMZ E; 2Z1C A; 3OS5 A; 3J81 j; 3H15 A; 2F0I A; 3FPU B; 5JEA H; 1Y1W G; 4X65 L; 1OXT A; 4GOP A; 2DFU A; 4XAX B; 5IYB G; 4HID A; 4P72 A; 1M8V A; 3OYJ A; 3G71 T; 3AMU A; 3S1M B; 4ILW A; 1TXQ A; 1Q46 A; 1XTC D; 4A3D B; 4KV6 A; 4DPG A; 2DUD A; 1MKH A; 3W3A A; 4X3T A; 5E4X A; 2DZ9 A; 2JM9 A; 3HP3 A; 2JA8 H; 2K3B A; 2O01 E; 1XXG A; 1L1O C; 1JB0 E; 3RN5 A; 4AH6 A; 3JCM J; 3CU7 A; 3GIB A; 3SWN C; 2NWG A; 1PH6 A; 4RWS C; 1WJS A; 2DL3 A; 2CUB A; 1RCV D; 4GLK A; 2Z6K A; 4EMK B; 3CCM Q; 2SDF A; 4QVD A; 2NVX B; 1QVC A; 3M3Y H; 4BY6 B; 1KJW A; 2G6F X; 1ILP A; 1JOQ A; 3GF5 A; 2RJF A; 2UUA L; 2FJ2 A; 2KUM A; 5GAN 3; 2H4O A; 3QOL A; 5D65 A; 1ZYR D; 3T9N A; 2J0T D; 3JB9 F; 3JSC A; 3X24 B; 1CKN B; 3GV3 A; 3K0Y A; 1WHL A; 1WXB A; 1W4S A; 4C3I H; 1ASY A; 2NUZ A; 2VWF A; 1WJJ A; 1P8L A; 2NTT A; 1K9M U; 4WOR A; 1SFO H; 4BBR H; 4P5N A; 2MO0 A; 1M4Z A; 5DM7 3; 2F0D A; 2EOT A; 5I2S A; 2RHI A; 5GAO r; 2VGN A; 3CME A; 2KRN A; 5IH2 A; 1CKM A; 3KF8 A; 4DNR B; 4IKF A; 1YNX A; 5BN3 A; 1PRT C; 4C0Z A; 2HNI A; 3TZU A; 5AJ3 L; 1L1O B; 3F2D A; 4DBH A; 3S1N H; 3C95 A; 1G6P A; 1QVG A; 1CK1 A; 2V8J A; 1QG7 A; 3A54 A; 3SWN B; 2XIC A; 4C92 G; 4A3K H; 4F88 A; 1B2T A; 3G48 A; 5IWA L; 5GAM h; 3KDF A; 2DMO A; 4N8F A; 4ULL A; 1SBB B; 1YI2 Q; 1YNN C; 4CDF C; 2F0J A; 1OQK A; 3E9G A; 4ZGE B; 2VB3 X; 1JJ2 S; 4CC7 A; 3ERO A; 1I5F A; 2B2U A; 1K73 R; 3BW1 A; 3C46 A; 4GKK L; 4A3F H; 2JA8 B; 1AOJ A; 1I4H A; 1Q82 U; 2CZ0 B; 2X6L A; 2RO0 A; 1BVS A; 4RTT A; 2ENB A; 3C1F A; 3FKI H; 2VAS A; 3PUW A; 3HG9 A; 2E76 C; 4M77 G; 4BUG A; 2L5T A; 4R4O A; 1B7Y B; 4XU2 A; 2RQV A; 3LYI A; 4QIW B; 2PI2 E; 3NTH A; 2Z4I A; 4LFC L; 3W5O A; 3KBG A; 2YDL A; 1PRT B; 4FYW B; 1VA7 A; 5KYL A; 2R7Z G; 4JRP A; 3E5S A; 1BBZ A; 4MML A; 1CQV A; 1F9P A; 2EDG A; 3S9W A; 4C3I B; 4FO2 A; 4MZM A; 3HZX A; 5JAV A; 2LCS A; 1K73 C; 4Q5W A; 4R0K A; 2MPM A; 4NXN L; 1S5B D; 1ZUY A; 1SFO B; 3JCM f; 1WCM G; 3UE0 A; 3X20 B; 2WSE E; 2GFU A; 4DGZ A; 1BI0 A; 3HRT A; 2AZV A; 1UNS A; 1OZ2 A; 3OYL A; 2HSE B; 3I73 A; 2KED A; 3L3O C; 3NP8 A; 4IFD H; 4LF8 Q; 2ZAE A; 2DY7 A; 3FRN A; 4JUV A; 2JNG A; 1EUJ A; 4IFD J; 3AQQ A; 3S15 H; 3QJY A; 2A1A A; 4JCV E; 1VQ5 T; 1KQ2 A; 2KI7 A; 4MZT A; 1LTT D; 1LTG D; 2GVB A; 2LT1 A; 2E70 A; 2QEX Q; 4HMJ A; 2R92 H; 1KC8 U; 3TBI B; 2QQR A; 1WJR A; 1BB9 A; 1TVX A; 4A93 B; 1B07 A; 5GMK e; 2XDD A; 4IN3 A; 2UU9 Q; 4C0F A; 2ROL A; 4FCY A; 3A7L A; 1X2Q A; 1VQL Q; 2VL6 A; 2VB6 A; 2EAY A; 3EFS A; 3G4S T; 5LJ5 g; 3CD6 A; 1STN A; 4J9B A; 3V2T A; 2VQF Q; 3Q23 A; 1EX4 A; 3AYH B; 4A3F B; 1GCQ A; 1D0Y A; 4G7O C; 2KCC A; 4QW2 A; 3C3L A; 3KF6 A; 4HMI A; 5IYD H; 3D8G A; 4OIK A; 3FKI B; 5HKC A; 3KJP A; 3UAT A; 2XWB B; 1WTO A; 1AFP A; 1S5E D; 2BCO A; 4A3L H; 3OBY A; 2EQM A; 3RZD B; 3FJP A; 2JE6 I; 1OI1 A; 2R5M A; 1MMG A; 3TRE A; 1K8A R; 3QSU A; 2UXB Q; 1O78 A; 1SRM A; 1LUZ A; 3K80 C; 5DLQ E; 5M5W H; 4BA2 I; 3NUC A; 1WD1 A; 4XTZ A; 5C3E G; 4ZUJ A; 3JAM i; 1SYG A; 1NE8 A; 4AT1 B; 1YCS B; 1VQB A; 2I5H A; 3EFX D; 4F7X A; 5BN5 A; 4JBM A; 2ZTE A; 3HP9 A; 3VYS A; 1KIK A; 2C9O A; 4J9D A; 4PL6 A; 3P20 A; 1XPO A; 2XID A; 4BY1 G; 1SNP A; 3S17 H; 2FFK B; 2DXU A; 4MZP A; 3JCM e; 1K8A C; 1E3P A; 3QUI A; 1LAB A; 4NL2 A; 4PNO A; 3OYK A; 4ZK9 B; 1GO3 E; 5FLM G; 3CCQ T; 1B34 A; 2KOL A; 3MO8 A; 1PH1 A; 4R4Q A; 2G9H D; 3OA6 A; 3PO3 B; 4Y92 A; 2CT4 A; 1CSK A; 1IHV A; 3I7F A; 5M3F H; 2R92 B; 2KRS A; 2B2Y A; 5EG2 A; 2DTO A; 2YUO A; 1N36 L; 4JV5 Q; 4ZKB B; 1VQD A; 1FMW A; 4M7D G; 2OI3 A; 1GXI E; 2PZU A; 3K7A H; 3M4P A; 