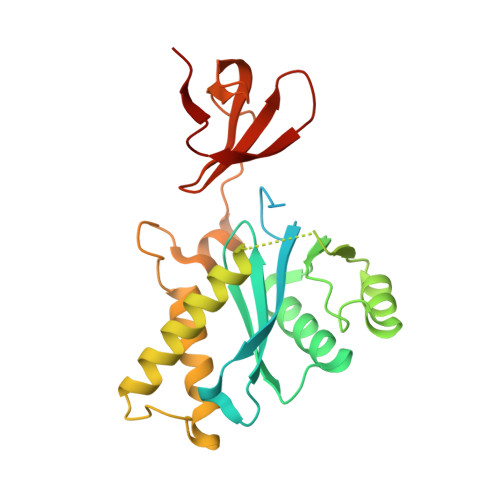
The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
69
|
sequence length |
218
|
structure length |
210
|
Chain Sequence |
PRGLGPLQIWQTDFTLEPRMAPRSWLAVTVDTASSAIVVTQHGRVTSVAVQHHWATAIAVLGRPKAIKTDNGSCFTSKSTREWLARWGIAHTTGAMVERANRLLKDKIRVLAEGDGFMKRIPTSKQGELLAKAMYALNHKERGENTKTPIQKHWRPTVLTEGPPVKIRIETGEWEKGWNVLVWGRGYAAVKNRDTDKVIWVPSRKVKPDI
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
A possible role for the asymmetric C-terminal domain dimer of Rous sarcoma virus integrase in viral DNA binding.
pubmed doi rcsb |
| molecule keywords |
Integrase
|
| molecule tags |
Dna binding protein
|
| source organism |
Rous sarcoma virus
|
| total genus |
69
|
| structure length |
210
|
| sequence length |
218
|
| chains with identical sequence |
B
|
| ec nomenclature |
ec
2.7.7.49: RNA-directed DNA polymerase. |
| pdb deposition date | 2012-06-29 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00552 | IN_DBD_C | Integrase DNA binding domain |
| A | PF00665 | rve | Integrase core domain |
| A | PF02022 | Integrase_Zn | Integrase Zinc binding domain |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Beta | Roll | SH3 type barrels. | Integrase, C-terminal domain superfamily, retroviral | ||
| Alpha Beta | 2-Layer Sandwich | Nucleotidyltransferase; domain 5 | Ribonuclease H-like superfamily/Ribonuclease H |
#chains in the Genus database with same CATH superfamily 1J9A A; 5HBM A; 1JLQ A; 4Q0B A; 2FBX A; 3VQD A; 2KTQ A; 2PZS A; 4FW2 A; 1WSE A; 4HUF A; 3HPH A; 1HQU A; 3UIQ A; 4CJK A; 4W5R A; 2IS3 A; 1Q9X A; 4QCL A; 1LAV A; 5HRP A; 4PWD A; 2Q3S A; 2Z1H A; 5HRO A; 2EX3 A; 3OS1 A; 1BI4 B; 1DTQ A; 3L8B A; 3IAY A; 2HHU A; 4FW1 A; 3OYB A; 3K5M A; 3V1R A; 2E6M A; 1X9W A; 4W5O A; 3TAG A; 2VG5 A; 4BAC A; 1TK5 A; 2B5J A; 1RNH A; 4Y1D A; 2DFF A; 5CZ2 A; 1S6P A; 4DFP A; 3RMD A; 4B08 A; 4F2R A; 4CEF A; 3TWH A; 3U6F A; 1BHL A; 1RTI A; 2OPR A; 4DQI A; 5C25 A; 1VSM A; 3CFO A; 4EZ9 A; 3CG7 A; 2IDO A; 4B9N A; 5KTQ A; 4KHQ A; 3K2P A; 3L2V A; 3EY1 A; 1TAQ A; 4CEO A; 1WSJ A; 1UA0 A; 2HHT A; 3T1A A; 1JXB A; 3QNN A; 4LSL A; 3SV3 A; 4F3T A; 3LP0 A; 2AJQ A; 2G8I A; 1S1T A; 3NH0 A; 3O4Q A; 1L3V A; 4CER A; 3TR8 A; 4LH5 A; 1XC9 A; 4OKK A; 1HPZ A; 1S1U A; 3PX0 A; 4B9S A; 1IKW A; 3LAN A; 2GV9 A; 3C95 A; 3TAQ A; 5J2P A; 1VSI