1FXX A; 4FM4 B; 2MWP A; 2I07 B; 5I9P A; 4LFC Q; 5JVG 3; 4C1N K; 2KA7 A; 3MXG A; 1CSP A; 1D5X C; 4AFZ C; 1HR0 L; 1I81 A; 4HSV A; 2FO0 A; 1QWF A; 4OMQ A; 1SE3 A; 2QAZ A; 5D6X A; 3REB B; 3ERQ A; 4R9W A; 4DV1 L; 3GLX A; 2ABL A; 4OMO A; 2Z9H A; 4DK3 C; 4R55 A; 4JOI A; 3Q24 A; 4NXN Q; 3HOV G; 4LUO A; 4M75 A; 5I2X A; 1A63 A; 4M78 F; 1GD7 A; 1M1G A; 3CPW A; 1AZE A; 4X64 Q; 4X66 L; 4AYB B; 5BN4 A; 2CKK A; 4Z88 A; 2EQS A; 4A4E A; 5IIF A; 5KNC A; 2B2V A; 3TSW A; 2A68 D; 2IHE A; 3J80 L; 3M7N A; 1UEB A; 4PFO A; 5IE8 A; 3OS0 A; 4ZNX A; 3OW7 A; 1NBW A; 1Q12 A; 1HZB A; 4M75 E; 4K6L A; 4DV4 L; 4AQY L; 1MMN A; 3VUB A; 1VDZ A; 4XU3 A; 1KXL A; 1UJY A; 3CD6 T; 4A4H A; 4X6A H; 3PUZ A; 5M3M G; 4BD3 A; 4L5Q A; 2QF7 A; 2EIF A; 2HQE A; 2YRV A; 1ZBX A; 2DY8 A; 1Z9F A; 2PUO B; 2OF1 A; 1SSH A; 1JOO A; 4B3M L; 2LJ3 A; 2QG9 B; 3S17 B; 1FMK A; 1EEF D; 2E3H A; 1J5E L; 3HOY H; 2RJE A; 1G91 A; 4FYV B; 1VMP A; 4J5Y A; 4A3C H; 2GN5 A; 4CDC C; 4DV6 L; 1QKW A; 4RMF A; 3NHH A; 3A8O B; 1TY2 A; 1JQY D; 2DZC A; 4M4Z A; 1Y96 A; 4TQU S; 2LDM A; 5M3F B; 4JI6 L; 2K6A A; 1B44 D; 2WAQ G; 5M5X H; 3JCM c; 2EYH A; 4H0L C; 4CKB A; 2TSS A; 2DAQ A; 1Q86 U; 2EBP A; 2PZ1 A; 1EU3 A; 4B3T L; 4OF1 A; 3ULR B; 3VA7 A; 2RK2 A; 1TS2 A; 2K3Y A; 3Q1J A; 4I7Z C; 2COY A; 2VUB A; 2UUC Q; 1D1I A; 2AUJ D; 3K5N A; 3K5O A; 4G7H C; 2R7Z H; 4U68 A; 1VQ5 Q; 1W2B S; 4F7U F; 4EIK A; 5GAO q; 1KQS A; 3IFT A; 4FF3 A; 4M8A A; 1Z47 A; 1YFN A; 2LSS A; 5LJ5 d; 2E5Q A; 2CFM A; 3P7J A; 1FYN A; 3CCR T; 2EY6 A; 2PNR C; 3I90 A; 2A69 D; 3EGD A; 3PTA A; 1NPP A; 1AEY A; 3UDI A; 2ZXP A; 1Q81 R; 2EJG A; 2YWW A; 3OYG A; 1BI2 A; 2K6D A; 4A3I B; 4A53 A; 4OB3 B; 3LWU A; 1K8G A; 2DTH A; 4NKL A; 4M78 C; 1F77 A; 2ZZD A; 2Q8R E; 2M16 A; 1WHK A; 2WB1 B; 4K6D A; 1WXU A; 3OYE A; 4A3E H; 5HK0 A; 3HRS A; 1N36 Q; 3CC7 Q; 1TS4 A; 1T5X D; 5DM6 3; 2FVU A; 3TDQ A; 1RJT A; 2LXJ A; 2DL4 A; 2OQK A; 1ESR A; 2F1L A; 4A3J B; 2F69 A; 1RI0 A; 2A6E C; 2KCM A; 3HOY B; 4FDB A; 4IUU A; 1S4Z A; 2EKO A; 2YUP A; 3FC3 A; 1UAP A; 4RKL A; 3H0G G; 4N0A A; 2L11 A; 1FNU A; 4DV1 Q; 1Q81 C; 2YLB A; 1SYB A; 4TVA B; 3I4M G; 1XMQ Q; 1VQJ A; 4OB2 B; 4M7A A; 3VYU A; 2L55 A; 3OYI A; 4K2L A; 2LC4 A; 4X66 Q; 5M5X B; 3J81 i; 1M3A A; 1TII D; 4JDS A; 4RUG A; 4LWC A; 1XNR Q; 4NPS A; 3QWY A; 1EEI D; 1VQN T; 3PIP 3; 4XXE A; 1BU1 A; 1KL9 A; 5HGQ A; 1HA6 A; 3E0J A; 3GD7 A; 1Y1V B; 4Y91 A; 3CCU Q; 1OOT A; 2F0V A; 4M78 B; 4APV A; 3CCQ Q; 4AQY Q; 4OWX B; 3HOW G; 4C3H G; 1SN8 A; 1SKU B; 3KOJ A; 3CHB D; 4RTX A; 4RTV A; 4LFB L; 2KRM A; 1NJI R; 2D0P A; 4MDW A; 3EH1 A; 4ABN A; 3NTK A; 1KD1 U; 3KLW A; 3P75 A; 2ASB A; 1DM0 B; 1D6E C; 4XYQ A; 2PU9 B; 1SF9 A; 2WKD A; 2R93 G; 1PSE A; 2ID0 A; 5GAN 6; 2HL3 A; 2NLU A; 2A28 A; 1NUC A; 1ROD A; 3N6R A; 4KD3 A; 2DO3 A; 2NN6 G; 4E6R A; 5CKB A; 4A3I H; 4JI4 L; 3CC4 T; 1N34 Q; 2F0U A; 3NK9 A; 3NBH A; 1JB7 A; 5C4J H; 2X4W A; 1VF7 A; 1NYF A; 1JCK B; 2FHT A; 2IX3 A; 4M7A D; 5FLM H; 3IAY A; 4JI6 Q; 4QCL A; 4RGT A; 4A3E B; 1A8V A; 3A0J A; 2HIX A; 3DB4 A; 3OYY A; 2YRC A; 1NJI C; 3A8J E; 2E5L L; 4O1N A; 4B3T Q; 2Y0S G; 4JRQ A; 1NTG A; 4NP5 A; 3H41 A; 3RR6 A; 2OTJ Q; 4ME7 A; 2PQA B; 2L3P A; 5CKH A; 1QE6 A; 4PJO A; 4FXK C; 2O5J C; 3DB3 A; 4R8I A; 2CW0 D; 2F1M A; 1NMG A; 2WAC A; 1V1P A; 3SGI A; 4LF7 Q; 3RZO H; 3PFS A; 1ARK A; 2RB6 A; 2GGR A; 2BOS A; 2KT1 A; 4OMP A; 4BAC A; 1YVC A; 2K4K A; 4K0K Q; 2LS0 1; 4FYD A; 2C5C A; 1JJ2 P; 1FR3 A; 4KHZ A; 2JTF A; 4F7U B; 1E2W A; 1EWH A; 4DBF A; 3LGL A; 5GAO l; 4DUY L; 3TS2 A; 3QDF A; 1VQ6 Q; 