A; 1FKO A; 4KTW A; 5EU7 A; 3OYH A; 2IGI A; 3DLR A; 1ZYQ A; 1NJZ A; 4L8R B; 3AO3 A; 4Z4C A; 4ELV A; 4DU3 A; 3NH2 A; 1VK0 A; 1ASV A; 4HTU A; 1S1V A; 2D0C A; 5DK5 A; 4CHZ A; 4KHW A; 3ZSX A; 2PYL A; 4CES A; 4DTR A; 3NGZ A; 1RBV A; 1W0H A; 1HMV A; 4DF8 A; 1GOC A; 3LZI A; 5SWM A; 3OYD A; 1NK0 A; 4FZY A; 1QSL A; 4OVL A; 3ZT0 A; 2G8U A; 5J2N A; 1NK8 A; 3VQ7 A; 1SKR A; 1D9D A; 3HO1 A; 2F96 A; 1TL1 A; 3HPG A; 4Y1C A; 3OYF A; 4ID5 A; 1WNS A; 4ID1 A; 3HYF A; 4Z4I A; 3VQP A; 2BAN A; 3AVL A; 3KLH A; 4BDZ A; 4DTJ A; 2PY5 A; 4CFB A; 4M3T A; 1NKC A; 1RT7 A; 3LP1 A; 3DOK A; 4OPJ A; 3LP2 A; 2DFH A; 1SV5 A; 2BE2 A; 4NYN A; 4RW7 A; 5CYQ A; 2HND A; 4CJS A; 4N41 A; 3CFR A; 1Z25 A; 1DLO A; 1C1C A; 4CJT A; 2KW4 A; 3V4I A; 2Y1J A; 3ZSZ A; 3NAE A; 4FK0 A; 4KI4 A; 4Z0U A; 4OJR A; 3D0P A; 5D7U A; 4KB0 A; 1LWF A; 3WNG A; 1VSF A; 1EX4 A; 1J53 A; 2X6S A; 3WNH A; 1TKX A; 1XWL A; 2JGU A; 1WSI A; 3U3Y A; 2YNG A; 3THV A; 1BQN A; 3L2R A; 4B9U A; 3QEP A; 1CZB A; 3MEC A; 1QTM A; 4FYD A; 1KVA A; 2HMI A; 3LWL A; 4M3Y A; 3VN5 A; 2ITG A; 1BGX T; 3LDS A; 4XIU A; 1LV5 A; 1HQE A; 4CJU A; 1UAX A; 2YV0 X; 5CYM A; 1U4B A; 3AVB A; 4C8N A; 3C94 A; 3KTQ A; 4QOZ B; 4CGJ A; 3HP9 A; 3ZT1 A; 4HUG A; 4RW4 A; 3SUQ A; 4DLG A; 1JXE A; 1C0U A; 1RIL A; 4AHV A; 4CFA A; 4M41 A; 1EET A; 3DLG A; 3HPO A; 4FJ9 A; 1ITG A; 4DQR A; 5I42 A; 1DTT A; 4GMJ B; 1NK5 A; 1W9H A; 1YT3 A; 2Z1G A; 3OYL A; 4CF9 A; 2KZM A; 1QQC A; 3HL8 A; 1LWC A; 3LAM A; 4B9M A; 2VWK A; 2A1R A; 3LPT A; 1U47 A; 3KK2 A; 3SI6 A; 1F21 A; 2A1S A; 4RW9 A; 2FBY A; 1U04 A; 3I0R A; 4DTU A; 4DFJ A; 1S5J A; 1BQM A; 1VRT A; 3AA4 A; 1RDB A; 1C1A A; 3LWM A; 4ZTJ A; 1NK7 A; 3PO5 A; 1RBT A; 1S9G A; 3AO2 A; 4CF0 A; 3V81 A; 2YHB A; 1ZBL A; 4LD0 A; 2ATQ A; 4MQ3 A; 4RW8 A; 3TAP A; 1NK9 A; 3QET A; 3OYN A; 3PX6 A; 2DFE A; 1LW0 A; 4CHP A; 5J2Q A; 2WOM A; 2GBZ A; 3AA5 X; 3QEV A; 1KVB A; 5HLF A; 1NJY A; 3MEE A; 3AVJ A; 4CGG A; 4AHU A; 1SL0 A; 4KRF A; 3MAQ A; 2BGG A; 1JL2 A; 1KFD A; 3ZSO A; 4CK1 A; 3K59 A; 1NOY A; 1A5X A; 1IKY A; 3RTV A; 4HCB A; 3AO5 A; 3ZSY A; 4R5P A; 3QNO A; 3HJF A; 5HP1 A; 4FZX C; 3U3G A; 4FJ8 A; 3V9Z A; 1U48 A; 3V1O A; 4CF8 A; 1C0T A; 4K8Z A; 3CQ8 A; 1CLQ A; 4I9L A; 3TAR A; 1NK6 A; 1IHW A; 2P5G A; 4KB1 A; 1TVR A; 3QIO A; 1RBS A; 3SPY A; 3HT3 A; 1NOZ A; 1T7P A; 4NCA A; 3B6P A; 1Y97 A; 5HRS A; 1I39 A; 1NJW A; 1HYZ A; 3ZCM A; 4E1M A; 1CXU A; 1IKV A; 3O4N A; 3OYM A; 4F2S A; 3MXM A; 4M42 A; 4OLB A; 4CGF A; 3AVF A; 5FRM A; 3RWU A; 1KVC A; 2D0A A; 3KJV A; 2HHW A; 1ASW A; 3CFP A; 2OYQ A; 1JL1 A; 1C6V A; 3NH1 A; 1B92 A; 2XHB A; 3VQ9 A; 3JYT A; 4FJK A; 4CK2 A; 4QAG A; 3AVG A; 2G8K A; 4Z4F A; 3QIN A; 2HVI A; 4YNQ A; 4OKJ A; 3L2W A; 4CJR A; 2RPI A; 1XHZ A; 4F3O A; 5D3G A; 3NF6 A; 4JRP A; 3HXM A; 4DS5 A; 3BDP A; 2O4I A; 3CYM A; 4BDP A; 3SUP A; 4M3W A; 1BIS A; 3O3G A; 4CIG A; 1N5Y A; 4J2D A; 4DTP A; 4FJG A; 4CEC A; 1TAU A; 4CJ3 A; 1BIU A; 2B6A A; 2OZM A; 1RDA A; 2VWJ A; 1HRH A; 4N76 A; 4KTQ A; 2IC3 A; 1HYS A; 1TK8 A; 3LP3 A; 4E7L A; 4Q5V A; 3VQQ A; 4OGC A; 4ELT A; 4M3U A; 4FJN A; 4NCG A; 4FLX A; 3NHG A; 3TAN A; 2FBV A; 3S9H A; 3P83 D; 1IO2 A; 2ZE2 A; 4I9Q A; 2HHX A; 4FK2 A; 4WE1 A; 3NE6 A; 3L2Q A; 3SYZ A; 3HM9 A; 1X9S A; 4M3Z A; 3ZSR A; 1RT2 A; 4FZZ A; 2RF2 A; 2B4J A; 4CJW A; 4W5T A; 3V6D A; 1NJX A; 2ZQB A; 4GVM A; 5K14 A; 2OPS A; 3SJJ A; 1RTH A; 1S9E A; 4HUE A; 4CJQ A; 4FXD A; 1WLJ A; 4O44 A; 4CFD A; 3OYA A; 4FJM A; 2ZD1 A; 4E19 A; 1B9D A; 4Z4E A; 4Z4G A; 4CHY A; 3TAE A; 3RMC A; 4RW6 A; 4FVM A; 1L3U A; 1HYV A; 1C1B A; 1C0M A; 1VSK A; 3DYA A; 5CY4 A; 4CEA A; 4O4G A; 4OGE A; 1UA1 A; 2X6N A; 4IBN A; 3AVI A; 1TGO A; 1ZBU A; 4GW6 A; 5KRS A; 5KGW A; 4JRQ A; 2P51 A; 1YVU A; 3VQC A; 1TK0 A; 3RR8 A; 1RT1 A; 3K5O A; 3OJU A; 1VSD A; 1UWB A; 3SUN A; 4N5S A; 4B8A B; 3KD1 E; 4EZ6 A; 1BCO A; 2VG7 A; 3NGI A; 4DSI A; 4E89 A; 1D5A A; 1RTJ A; 3OVN A; 5FRO A; 3S3M A; 2DTU A; 1A5V A; 3F73 A; 1LAW A; 1NK4 A; 3AV9 A; 4JS5 A; 1QSY A; 4HCC A; 4FK4 A; 2Y1I A; 5KI6 A; 1O1W A; 4CJP A; 1FKP A; 4CEQ A; 4HHT A; 4DQS A; 4CIF A; 4NCB A; 4C8K A; 3AA2 A; 1RT6 A; 3D45 A; 4DSK A; 1CXQ A; 4CK3 A; 4O0I A; 4CED A; 1YTA A; 3OYE A; 4DSE A; 3ITH A; 2YHA A; 3QER A; 4DTO A; 3VQA A; 1T05 A; 1RCH A; 3V9W A; 3SV4 A; 4J2A A; 2I5J A; 3HVR A; 1TKD A; 3M8R A; 1HJR A; 1KLN A; 3ULD A; 3QEX A; 4KO0 A; 5C24 A; 1D8Y A; 5J1E A; 3RMB A; 3RR7 A; 3MXJ A; 1SUQ A; 3NF9 A; 4FCY A; 3TI0 A; 4FJX A; 2FC0 A; 2G8H A; 1REV A; 1IKX A; 3OJS A; 3ZT2 A; 4FLZ A; 1S6Q A; 1K6Y A; 2YKN A; 1EP4 A; 3RMA A; 3DLB A; 4K8X A; 4PUO A; 3PY8 A; 4AHT A; 4J2B A; 4FJ5 A; 2XY5 A; 3E01 A; 4ELU A; 3KKR A; 4CHN A; 1QE1 A; 4AHC A; 4AH9 A; 3T3F A; 4DFK A; 2OPQ A; 3PX4 A; 3DRS A; 3RRG A; 4FM1 A; 2RN2 A; 3ISN C; 3G0Z A; 4B9L A; 1ASU A; 3EZ5 A; 5JS1 A; 3KIO A; 1RBR