3PIP A; 3GTG B; 1O75 A; 2A19 A; 2KBN A; 3QYG B; 4LF5 L; 3CCJ Q; 4JI8 L; 4C8Q A; 1MUZ A; 1DGS A; 2K7V A; 1SMX A; 1ZKO A; 1VQK A; 2B3G A; 2COZ A; 1HK9 A; 1YNJ C; 4WJW A; 4BI4 A; 2KCT A; 1JRI A; 4PV1 C; 2CT3 A; 3NTI A; 1BR9 A; 2EYP A; 2I0Q A; 2DA9 A; 3M4Y A; 4A54 A; 1O80 A; 3BVG A; 4PJO D; 4AG1 C; 2RA2 A; 2D0O A; 4HVU A; 1WQ7 A; 4JRK A; 3TVT A; 4B9X A; 1YHQ Q; 4XQN A; 5C4J B; 5KYX A; 2E2J B; 2RDG A; 3FLG A; 1VQ8 Q; 2ES2 A; 3V7C A; 1A3U A; 3T1Y L; 2QQA A; 3FEW X; 2F3I A; 1GRI A; 2J6K A; 3QPH A; 3CCR Q; 1S1N A; 3QOJ A; 1BO0 A; 2FXQ A; 2WIN B; 4R8N A; 1V1C A; 4PKC C; 4PJM A; 2JA6 H; 1M5Q 1; 2BTT A; 3ZEH A; 3RUZ A; 1S5D D; 4XDX A; 1TOV A; 1IBK L; 2VC8 A; 2EE1 A; 2LRQ A; 3H0I A; 5IGG A; 2QQ9 A; 4ME3 A; 4DR7 Q; 3CCV Q; 3FJ5 A; 1EYA A; 2AMC B; 4FP5 D; 4LF7 L; 4LFB Q; 3E2U A; 1UE9 A; 5E4W C; 3VR2 A; 1U9R A; 4NDX A; 4DQ2 A; 4C8Q D; 2F2W A; 4J9G A; 2VYT A; 3IFD A; 3LGF A; 1UWV A; 4QUC A; 1WYX A; 3PF4 A; 4A3D G; 3I56 Q; 1WTV A; 4HJ8 A; 1LT5 D; 4DR2 Q; 5LMU Q; 3OSS C; 3CXC A; 4GOP C; 3E3X A; 1WZO A; 2EQI A; 4RKU E; 5GAN 2; 1M27 C; 3THK A; 1U1T A; 1QW1 A; 1VQP T; 5C44 G; 2D9U A; 4JI4 Q; 4B6M A; 4GY5 A; 4KLK A; 3MEU A; 1FJG Q; 3AFQ A; 2KHJ A; 2OT2 A; 3ZJY C; 5KH6 A; 4GNX A; 1SE2 A; 3QYH B; 2DXT A; 3F2U A; 4S3S A; 4DV5 Q; 3BYY B; 3QHS A; 3NPF A; 1W6K A; 2EWN A; 2EJF A; 3PMF A; 2PMZ B; 3O13 A; 3AEV A; 4HCS A; 5KE3 A; 3Q6S A; 2Z6K C; 1E2Z A; 1VIE A; 3ULP A; 2O31 A; 2FEI A; 5L8R E; 4OO1 H; 1ONL A; 3Q22 A; 1VQ7 Q; 3GTQ H; 4PJL A; 1M8M A; 2PNI A; 2RM4 A; 3A8K E; 4KD4 A; 1M30 A; 1SNQ A; 3MEF A; 2LQN A; 1HTL D; 1Z7T A; 3A5U A; 3GTJ H; 4B3R L; 1YHA A; 4EMH A; 2MQH A; 2KR3 A; 4Q0F A; 4GOP B; 1I3Q B; 5FJ8 H; 2LMC B; 2RK1 A; 2JA6 B; 4DAM A; 1TS5 A; 4ZLT F; 1I07 A; 3J7A 5; 1QCN A; 3VS3 A; 3VYH B; 2DJF C; 4P2C B; 2BE7 D; 2L0A A; 3VV0 A; 5JEA G; 3S6W A; 1OZ3 A; 2ZPH B; 3M0S A; 3CC4 Q; 4JJN K; 1M2O A; 1WNL A; 1A62 A; 3CCE Q; 1EY8 A; 1OXS C; 2A74 C; 3HOU H; 4PLI A; 3RBB B; 2MGS A; 4LF5 Q; 4LAA A; 5J1Z A; 4JYA Q; 4JI8 Q; 1U4M A; 1Q95 G; 1K1Z A; 1C0M A; 1ET6 A; 2Y4Y A; 2AHO B; 1VYV A; 3A56 A; 1LPL A; 4L63 B; 2EQU A; 2VZA A; 3KFU A; 2AHJ B; 3BE3 A; 1UB4 A; 2KT8 A; 5D6Y A; 3ASK A; 3I8Z A; 4D8K A; 3PGG A; 4C92 A; 1G3T A; 2JA8 G; 4KJO A; 2XU8 A; 2AQ3 B; 4GLX A; 4V2S A; 1UE7 A; 3A55 A; 3CEY A; 4ZTF A; 1UEA B; 3H91 A; 4UDU C; 3SEB A; 4E7Z A; 4EQP A; 5KGU A; 2COW A; 3KJO A; 2JN0 A; 2B2U C; 4F16 A; 1YIT Q; 1XNQ L; 2KAF A; 1IHZ A; 1IL2 A; 3S2H B; 2QTV A; 3K0X A; 4MDX A; 3KUF A; 1G3S A; 1UGP B; 4CRI A; 2HBW A; 2B63 H; 2PTK A; 2RJD A; 1D1B A; 2N55 A; 1MGQ A; 1AP0 A; 2CRE A; 4C3I G; 2F0S A; 3U9O A; 5HCK A; 4M77 A; 1QVG S; 1X32 A; 3DMU A; 5FK9 C; 1SHF A; 1SRU A; 2WP8 J; 3DEO A; 4BBR G; 3RLN A; 2CHB D; 3CP0 A; 4ALP A; 2YUQ A; 1RTO A; 5A2Q E; 3STB D; 4JC0 A; 4X1V A; 4PFP A; 1SHG A; 1QKX A; 1CI3 M; 2EQJ A; 2HCK A; 1NMF A; 4J15 A; 5DNF A; 3SK5 A; 3GTP H; 5IP9 H; 2LY7 A; 3GTJ B; 2SNM A; 2L2P A; 2YTX A; 5GMK h; 4ID6 A; 3EFO A; 4GS3 A; 4FW1 A; 4C92 D; 4CC3 A; 2PZW A; 4KN4 C; 4JI0 Q; 3L9A X; 2K33 A; 2BH2 A; 4K8J A; 4A3K G; 2F52 A; 1IBM Q; 4NMZ A; 2PCF B; 2RPN A; 2V1R A; 3QG1 A; 2OTL Q; 3FDT A; 3HOU B; 3D3R A; 5IT9 L; 5GAN g; 4Y7N H; 1NEG A; 4J7F A; 2GQV A; 4KY7 A; 4P71 A; 3VR4 A; 1XPR A; 1IKM A; 4UP9 A; 4DUZ Q; 1UUP A; 4ZRZ A; 4FIB A; 2VUM H; 5I6Y A; 3JRZ A; 4M75 F; 1HZ9 A; 4A3F G; 1EYD A; 1JEG A; 3FHW A; 3ME9 A; 4RAU C; 3RMQ A; 4M77 D; 3FKI G; 4WRD A; 1PC0 A; 2K79 A; 1X75 C; 1PYB A; 2HDL A; 2LX7 A; 4KUL A; 1RI9 A; 3VJP A; 3OMC A; 3X3M A; #chains in the Genus database with same CATH homology 3QJ6 A; 2OAU A; 3GTK H; 1G51 A; 2CYZ B; 2BIV A; 2MI6 A; 1QUQ B; 3ER0 A; 4ZN1 A; 4B3S Q; 2UU9 L; 2JEA I; 1G29 1; 3CF5 A; 4IFD G; 4OP0 A; 3S1M H; 3TU4 K; 4BDZ A; 3HPW A; 2R92 G; 1EQR A; 4O5V A; 1AH9 A; 3RIR A; 3JCM W; 2M1H A; 4IUT A; 3VYG A; 2ZPB B; 1HZC A; 2PUK B; 2M0O A; 3S1Q H; 2XTI A; 3BJU A; 2P0P A; 4XTU A; 4PZ6 A; 4M7D A; 4A3L G; 2LT4 A; 3BDL A; 1XQH A; 2EYQ A; 4JOI C; 4M75 C; 2QXF A; 1VQ4 A; 5E7N A; 4XQ3 A; 5ELO A; 5LJ5 h; 2QA4 A; 4LD9 K; 3BY7 A; 4HCB A; 4L5T A; 4A3G H; 3CBO A; 1YJN A; 4II1 A; 3G6E A; 2LQK A; 2K2V A; 2F4V Q; 3RES A; 2E1H A; 4UC4 A; 1YIJ A; 2B29 A; 4HIK A; 5JEA J; 4OWT B; 2AWN A; 2K75 A; 1JMC A; 1O7I A; 2ZTC A; 4LFA Q; 5M3F G; 2JVV A; 3OYF A; 4M7D D; 4LF8 L; 3LWI A; 2HT1 A; 2VY3 A; 1VQ9 A; 3H3V I; 3CPW S; 4RMO A; 3OS1 A; 3NEN A; 4M75 B; 3TQY A; 1GKH A; 1EQQ A; 5FLX E; 3HFO A; 5GAN 7; 1E7Z A; 4LW1 A; 3RKY A; 5GAN d; 2DD4 A; 3GPL A; 4DV0 L; 4YHH L; 4XU0 A; 1U0L A; 2VQE L; 2UU9 Q; 2MF2 A; 1N8R C; 4XIG S; 3TS0 A; 2CZ1 B; 4BE0 A; 4PG3 A; 2E8G A; 1TH7 A; 3UWN A; 4FT2 A; 3R6O A; 3HL8 A; 5I2M A; 3HKZ E; 2UXB L; 2YLC A; 5BR8 Q; 2WB1 G; 2DPP B; 1VQI A; 1R8O B; 4L5R C; 4A3J G; 1E22 A; 3HOY G; 4A93 H; 4LF9 L; 1LTL A; 4A3C G; 2RHU A; 3L2P A; 3J80 c; 2Q2U A; 1M90 U; 3BZC A; 1N33 Q; 4JYA L; 1XMO L; 5AYF A; 5HY6 A; 1VQ4 T; 2JEB I; 3JCM g; 3M5A A; 4CI0 C; 1QVP A; 3FIF A; 4N0A C; 2QA4 T; 2YV5 A; 3MHX A; 3V8K A; 2XDP A; 3RZD H; 4JV5 L; 1YEZ A; 1YJN T; 3DLL A; 2PNH A; 3G6E T; 1Y1V G; 4C0D C; 4M78 G; 2K5V A; 3JB9 H; 3CC2 A; 4KHP Q; 1HH2 P; 4C8Q F; 1D7Q A; 1YIJ T; 4IOC A; 2RI3 A; 2RF4 A; 4J6Y A; 5A0L A; 3OYB A; 4E7I A; 2V8I A; 4IPH A; 4X64 L; 1WGS A; 2RHZ A; 4J7I A; 1B70 B; 1VQO A; 1VQ9 T; 1FGU A; 4P74 A; 3U58 A; 2EXD A; 1UGS B; 2I2L A; 4A3E G; 3PO3 H; 5DLO A; 1VQH A; 1VUB A; 3J80 E; 4OMF B; 2L3R A; 1FVI A; 5M5Y H; 2HBP A; 2ZPE B; 4R56 A; 3UE1 A; 2Y90 A; 1P92 A; 4DV0 Q; 4YHH Q; 1BBU A; 3JCM X; 1X3G A; 3OTO L; 3X28 B; 5FRN A; 2VQE Q; 2VNU D; 5IN1 A; 3CME T; 2JJQ A; 1Y7X A; 2K5N A; 4DOW A; 2C06 A; 1X9N A; 2YTY A; 3ZFJ A; 4F7U G; 2VW9 A; 1KQ1 A; 1JE5 A; 1VIF A; 3INZ A; 2HQX A; 3CBP A; 4ZN3 A; 3QZ5 B; 2NN6 I; 2IT1 A; 5LJ3 f; 3OYD A; 4YWK A; 2UUC L; 3FDR A; 5FU6 C; 2ZJQ A; 1KHI A; 3WVD B; 4LF9 Q; 3I55 A; 3CCS A; 1IZ6 A; 4A3I G; 4C8Q C; 4HT8 A; 1XMO Q; 3M57 A; 3CMA A; 1HNX Q; 3CCJ A; 3PMT A; 2WSC E; 2A8V A; 2BH8 A; 1RIP A; 1X6O A; 4Y28 E; 1BI1 A; 3V7R A; 4HCZ A; 3TEH B; 5A0D A; 3OYN A; 1VQO T; 4YY3 L; 2VV5 A; 5FU6 B; 1XMQ L; 3OYC A; 3WXM B; 1XJV A; 4EX5 A; 1X01 A; 1XNR L; 3JCM d; 3QDR B; 4C5E A; 2IHF A; 1YZ6 A; 3K7U C; 3T1H Q; 2GF7 A; 2UUB L; 1K73 U; 1U1S A; 4GNX C; 1XTD A; 5LJ3 e; 5KE2 A; 2OCE A; 2BUD A; 3A8M B; 2E65 A; 2O4X B; 1OTC A; 1KD1 C; 4A3J H; 1PH3 A; 2RHS B; 4IUV A; 2FYK A; 3OTO Q; 5GMK l; 5CR2 A; 1I4K 1; 2XK0 A; 4IUP A; 3M56 A; 1WPY A; 2JA6 G; 2KK4 A; 4HJ5 A; 3E0E A; 3C12 A; 1Y1V H; 3I55 T; 3CCS T; 4OXP A; 5GAM f; 3OYM A; 3CMA T; 4E7J A; 2ZPG B; 5GAO m; 5C0M A; 2GVA A; 4DK6 C; 4L5S A; 2DIG A; 3C94 A; 3W1B A; 2K5H A; 4K0K L; 1K3R A; 3GO5 A; 3F2C A; 4BI8 B; 2IVW A; 2ZGW A; 2BA1 A; 4NB3 A; 1IRE B; 1I6H H; 3GTL H; 3QII A; 3MEV A; 3SB2 A; 5IY0 A; 3ZBD A; 4OO1 G; 4XTV A; 3BFM A; 1K8A U; 5KCO A; 1YIT A; 2RI5 A; 5JJK A; 3UT1 A; 4YY3 Q; 1N34 Q; 2IG0 A; 1WOC A; 4NXM Q; 4FL6 A; 4UPA A; 3RN2 A; 5C4X H; 1PA6 A; 5FJ8 G; 3U50 C; 3CPF A; 1N9S A; 4DV7 L; 4IPG A; 2N78 A; 2JZ2 A; 3HOU G; 2X35 A; 1YCY A; 4M7A E; 3H6Z A; 2UUB Q; 4DR7 L; 4HJ9 A; 1C9O A; 3GTG H; 3JAM c; 3M58 A; 