A; 1IHV A; 2X78 A; 3QO9 A; 2WON A; 4E1N A; 3DM2 A; 3SPZ A; 3HP6 A; 4FJI A; 1I3A A; 5KGX A; 1JLC A; 3AVH A; 1HNV A; 3MEG A; 2KFN A; 1JLG A; 4PY5 A; 4OLA A; 4B9V A; 3H08 A; 1WAJ A; 4NYF A; 2P5O A; 3HST B; 1JLE A; 4IKF A; 3S3N A; 1RTD A; 3FFI A; 3VQ4 A; 1TKT A; 3B6O A; 1U45 A; 1JLA A; 3KLE A; 3AVC A; 3QEI A; 5FRN A; 4BWM A; 2HNZ A; 3V1Q A; 3HVT A; 1IH7 A; 3SQ0 A; 5FDL A; 3SNN A; 2OA8 A; 4H4M A; 3DLK A; 3KLI A; 4HVJ A; 4KFB A; 4DTN A; 1T8E A; 4DG1 A; 4O55 A; 4DSL A; 2HW3 A; 4CEB A; 4O5B A; 3KLF A; 1R0A A; 1GOB A; 3EYZ A; 1JKH A; 4CHQ A; 3KK3 A; 1BI4 A; 3K5L A; 2G8F A; 3HK2 A; 4F4K A; 1Q8I A; 3VQ8 A; 4DQQ A; 4KI6 A; 3OS0 A; 4I2Q A; 4TSX A; 4J2E A; 4ICL A; 1FXX A; 4DLE A; 5CZ1 A; 4KHU A; 4LH4 A; 4B3O A; 3PUF A; 3OS2 A; 3K57 A; 1SKS A; 2QKB A; 3MXI A; 4KV8 A; 3TAB A; 4JS4 A; 1HNI A; 4RG8 A; 1NKB A; 4I7F A; 2XO7 A; 1RDD A; 4FLT A; 1BCM A; 1QS4 A; 3I8D A; 1ZBF A; 1D9F A; 2F8S A; 4I2P A; 4B8C A; 2LSN A; 2W42 A; 2D0B A; 1L3T A; 3MED A; 1Q9Y A; 4E3S A; 4KHS A; 1WSG A; 4FJ7 A; 1KCF A; 4CEE A; 4DU1 A; 3M8Q A; 5I3U A; 4DSJ A; 2DY4 A; 4ZHR A; 1CZ9 A; 4W5Q A; 1S1W A; 4DTM A; 2IOC A; 3VA0 A; 4E0D A; 4Z4H A; 4KTZ A; 3OYG A; 2KZZ A; 3C6T A; 1WN7 A; 3IS9 A; 3PV8 A; 2EHG A; 3F9K A; 3AA3 A; 4IDK A; 4CE9 A; 4FJH A; 5T7B A; 1NKE A; 4CFC A; 3I0S A; 2E4L A; 3NF8 A; 3V9S A; 2X74 A; 4BE0 A; 4FJJ A; 4N47 A; 5KRT A; 3NDK A; 4G1Q A; 3SQ2 A; 3ZSQ A; 2PYJ A; 4UQG A; 4W5N A; 5C42 A; 4IG0 A; 2YNF A; 1TV6 A; 4OPK A; 3V9U A; 3AVM A; 2QXF A; 4CGH A; 5E41 A; 1KRP A; 1IG9 A; 2HNY A; 3G10 A; 3OYJ A; 1VSH A; 4PA1 A; 4CJE A; 4FM0 A; 1EKE A; 3P1G A; 4B9T A; 4DWI A; 2OPP A; 1X1P A; 3CM5 A; 1KLM A; 4CCH A; 1SL2 A; 3ALY A; 4IG3 A; 1TL3 A; 3KLG A; 4YFU A; 4H8K A; 4FM2 A; 4BE2 A; 4FLU A; 1XI1 A; 1N6Q A; 3QES A; 4KHN A; 1QHT A; 1A5W A; 4CEZ A; 2QK9 A; 1WSH A; 3DOL A; 1ZBI A; 4EP4 A; 1X9M A; 4ZTF A; 4FLW A; 2NUB A; 3LAK A; 4C8L A; 3M8S A; 3QIP A; 4F8R A; 2R7Y A; 3VA3 A; 2RKI A; 3DMJ A; 3OYK A; 3DLH A; 3AVN A; 3NF7 A; 3ZT4 A; 2P1J A; 4AHS A; 3NFA A; 4DFM A; 2QKK A; 1BL3 A; 1ZBH A; 4KAZ A; 1L3S A; 3IRX A; 3KKS A; 4CJL A; 3VQ5 A; 1TKZ A; 1VRU A; 1Z26 A; 2ETJ A; 3ASM A; 3M8P A; 2D5R A; 3DRR A; 3WNF A; 5DMR A; 2BDP A; 2HHS A; 1EXQ A; 4LY7 A; 1J54 A; 2F8T A; 1VSE A; 3SQ4 A; 2IAJ A; 3VQ6 A; 1B9F A; 3K58 A; 4BWJ A; 4KRE A; 3SQ1 A; 3QLH A; 4DQP A; 3A2F A; 2XY8 A; 3WNE A; 4IFV A; 3DLE A; 3V9X A; 1J5O A; 4DS4 A; 4CHO A; 2XRI A; 3O3H A; 1RBU A; 3ZT3 A; 2YNH A; 1RT3 A; 1GOA A; 1VSL A; 1RT5 A; 3AO1 A; 3L2U A; 2VG6 A; 1JLB A; 2XY6 A; 3AO4 A; 3AVK A; 3DRP A; 3NCI A; 1YTU A; 3NBP A; 1KSP A; 1RT4 A; 1XHX A; 4N56 A; 2HB5 A; 2Z1I A; 3SUO A; 2JLE A; 4AIL C; 3NGY A; 4FLY A; 1RDC A; 4EP5 A; 3OYI A; 3QEW A; 4CGD A; 1QMC A; 1WAF A; 1L5U A; 1WSF A; 5HRN A; 4PQU A; 5HRR A; 3CM6 A; 3PO4 A; 4M3R A; 1QSS A; 1UOC A; 3P5J A; 2G8V A; 1LW2 A; 1S1X A; 3KD5 E; 4MFB A; 3O3F A; 2HVH A; 2Z1J A; 4CJ4 A; 3OY9 A; 3ZSW A; 1SKW A; 4M45 A; 4CGI A; 4KHY A; 2FBT A; 2HHV A; 3AVA A; 4CJV A; 5SZT A; 3DI6 A; 4O0J A; 3T19 A; 3K5N A; 4DF4 A; 3VQB A; 3L3U A; 5JS2 A; 4AHR A; 3RRH A; 4CJ5 A; 4OKE A; 4DSF A; 4KPY A; 1VSJ A; 4DMN A; 2E6L A; 4E7H A; 3IG1 A; 4E7K A; 4DTX A; 3TAF A; 3OYC A; 4CJF A; 4CF1 A; 1SL1 A; 4DTS A; 1KFS A; 2YKM A; 3S3O A; 4KKO A; 4FLV A; 2YNI A; 3LZJ A; 1C6V X; 2KFZ A; 3SCX A; 3BGR A; 1T03 A; 1G15 A; 4C8O A; 2OZS A; 2KQ2 A; 3LPU A; 2O4G A; 4DU4 A; 1LWE A; 4IFY A; 2XY7 A; 4BDY A; 4BE1 A; 1FK9 A; 1KTQ A; 1MU2 A; 3C6U A; 3TAM A; 3VQE A; 2HHQ A; 4E7I A; 4E7J A; 2GUI A; 1JLF A; 4C8M A; 3JSM A; 4H4O A; 4CF2 A; 4FJL A; 4M3X A; 5J2M A; 2G8W A; 4LSN A; 1BIZ A; 4JLH A; 3SZ2 A; 3ZSV A; 3L3V A; 4CIE A; 3KK1 A; 1U49 A; 3LAL A; #chains in the Genus database with same CATH topology 1K8A C; 5HBM A; 2GWJ A; 2A3Z A; 4BR2 A; 2FBX A; 4WYB A; 3EHQ A; 1GL5 A; 2ZJQ L; 4IOC L; 2A3L A; 4ME7 A; 1M30 A; 1Q9X A; 5HRP A; 1EO1 A; 1GLF O; 2IIM A; 4CZI A; 1XMO K; 4C92 F; 3QIC A; 5FPN A; 5BN8 A; 1YIT N; 1X2Q A; 3V1R A; 2RPN A; 1K73 R; 4W5O A; 4RWT A; 3ENH A; 1N6A A; 5LMU K; 3PVL A; 3CJ9 A; 3U6F A; 1J6Z A; 5FU7 B; 1RTI A; 1VQ8 N; 4DQI A; 1TH7 A; 4DV7 K; 4QVC A; 3HZ6 A; 4DV5 K; 3CFO A; 4BI8 B; 2RNA A; 5GMK h; 2COW A; 2GNC A; 1P90 A; 2IDO A; 2EPD A; 4FOE A; 5AQK A; 3EY1 A; 1NGG A; 3FJP A; 1NYG A; 3MJX A; 4CD3 A; 2PTK A; 3LDQ A; 1QCO A; 1D1B A; 2AT1 B; 3Q7C A; 4NLB A; 2KT1 A; 3B8A X; 3BF1 A; 3MWP A; 4LSL A; 3SV3 A; 2NUP B; 5I9E B; 2G8I A; 4DQ2 A; 1S1T A; 2MAM A; 1OH8 A; 1HYO A; 3D2E A; 3CCQ Q; 2DXT A; 5KYN A; 1HPZ A; 2WUS A; 1KK7 A; 4AFZ C; 1G3W A; 4KTW A; 4M7A C; 3OYH A; 4WFB L; 5IXF A; 1AWX A; 5GAM f; 1VK0 A; 2O8B B; 4DQ8 A; 2HL5 C; 3KWI A; 4KHW A; 4V2S A; 4NB4 A; 1HKB A; 4E1J A; 3HTR A; 4JI8 K; 3NGZ A; 9ATC B; 1QZ6 A; 2QWL A; 3AGK A; 1HMV A; 3EG2 A; 3ULR B; 1GOC A; 4DF8 A; 