5GAH C; 4IUR A; 5GAM e; 4DR2 L; 4JI7 L; 5LMU L; 2KMT A; 4DR3 Q; 4X9D A; 3L2U A; 4DR6 Q; 1FJG L; 4A3M G; 2YYZ A; 5C0X J; 3M9B A; 2E2J H; 2B63 G; 1QVF A; 3FEO A; 1OXX K; 3E9I A; 1VQM A; 4DV5 L; 5CCA A; 4OON A; 5CQY A; 4XTC S; 2HQL A; 2ZPF B; 1X3E A; 1JT8 A; 3HFZ B; 4J83 A; 3UDC A; 3HOZ G; 2ZJP A; 2L5Q A; 3OYA A; 5IP9 G; 3S3O A; 1M1F A; 1M1K U; 3SD4 A; 1TWG H; 1GM5 A; 3L2R A; 4Y52 H; 2N8N A; 4QJF A; 1NPR A; 5IYC H; 2KHI A; 3EFR A; 4PJO E; 3JAM E; 4H02 A; 3GOX A; 1NE3 A; 4UP8 A; 4PMW A; 2VUM G; 4DR4 Q; 4P73 A; 1FX7 A; 2E6N A; 1VQA A; 4UP7 A; 3L42 A; 4YCU A; 1M9S A; 4GLV B; 2RD1 A; 3W1G A; 3M53 A; 1G3W A; 5GAN h; 5GMK j; 4PZ8 A; 3JCM P; 4C8Q E; 2XGT A; 4C5I A; 1PV4 A; 4P75 A; 4HKE A; 4E7K A; 3L2Q A; 4IFD I; 3LH0 A; 2HAX A; 1I3Q H; 4DV7 Q; 1LJO A; 3OW2 S; 4E47 A; 2LCL A; 1PYS B; 2D0Q B; 2KDS A; 1LOJ A; 2K3X A; 3D31 A; 1Q81 U; 2WFW A; 3TCJ A; 4GLA C; 1SRO A; 5LL6 S; 4JI7 Q; 3VUZ A; 2XG8 D; 4JI0 L; 1UGR B; 1VQM T; 3RER A; 1IBM L; 1TWC H; 3S3M A; 3UDF A; 3PNW C; 5D6W A; 4GN4 B; 3S2H H; 2E64 A; 3OS2 A; 2RHQ B; 4DUZ L; 2HD0 A; 5GAN 5; 2OQ0 A; 3DM3 A; 2DK3 A; 3HKS A; 4BB7 A; 3IRB A; 5DY9 A; 3RLF A; 1Y07 A; 1HJP A; 1HZA A; 3K8A A; 2R57 A; 3CCM A; 4A3G G; 1NJI U; 3J81 c; 1W2B A; 3I2Z A; 3I4N H; 2KQW A; 1VQK T; 3HRU A; 4B3S L; 2DVE A; 1KIX A; 2LTO A; 1CKO A; 3MET A; 5CO7 A; 1UE1 A; 4POF A; 4HJ7 A; 3TEE A; 1SR3 A; 2JA7 G; 3DKM A; 1TWH H; 1D3B A; 5LJ5 j; 4X9C A; 1U8R A; 2NZH A; 1YI2 A; 4A3M H; 3PSI A; 1LNX A; 4C92 E; 1X55 A; 2I5L X; 4LDW A; 3LX7 A; 4JG2 A; 4MZ9 A; 3JCM Q; 3RR5 A; 3GTO H; 3KF8 B; 5DM7 R; 3S2D H; 2EJ9 A; 4L5G A; 4DR1 Q; 2MNA A; 3A8L B; 3RNU A; 2KVQ G; 4M78 A; 3J81 E; 3A5Z B; 2CZ6 B; 2M9U A; 4VUB A; 3KDF B; 4FU6 A; 4ZGJ B; 5GAG C; 2VS1 A; 4M77 E; 2BA0 A; 3UDG A; 1LYL A; 3QXE B; 1H3Z A; 3JC6 6; 3L2Z A; 4A93 G; 4XTX A; 1I5L A; 2JQO A; 2K5I A; 2PVO B; 2F4V L; 4EMG A; 3L2V A; 1BBW A; 3CCM T; 4JRI A; 3PCO B; 4QGU A; 2K52 A; 4JI3 Q; 3MEA A; 1L0W A; 1G3Y A; 2N49 A; 4LFA L; 2QEX A; 4JLI A; 4JS4 A; 2PQA A; 4M78 D; 1PH2 A; 2XD0 A; 3JB9 G; 2PVD B; 3MCA B; 1VQL A; 5FRO A; 1I94 L; 1OXV A; 4A4K A; 1ZQ1 A; 1WQW A; 2DJZ A; 3FP9 A; 4ATO A; 3JCM h; 4NL3 A; 4HA8 A; 4L59 A; 1PFS A; 4BXX H; 3D0F A; 4BDY A; 4BA1 I; 4F7U A; 3PUX A; 1FWZ A; 1Q7Y U; 2R5A A; 3A8H B; 4DNI A; 1K9M C; 1V29 B; 4WJ4 A; 3C4S A; 5BR8 L; 1YI2 T; 3PO3 G; 2ZCF B; 5GQO A; 4ELY C; 2CS7 A; 3UPU A; 2UXC L; 1S3O A; 2GFA A; 2KY9 A; 4FT4 A; 3EBE A; 2P0Q A; 3KF6 B; 4HIM A; 2K5F A; 1NY4 A; 5I21 A; 3JCM b; 4YWL A; 2PQW A; 2P0K A; 4QL5 A; 2CCZ A; 1N33 L; 3M4O H; 1M90 C; 1AHJ B; 5GAO p; 4KUI A; 1B8A A; 4AYB G; 3E19 A; 4RG8 A; 4XK8 E; 1PH4 A; 4MNO A; 4LD6 A; 1Q82 C; 2XDB A; 1N32 L; 1WI5 A; 2QTX A; 5LJ3 h; 3S14 H; 4BI3 A; 4M7D E; 4KHP L; 3N5B B; 4RG2 A; 2LX9 A; 5CK9 A; 1LM0 A; 1B34 B; 4MTN A; 4LDV A; 2CWA A; 2RCN A; 4A75 A; 4LDX A; 1JJG A; 2YHT A; 2QEX T; 4LF4 Q; 4M7A F; 4PLL A; 1JJC B; 4HIO A; 1MJE A; 1V43 A; 5GAN 8; 1VQL T; 1PVO A; 3E1S A; 4BY6 C; 1QZH A; 4KUD K; 2LNV A; 4FSX A; 4HT9 A; 2JA7 H; 1I94 Q; 3URG A; 2QGQ A; 1H64 1; 1KC8 C; 3UDX A; 2ZM6 L; 2QQS A; 3VKI A; 3S1R H; 3QKJ A; 2QDY B; 3Q8D A; 1NZ9 A; 4R1J A; 1OYX A; 5LMN W; 2JA5 H; 1SE8 A; 1V9P A; 2HHH L; 3HOX H; 2F5K A; 2XKO C; 4DR5 L; 2NZO A; 2F5T X; 2WAQ E; 4BBS G; 3GDE A; 3F1Z A; 1VQ5 A; 3PO2 H; 2UXC Q; 2AKW B; 5JJI A; 5GMK i; 1Y96 B; 3P8H A; 2PVG B; 3P8D A; 4OGE A; 3AHU A; 1BYM A; 4PJO F; 5GAN 4; 3G4S A; 3VU3 C; 3HFN A; 3L1A A; 2LCC A; 3MEW A; 5K36 G; 3HHT B; 3OY9 A; 4XU1 A; 4WJ3 M; 3PUY A; 2D62 A; 1KQS S; 4IPC A; 5H1S E; 3CC7 A; 4DV2 Q; 1HNX L; 1KRS A; 1N32 Q; 5GAD C; 4JBJ A; 1H95 A; 5A0G A; 4M7A C; 1G5V A; 3F2B A; 4R4C A; 2OWO A; 5KCH A; 4EQ5 A; 3PT9 A; 3JB9 J; 1JBM A; 1C7Y A; 3LGJ A; 4YTL A; 5IP7 H; 2PI2 A; 1N6A A; 4C3J H; 4ZTJ A; 2RHY A; 4IUQ A; 2E10 A; 3WVE B; 2X4Y A; 2CWP A; 3CCU A; 1VQF A; 3OMG A; 3CCQ A; 4OO1 I; 2ATW A; 2H3J A; 4JI2 L; 3O8V A; 1VQC A; 1E1T A; 2E12 A; 4N4G A; 3T1H L; 4M7A B; 5GMK m; 3VDY A; 1PHJ A; 4C0D B; 2D9T A; 1PH7 A; 2B8A A; 4X65 Q; 4OGC A; 5LJ3 g; 2ZM6 Q; 2Q2I A; 3PUV A; 3V8L A; 1QUQ A; 5C4X G; 2EQK A; 4KI0 A; 3M55 A; 3M85 A; 3PMI A; 3RLO A; 2NVT H; 4PJO C; 1AE3 A; 2L8D A; 3ERS X; 2HHH Q; 5GAE C; 4E7H A; 3U4V A; 2OTJ A; 1MJC A; 4DR5 Q; 3PF5 A; 3V8J A; 1E24 A; 1Q86 C; 3BZK A; 4GN5 A; 2Y0S E; 2UUA Q; 1QVF S; 4OB1 B; 4AIM A; 4POG A; 3NBH B; 1N8R U; 4A76 A; 2DKG A; 3JCM V; 4BY7 G; 3CC7 T; 5C0X G; 1VQ6 A; 2R58 A; 2N3S A; 3L7Z C; 4DV3 L; 4PJO B; 4OB0 B; 3CC2 T; 5AJ3 Q; 1E1O A; 4A3B H; 1XOV A; 3H8Z A; 5LJ5 f; 1PH9 A; 5IWA Q; 5IAY A; 1U5K A; 2WSF E; 3M54 A; 1YHQ A; 3PV0 A; 2A3L A; 1Y1W H; 1VQ8 A; 4PZ7 A; 3CCU T; 4C92 F; 5IYB H; 1N34 L; 1C0A A; 3NFC A; 4NXM L; 4GKK Q; 1X54 A; 1QP2 A; 1N9R A; 4KBM B; 3CCR A; 4IJL A; 5IYC G; 2I5M X; 4LNQ A; 4C8Q B; 2YTV A; 1IYJ B; 4JS5 A; 4N4I A; 2XHC A; 4C0G A; 3PIO A; 3MP8 A; 3CCV A; 2L15 A; 4JI2 Q; 2QI2 A; 3OB9 A; 1TXY A; 4BBS H; 2R6G A; 4GN3 B; 4A4F A; 4QQG A; 3HSB A; 3RKX A; 3I56 A; 1VQG A; 4M77 F; 5CAI A; 4R4I A; 2K14 A; 1SSF A; 1Q1E A; 5GAO n; 3BU2 A; 4EMK A; 5GAM g; 1D8L A; 2K50 A; 4DR3 L; 5CIU A; 2OTJ T; 4DR6 L; 5J3B A; 3ULL A; 2FB7 A; 1CKN A; 2JTM A; 1WJQ A; 3F8T A; 5JJL A; 1VQN A; 3CXC S; 2JK8 A; 4XTW A; 5CKD A; 1UE5 A; 5LJ5 e; 3FB9 A; 1UGQ B; 1VQ7 A; 5CVI A; 1A0I A; 3L0O A; 1L1O A; 1VQ6 T; 1K83 H; 4GNX B; 3M9Q A; 5LJ3 d; 1SLJ A; 3SWN A; 3STB C; 4JBK A; 4M75 D; 3HKZ G; 1M1K C; 1T9H A; 2X4J A; 4QVC A; 3CCJ T; 5GAN j; 4OX9 L; 3CC4 A; 3CCE A; 4C92 C; 4LUZ A; 2RI2 A; 4DV3 Q; 4RCB A; 1JJ2 A; 4DR4 L; 1KG1 A; 1YHQ T; 4JI1 L; 3MP6 A; 4A4I A; 1PH8 A; 4QQD A; 1VQ8 T; 1L2F A; 4MMK A; 4RG1 A; 1YHB A; 1AE2 A; 1Y14 B; 4Z02 A; 1WCM H; 2IX0 A; 1PH5 A; 4M77 C; 3QO3 A; 1RL2 A; 3CCV T; 3E9H A; 3I4N G; 3X25 B; 4QIW E; 3KXT A; 3I56 T; 4C92 B; 1DJ7 B; 2G3R A; 4LF6 L; 1IXR A; 2JA9 A; 1HR0 Q; 4UY8 C; 1D3B B; 1BI3 A; 2LXK A; 4M7D F; 1BIA A; 2OTL A; 4BY7 H; 2LQ8 A; 4M77 B; 2RHX A; 4DV4 Q; 4YCV A; 1VQ7 T; 3OBW A; 1QZG A; 1HNZ L; 4WF2 A; 4OWW B; 1X3F A; 2R7D A; 5C3E H; 2DXC A; 2DD5 A; 1IBL L; 3L2W A; 4B3M Q; 1HXD A; 4LUV A; 1J5E Q; 3ICE A; 2ALY B; 2NVQ H; 5CQX A; 4DV6 Q; 4BY1 H; 4JI5 L; 3CCE T; 5GAM d; 1PSF A; 4JLG A; 4OX9 Q; 3U4Z A; 1VQP A; 4LDY A; 3B6Y A; 5FRM A; 4C2M H; 2YU9 H; 2E2I H; 2E6Z A; 4KT0 E; 4JI1 Q; 3AFP A; 2EFI A; 1YIT T; 2L89 A; 2IY5 B; 4NOY A; 3JB9 I; 2LVM A; 3OQ5 A; 3H3V H; 3HOV H; 3JB9 D; 4OVA A; 2JUF A; 3KYH C; 5IT9 c; 1VS0 A; 4M7D C; 3WWV A; 2IX1 A; 3SDZ A; 2ZPI B; 3NEL A; 4BE2 A; 3A8G B; 1V1Q A; 1HR0 W; 1Q1B A; 4BXX G; 4D6W A; 5GRO A; 2C35 B; 4DA4 A; 5ELN A; 1I50 H; 2D6F A; 4LF6 Q; 5M3M H; 2UXD L; 1O9S A; 1R9T H; 1YBY A; 4DR1 L; 2CZ7 B; 3K81 C; 2R7F A; 4J6X A; 2OTL T; 1OXU A; 3I4O A; 4DKA C; 4AYB E; 4C5G A; 1TWA H; 4IO9 A; 2O4X A; 4GKJ L; 2P4T A; 4O6J A; 1X56 A; 4M75 G; 4M7D B; 3S24 A; 2ZJR A; 2X4X A; 1HNZ Q; 5HK3 A; 1FL0 A; 4BE1 A; 5IT9 E; 1YJ9 A; 1IBL Q; 3GNF B; 4JI3 L; 3ULJ A; 1WYD A; 3QZ9 B; 3F70 A; 3CW2 C; 4O2D A; 5CKF A; 4JI5 Q; 2GCX A; 4ZGD B; 3J7Z C; 2CKZ B; 1YJW A; 3M4G A; 1KAW A; 2HIV A; 2E5L Q; 4JBW A; 5EAY A; 2VGM A; 4O0A A; 3QBY A; 