3G6E Q; 3OYD A; 5SWM A; 4YHH K; 1B3Q A; 3CCJ A; 3ZT0 A; 5J2N A; 5IH2 A; 5M1P A; 3VQ7 A; 3HO1 A; 4IZ9 A; 2MWO A; 5FLX O; 3OYF A; 3CI5 A; 4BDZ A; 3QDF A; 1FMW A; 2JM8 A; 5AQW A; 3VQP A; 4CFB A; 5KPT A; 2X3X D; 3IDH A; 3LP1 A; 1M27 C; 2LVM A; 4OPJ A; 2DLP A; 3J81 O; 2DFH A; 1HJO A; 2W41 A; 1RAA B; 5CYQ A; 3H9N A; 3DLM A; 3RER A; 4RTF D; 3V4I A; 3NAE A; 1TYQ A; 5GAM j; 3ZBD A; 5D7U A; 3GE1 A; 4M7Y A; 2X4J A; 1ZSG A; 1J53 A; 2YWW A; 3PVQ A; 1XWL A; 3IUC A; 2Q1N A; 2JGU A; 2FZC B; 2YNG A; 3OQ5 A; 3L2R A; 4B9U A; 1C0F A; 1QTM A; 3CCV A; 4FYD A; 1VQK Q; 1YJ9 N; 3VES A; 1OX8 A; 3DWL A; 3LWL A; 4CW7 B; 4M3Y A; 1JCE A; 2B8A A; 2PNI A; 3LDS A; 4DR7 K; 2QQR A; 2E5K A; 3H41 A; 1VQN N; 3ENB A; 1HQE A; 1KQ2 A; 1HJD A; 3BUZ B; 4C92 E; 5CYM A; 2QWN A; 5AQU A; 3C94 A; 4DBQ A; 3SBT A; 4QOZ B; 2P9U B; 2DVE A; 4HVW A; 1OH5 A; 4JI0 K; 4RW4 A; 2MIO A; 2LQW A; 1JXE A; 1RIL A; 1QQN A; 4CFA A; 1VQM T; 1ZXO A; 4J9B A; 3DLG A; 1YFN A; 3A5O C; 4L5G A; 2IVP A; 3G71 N; 3FEO A; 3E9L A; 4HVV A; 3VYG A; 3VEV A; 2A1R A; 3CJ7 A; 5KYU B; 3ID8 A; 3WXI A; 3MP6 A; 3SI6 A; 3KUF A; 1U47 A; 1I94 K; 2A1S A; 4RW9 A; 3OS5 A; 2QQ9 A; 3WVE B; 1YIT Q; 1MWK A; 1VRT A; 4BRC A; 1C1A A; 4ZTJ A; 2F2V A; 1SHF A; 1RBT A; 1UDL A; 4QT7 A; 3GNF B; 2YHB A; 3EG9 A; 5H1S Q; 1CZA N; 4MQ3 A; 5M1K A; 4PK4 A; 1LPL A; 3TAP A; 2EW3 A; 1NK9 A; 2CP2 A; 5C2F A; 3PX6 A; 1VQ8 Q; 2DFE A; 3I9Q A; 4CHP A; 4LD6 A; 4DV1 K; 5C12 A; 3QEV A; 3DB3 A; 5LJ5 g; 4BG9 A; 2QEX N; 1SHG A; 1H3Z A; 1M2O A; 2YLC A; 5C15 A; 2VGM A; 1JL2 A; 4CK1 A; 3MAQ A; 1YIJ A; 3K59 A; 1QVG S; 1WX6 A; 3UKR A; 2OAU A; 4CC2 A; 1KQS P; 4FZX C; 2O8D A; 3CCL N; 3V7S A; 4J9D A; 3V1O A; 4CF8 A; 4MZM A; 2RHU A; 4K8Z A; 5CQY A; 2V51 B; 4IXC A; 1M8M A; 1NWK A; 2EQM A; 5IN1 A; 2RJE A; 1WNL A; 3CQ8 A; 2CZ7 B; 3QPH A; 1JJ2 A; 5CR4 A; 1NK6 A; 1IHW A; 4KB1 A; 3QIO A; 1T7P A; 4M7X A; 2BZ8 A; 5KQD A; 1WJS A; 1RGI A; 4OMN A; 1VQ9 A; 1HYZ A; 4E1M A; 4M42 A; 3VEZ A; 1F1B B; 4OLB A; 2P4R A; 2JNG A; 2CKK A; 3TU5 A; 1AHJ B; 4DR1 K; 2D0A A; 1ASW A; 1BDG A; 4KUI A; 1JL1 A; 1C6V A; 2YRD A; 4FJK A; 5AT1 B; 1HK9 A; 3JYT A; 4BR9 A; 4CK2 A; 2IG0 A; 4QAG A; 2KE9 A; 1TUU A; 2G8K A; 2BZX A; 4CBX A; 2V8K A; 2NLX A; 1VA7 A; 1Q82 O; 3INZ A; 4RCC A; 3L2W A; 1XHZ A; 3E1Y A; 5KYY B; 5AR0 A; 4XTU A; 3NF6 A; 4BY9 C; 1NPR A; 4BDP A; 2NUP A; 3JCM g; 3ENO A; 3O3G A; 1U06 A; 4FJG A; 4BC3 A; 5M9O A; 1RDA A; 2IC3 A; 1PKT A; 4ATO A; 2VCP A; 4OGC A; 5LMN K; 4FJN A; 4NCG A; 1AVZ C; 4MMK A; 3NHG A; 3TAN A; 2ZE2 A; 2AZS A; 1PRM C; 4QVD A; 1FW6 A; 1DJ7 B; 3SYZ A; 3VGK A; 4M3Z A; 1RT2 A; 5AJ3 K; 1YJ9 Q; 3MMV A; 3LW5 E; 5C2D A; 2VXF A; 5KYX B; 5K14 A; 4BB7 A; 1JJ2 M; 1S9E A; 4LDU A; 4F7U E; 4CJQ A; 4HUE A; 1VQN Q; 4O44 A; 2HDA A; 1VQ5 A; 4NLC A; 2G6F X; 5EX5 A; 4Z4E A; 2YUQ A; 3I56 Q; 3RMC A; 2QWM A; 4HTL A; 4RW6 A; 4JK7 A; 1C1B A; 3CJA A; 4JJC A; 3QEP A; 4CEA A; 4QKU A; 4YY3 K; 2P9P B; 2DIQ A; 2RHX A; 3EGX B; 4DEY A; 1YVU A; 3RR8 A; 1EWQ A; 3G71 Q; 3T1A A; 2KT8 A; 2F5K A; 4BI4 A; 1UWB A; 3HRG A; 3CPW P; 1K0X A; 4F17 A; 1BCO A; 1YI2 N; 4DSI A; 4BY6 C; 3E9P A; 4JKH A; 2LQN A; 3QBX A; 3LM9 A; 2EKO A; 2JMA A; 3F73 A; 3LWI A; 3G4S N; 3NTK A; 4JS5 A; 3LM2 A; 1NK4 A; 5IHK A; 1M1K C; 2EGA A; 2QWR A; 1QSY A; 4FK4 A; 4XH5 A; 1K8A R; 1O1W A; 1FKP A; 3EG0 A; 1QVG A; 3C0B A; 4CEQ A; 4C8K A; 4OO1 J; 1NJI O; 2JS0 A; 4H5T A; 4DSK A; 2QTX A; 1U6Z A; 1CKA A; 3OYY A; 2QEX Q; 4O6J A; 3VS7 A; 5LY3 A; 2K57 A; 5LJ5 h; 1NM7 A; 1BYM A; 5GMK i; 5GAO l; 5CHI A; 1RCH A; 3DLL A; 3VS2 A; 3MEA A; 4DR6 K; 5HL7 L; 1WXB A; 3R6O A; 5A0L A; 4J2A A; 1GFC A; 3CD6 A; 1TUD A; 3CCL Q; 4FCY A; 1BA0 A; 1SUQ A; 1TUY A; 2JXB A; 3MXJ A; 2FC0 A; 3TI0 A; 1YHQ N; 3OJS A; 4FLZ A; 2JUF A; 3GPL A; 3CCS N; 3QFU A; 3DLB A; 2DFU A; 4XTX A; 2DO3 A; 4PUO A; 3PY8 A; 2VYP A; 4AHT A; 3TSQ A; 4ILJ A; 1RAG B; 3O9R A; 1QE1 A; 4AHC A; 4Z88 A; 4FM1 A; 2RH2 A; 3I5R A; 2LTO A; 3ISN C; 3G0Z A; 3EZ5 A; 3KIO A; 2A40 A; 1AEY A; 3CC2 T; 4FJI A; 1I3A A; 2YUO A; 2O8E B; 1HNV A; 3MEG A; 2E88 A; 4ZN1 A; 4B9V A; 1RI0 A; 1WAJ A; 2O2O A; 5GAN 5; 1ZX6 A; 4N0A C; 2DVJ A; 1YP5 A; 3CCR T; 1OEB A; 2FRW A; 3X20 B; 4HT9 A; 3F5T A; 3P4I A; 3JCM d; 1NG9 A; 4AMC A; 4LF6 K; 4C0F A; 3M4Y A; 1IH7 A; 3V1Q A; 3SNN A; 4M7A G; 2KBT A; 3DLK A; 4FPA A; 4RG2 A; 4KFB A; 1OYX A; 1I5L A; 1T8E A; 4DSL A; 1ZSZ B; 2DL8 A; 4CEB A; 4M7A A; 1VQ7 N; 3KK3 A; 5FWC A; 3X24 B; 1O13 A; 3SAG A; 2FVU A; 2K79 A; 3OS0 A; 4TSX A; 1MMD A; 4KHU A; 4DLE A; 4IOX A; 4C0G A; 2KXD A; 5AFU A; 3PUF A; 3OS2 A; 5IWA K; 4WF9 L; 3K57 A; 4M77 G; 3MXI A; 1VQ6 N; 2JW4 A; 3VEO A; 4JI6 K; 1N8R O; 4HKE A; 1FJG K; 2LZ6 B; 2KLA A; 1RDD A; 4XH4 A; 4M77 A; 1QS4 A; 1SKU B; 1ZBF A; 1OVY A; 1WPY A; 4A62 A; 2W42 A; 5KE2 A; 1L3T A; 3TVT A; 1YJW T; 1BA1 A; 