2DXB A; 2AWO A; 4L6V 5; 3MP1 A; 1S72 A; 4NS5 A; 1Q7Y C; 2A7Y A; 4QY7 A; 2K57 A; 2CQO A; 3H0G H; 1XPU A; 5LMN L; 3I4M H; 2MO1 A; 3MXN B; 5LJ3 j; 4HJA A; 4IOA A; 4DUY Q; 1XNI A; 2WB1 E; 1I8F A; 3EAE A; 3TRZ A; 3J7A F; 3LW5 E; 1K0R A; 3HOW H; 4C3H H; 3GP8 A; 1Q8K A; 2MFI A; 2FL7 A; 2JA5 G; 2UXD Q; 5J39 A; 2DIQ A; 4XTY A; 4IJH A; 3CQZ H; 2Q2T A; 3OSV A; 3HOX G; 5J26 A; 4X62 L; 2R93 H; 3CCL A; 3RMS A; 2ZTD A; 4EDU A; 1P16 A; 1S40 A; 1VQE A; 3SFM A; 3T1Y Q; 3P8B B; 3PO2 G; 3GNG A; 1EFW A; 1X65 A; 4GKJ Q; 2Z0S A; 1UE6 A; 4M78 E; 4M7A G; 2MMP A; 1VCI A; 1YJ9 T; 1IBK Q; 4DAY B; 4LF7 Q; 2RH2 A; 2QQB A; 2K5L A; 4LF4 L; 3NEM A; 4TVR A; 3BDU A; 2DEQ A; 1GVP A; 3X26 B; 4HCC A; 1Y71 A; 1YJW T; 1W3S A; 2JOY A; 4QQ6 A; 1HNW L; 4DOV A; 2BKD N; 2FHD A; 4A4G A; 1EIY B; 3J80 i; 1J6Q A; 1S72 T; 5IP7 G; 3RKW A; 4YCW A; 2VXF A; 1MHN A; 4ELZ C; 3GTM H; 1QP3 A; 4QVZ A; 1EIF A; 3S3N A; 4EPC A; 2DTI A; 5FU7 C; 4E7L A; 1N9W A; 1BIB A; 3G71 A; 2Q2H A; 3CBM A; 5LMN Q; 2H5X A; 5GAN f; 4PJO G; 4B3R Q; 3EN2 A; 4F7U E; 3A74 A; 1ZHI A; 3M4Q A; 3S16 H; 2Y4X A; 4C5H A; 1CUK A; 4RCC A; 5GAM j; 1CSQ A; 4J8O A; 3EIV A; 2CGH A; 2KEN A; 2MAM A; 3E9F A; 3LLR A; 3RUX A; 3LWH A; 3CCL T; 4J6W A; 1MIU A; 5M9O A; 5C6S A; 2V8K A; 2RJC A; 2VXE A; 4LDU A; 4A3D H; 1KHC A; 3OW2 A; 4R4T A; 4DV2 L; 4C8Q G; 4X62 Q; 1EOV A; 5FU7 B; 1ASZ A; 2V1C C; 1XNQ Q; 3CAM A; 5C44 H; 1EYG A; 4IPD A; 4N4H A; 3PGZ A; 2MWO A; 3V7S A; 2DGY A; 5C0X I; 4B9W A; 2NVX H; 1TWF H; 1KRT A; 5IYD G; 5FLX c; 2E2H H; 2KSS A; 1N27 A; 3G7Z A; 5CKE A; 3DLM A; 1EWI A; 4X34 A; 2E41 A; 4EMK C; 4QQB X; 4A3B G; 4YWM A; 5GAN e; 1BKB A; 1WFQ A; 1HNW Q; 2K4Y A; 3M59 A; 3OYH A; 2XI9 A; 2PMZ E; 3OS5 A; 3J81 j; 3H15 A; 1Y1W G; 4X65 L; 1OXT A; 4GOP A; 4XAX B; 5IYB G; 4HID A; 4P72 A; 1M8V A; 3OYJ A; 3G71 T; 1Q46 A; 4DPG A; 2DUD A; 1MKH A; 2DZ9 A; 2JA8 H; 2O01 E; 1L1O C; 1JB0 E; 3RN5 A; 4AH6 A; 3JCM J; 3GIB A; 3SWN C; 1PH6 A; 1WJS A; 4GLK A; 2Z6K A; 4EMK B; 4QVD A; 1QVC A; 3M3Y H; 4BY6 B; 3GF5 A; 2RJF A; 2UUA L; 5GAN 3; 2VQF L; 3T9N A; 3JB9 F; 3JSC A; 3X24 B; 1CKN B; 4C3I H; 1W4S A; 1ASY A; 1WJJ A; 1P8L A; 1K9M U; 1SFO H; 4BBR H; 4P5N A; 2MO0 A; 1M4Z A; 5I2S A; 2RHI A; 5GAO r; 2VGN A; 3CME A; 1CKM A; 4IKF A; 1YNX A; 4C0Z A; 2HNI A; 5AJ3 L; 3F2D A; 1L1O B; 3S1N H; 1G6P A; 3C95 A; 1QVG A; 2V8J A; 3SWN B; 2XIC A; 4C92 G; 4A3K H; 3G48 A; 5IWA L; 5GAM h; 3KDF A; 3E9G A; 4ZGE B; 1JJ2 S; 1I5F A; 3BW1 A; 4GKK L; 4A3F H; 1Q82 U; 2CZ0 B; 2RO0 A; 1BVS A; 3FKI H; 3PUW A; 4M77 G; 4BUG A; 4R4O A; 1B7Y B; 4XU2 A; 3LYI A; 2PI2 E; 3NTH A; 4LFC L; 3W5O A; 3KBG A; 2R7Z G; 4JRP A; 4MML A; 4MZM A; 1K73 C; 4Q5W A; 4NXN L; 3JCM f; 1WCM G; 3UE0 A; 3X20 B; 2WSE E; 2GFU A; 1BI0 A; 3HRT A; 1OZ2 A; 3OYL A; 4LF8 Q; 3FRN A; 4JUV A; 2JNG A; 1EUJ A; 4IFD J; 3AQQ A; 3S15 H; 3QJY A; 2A1A A; 4JCV E; 1VQ5 T; 1KQ2 A; 4MZT A; 2GVB A; 2LT1 A; 2E70 A; 2R92 H; 1KC8 U; 2QQR A; 1WJR A; 5GMK e; 2XDD A; 4C0F A; 2VL6 A; 2EAY A; 3EFS A; 3G4S T; 5LJ5 g; 3CD6 A; 2VQF Q; 3AYH B; 4QW2 A; 3KF6 A; 5IYD H; 5HKC A; 3KJP A; 4A3L H; 3OBY A; 2EQM A; 3FJP A; 2JE6 I; 1OI1 A; 2R5M A; 3TRE A; 3QSU A; 2UXB Q; 1LUZ A; 3K80 C; 5DLQ E; 5M5W H; 4BA2 I; 4XTZ A; 5C3E G; 3JAM i; 1NE8 A; 1VQB A; 2I5H A; 4JBM A; 2ZTE A; 3HP9 A; 4PL6 A; 1XPO A; 2XID A; 4BY1 G; 3S17 H; 2DXU A; 4MZP A; 3JCM e; 1K8A C; 1E3P A; 3QUI A; 1VQJ A; 4NL2 A; 4PNO A; 3OYK A; 1GO3 E; 5FLM G; 3CCQ T; 1B34 A; 3MO8 A; 1PH1 A; 4R4Q A; 3OA6 A; 3I7F A; 5M3F H; 5EG2 A; 2DTO A; 1N36 L; 4JV5 Q; 1VQD A; 4M7D G; 1GXI E; 3K7A H; 3M4P A; 1FXX A; 4FM4 B; 2MWP A; 4LFC Q; 1CSP A; 1HR0 L; 1I81 A; 5D6X A; 4DV1 L; 3GLX A; 4DK3 C; 4R55 A; 4JOI A; 3HOV G; 4NXN Q; 4LUO A; 4M75 A; 1A63 A; 4M78 F; 1GD7 A; 1M1G A; 3CPW A; 4X64 Q; 4X66 L; 2CKK A; 2EQS A; 4A4E A; 2IHE A; 3M7N A; 1UEB A; 5IE8 A; 3OS0 A; 1NBW A; 1Q12 A; 1HZB A; 4M75 E; 4DV4 L; 4AQY L; 3VUB A; 4XU3 A; 1KXL A; 3CD6 T; 4A4H A; 4X6A H; 3PUZ A; 5M3M G; 4BD3 A; 4L5Q A; 2EIF A; 2HQE A; 2YRV A; 1ZBX A; 1Z9F A; 2PUO B; 4B3M L; 1J5E L; 3HOY H; 2RJE A; 4J5Y A; 4A3C H; 2GN5 A; 4DV6 L; 4RMF A; 3A8O B; 2DZC A; 4TQU S; 1Y96 A; 2LDM A; 4JI6 L; 2WAQ G; 5M5X H; 3JCM c; 2DAQ A; 1Q86 U; 4B3T L; 4OF1 A; 2RK2 A; 2K3Y A; 3Q1J A; 2VUB A; 2UUC Q; 2R7Z H; 1W2B S; 4F7U F; 1KQS A; 5GAO q; 1Z47 A; 2LSS A; 5LJ5 d; 2E5Q A; 2CFM A; 3CCR T; 3PTA A; 1NPP A; 3UDI A; 2EJG A; 3OYG A; 1BI2 A; 4A53 A; 4OB3 B; 1K8G A; 2DTH A; 4M78 C; 2ZZD A; 2M16 A; 4A3E H; 3OYE A; 5HK0 A; 3HRS A; 1N36 Q; 2FVU A; 2LXJ A; 2OQK A; 2F69 A; 1RI0 A; 2KCM A; 4FDB A; 4IUU A; 2EKO A; 3FC3 A; 3H0G G; 4N0A A; 4DV1 Q; 1Q81 C; 2YLB A; 4TVA B; 3I4M G; 1XMQ Q; 4OB2 B; 4M7A A; 3OYI A; 2LC4 A; 4X66 Q; 3J81 i; 4JDS A; 4LWC A; 1XNR Q; 4NPS A; 1VQN T; 5HGQ A; 1KL9 A; 3GD7 A; 4Y91 A; 4APV A; 4AQY Q; 4M78 B; 3HOW G; 4OWX B; 1SN8 A; 3KOJ A; 4LFB L; 2D0P A; 4MDW A; 3NTK A; 1KD1 U; 3KLW A; 2ASB A; 2PU9 B; 2R93 G; 1PSE A; 2ID0 A; 5GAN 6; 2NLU A; 2DO3 A; 2NN6 G; 5CKB A; 4A3I H; 4JI4 L; 3CC4 T; 1JB7 A; 5C4J H; 2X4W A; 4M7A D; 5FLM H; 4JI6 Q; 1A8V A; 3A0J A; 2HIX A; 3DB4 A; 3OYY A; 1NJI C; 2E5L L; 4B3T Q; 2Y0S G; 4JRQ A; 1NTG A; 4ME7 A; 2PQA B; 5CKH A; 4PJO A; 3DB3 A; 1NMG A; 2WAC A; 3SGI A; 3RZO H; 3PFS A; 2RB6 A; 1YVC A; 4BAC A; 2K4K A; 4K0K Q; 4KHZ A; 2JTF A; 4F7U B; 4DUY L; 3LGL A; 5GAO l; 3TS2 A; 3PIP A; 1O75 A; 2A19 A; 2KBN A; 3QYG B; 4LF5 L; 4JI8 L; 4C8Q A; 1DGS A; 1SMX A; 2B3G A; 1VQK A; 1HK9 A; 4BI4 A; 2KCT A; 1JRI A; 3NTI A; 2I0Q A; 4A54 A; 3M4Y A; 4PJO D; 2D0O A; 2RA2 A; 4B9X A; 1WQ7 A; 4JRK A; 3FLG A; 2ES2 A; 3V7C A; 3T1Y L; 2QQA A; 2F3I A; 3QPH A; 2FXQ A; 4PKC C; 2JA6 H; 1M5Q 1; 3ZEH A; 1IBK L; 2VC8 A; 2LRQ A; 2QQ9 A; 4ME3 A; 4DR7 Q; 2AMC B; 4LFB Q; 4LF7 L; 4DQ2 A; 4C8Q D; 2VYT A; 4A3D G; 3PF4 A; 3LGF A; 1UWV A; 4HJ8 A; 4DR2 Q; 5LMU Q; 3OSS C; 3CXC A; 4GOP C; 5GAN 2; 4RKU E; 1VQP T; 1U1T A; 1QW1 A; 5C44 G; 4JI4 Q; 4GY5 A; 4KLK A; 3MEU A; 1FJG Q; 3AFQ A; 2KHJ A; 3ZJY C; 5KH6 A; 4GNX A; 3QYH B; 2DXT A; 4DV5 Q; 3QHS A; 2EWN A; 2EJF A; 3AEV A; 5KE3 A; 2Z6K C; 1VIE A; 3ULP A; 5L8R E; 3GTQ H; 2RM4 A; 3MEF A; 3A5U A; 3GTJ H; 4B3R L; 1YHA A; 4EMH A; 2MQH A; 4GOP B; 5FJ8 H; 2RK1 A; 4DAM A; 3J7A 5; 3VYH B; 3VV0 A; 5JEA G; 3S6W A; 1OZ3 A; 2ZPH B; 4JJN K; 1OXS C; 1A62 A; 1WNL A; 3HOU H; 4PLI A; 4LF5 Q; 4JYA Q; 4JI8 Q; 2Y4Y A; 2AHO B; 2EQU A; 2VZA A; 3KFU A; 2AHJ B; 1UB4 A; 5D6Y A; 3ASK A; 3PGG A; 4C92 A; 1G3T A; 2JA8 G; 2XU8 A; 4GLX A; 4V2S A; 1UE7 A; 3CEY A; 4ZTF A; 3KJO A; 2JN0 A; 1XNQ L; 2KAF A; 1IL2 A; 3K0X A; 4MDX A; 3KUF A; 1G3S A; 1UGP B; 4CRI A; 2B63 H; 2RJD A; 1MGQ A; 4M77 A; 1QVG S; 4BBR G; 1SRU A; 2WP8 J; 3RLN A; 3CP0 A; 4ALP A; 3STB D; 5A2Q E; 4JC0 A; 3HOZ H; 2EQJ A; 4J15 A; 1NMF A; 3GTP H; 5IP9 H; 2YTX A; 5GMK h; 4GS3 A; 4C92 D; 4JI0 Q; 2BH2 A; 4A3K G; 2F52 A; 1IBM Q; 3QG1 A; 5GAN g; 4Y7N H; 4J7F A; 2GQV A; 4P71 A; 1XPR A; 4UP9 A; 4DUZ Q; 2VUM H; 4FIB A; 3JRZ A; 4M75 F; 1HZ9 A; 4A3F G; 3FHW A; 3ME9 A; 3RMQ A; 4M77 D; 3FKI G; 1X75 C; 1PYB A; 4KUL A; 2NVY H; 3VJP A; 3OMC A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...