1SS6 A; 3R3L A; 1IO6 A; 4EHT A; 5A2Q O; 4EMG A; 1KAX A; 1HNW K; 4CEE A; 4DSJ A; 4ZHR A; 4C5H A; 4W5Q A; 2H3G X; 3VV0 A; 2W0Z A; 5GMK e; 1YUW A; 3VA0 A; 4Z4H A; 4KTZ A; 5KPR A; 2YRC A; 5KYY A; 1T3S A; 1DEJ A; 2FZK B; 3FC3 A; 3EL2 A; 1ATR A; 2GTD A; 3DKM A; 3IS9 A; 5AQP A; 3EKU A; 3U8X A; 4B1X B; 1UHF A; 2F69 A; 1HR0 K; 2KR3 A; 2FG0 A; 3SB2 A; 1QP2 A; 4H0X B; 4M7A F; 4LFA K; 2QM1 A; 1YI2 Q; 3NDK A; 4G1Q A; 2XID A; 3S6W A; 4JKC A; 4CZJ A; 1U2V B; 3VGM A; 3V9U A; 5GAN g; 3G4S Q; 4PJN A; 2QWQ A; 2C0T A; 2HNY A; 3OYJ A; 3QB0 A; 4FM0 A; 1EKE A; 4FL6 A; 2E70 A; 5CO7 A; 3DXK A; 4FWK A; 4M77 F; 1X1P A; 3CM5 A; 5KYX A; 1WI7 A; 2L0A A; 3H0H A; 4XU1 A; 4IG3 A; 3CME N; 4M78 C; 4AFS C; 1TL3 A; 3M5R A; 4H8K A; 2YLB A; 4RTU A; 4MZT A; 4AQY K; 1N6Q A; 4YOV A; 3QES A; 4FU6 A; 1QHT A; 1PKS A; 3JCM X; 2JTE A; 4CEZ A; 4KHN A; 4WCI A; 2RHZ A; 1LOT B; 1WSH A; 7AT1 B; 1MDU B; 4OPH A; 3PIO A; 1X9M A; 4FLW A; 2AIR B; 2OTJ N; 2HBM A; 3LAK A; 3QIP A; 3CBO A; 2RE2 A; 3VA3 A; 2A37 A; 2KRN A; 1YHQ Q; 2NRW A; 1RAI B; 2BE7 D; 4AHS A; 2WTU A; 1P92 A; 4E7S A; 1BL3 A; 2NRT A; 5AQO A; 4BRK A; 3KKS A; 3CCS Q; 2JWK A; 3GP8 A; 3G6E A; 3M0U A; 3M8P A; 2ETJ A; 3ASM A; 3JCM P; 3DRR A; 3O9U A; 3N5B B; 4IUU A; 2I0N A; 4PL8 A; 5DMR A; 2HHS A; 5JVH L; 2XK0 A; 5DM7 L; 3BE3 A; 4BR7 A; 5ELZ A; 2JOY A; 4KRE A; 2DF6 A; 4BWJ A; 3VRY A; 1KJW A; 2J6O A; 3A5L C; 3MHX A; 2XY8 A; 1Q7Y O; 5FU6 C; 2KYB A; 3BZC A; 3V9X A; 3PLA A; 4ZN3 A; 1J5O A; 4L6V 5; 2MI6 A; 5AQR A; 4AFQ C; 4JEP A; 3ULE B; 3SAH A; 4DS4 A; 4OP0 A; 4CHO A; 1Y71 A; 1RBU A; 3L1A A; 1GCQ C; 1VSL A; 4M75 C; 4Z8A A; 2YNH A; 2ZPI B; 4IEI A; 2HL3 A; 2GF7 A; 5D6W A; 1XHX A; 2D0P A; 2HB5 A; 2Z1I A; 3VF6 A; 2NRH A; 1E3M A; 2DL3 A; 2LQK A; 3CC4 T; 2H8H A; 3NMU A; 1H64 1; 5EY4 A; 2GGR A; 3QEW A; 1WAF A; 4ZNX A; 3LX7 A; 3T7Z A; 3PO4 A; 4M3R A; 1QSS A; 3CCE N; 3P5J A; 2G8V A; 5DY9 A; 4E2F B; 1TWB A; 3QZ9 B; 1VQK A; 3QYG B; 1UHC A; 1VQ7 Q; 2ZGZ A; 4MO4 A; 2Q31 A; 3DXM A; 4MFB A; 5GAN 7; 1QVP A; 3BY7 A; 4DV0 K; 3PE0 A; 5B8H A; 1H8K A; 2P0K A; 4IIM A; 3AVA A; 2CS7 A; 4E7Z A; 4CJV A; 2KQW A; 1WJQ A; 4LFB K; 4KUD K; 1VQ6 Q; 1NGD A; 1U8R A; 3VQB A; 3RRH A; 4OKE A; 1K9A A; 1OPL A; 2DMO A; 3E9F A; 4JKG B; 4E7H A; 2DJZ A; 4DBF A; 3JB9 I; 2P9K A; 4H03 B; 4DTS A; 1IBK K; 2LJ0 A; 2HKQ B; 4KKO A; 4FLV A; 2O8E A; 3LZJ A; 1C6V X; 2ZCF B; 3ZYY X; 1JJ2 P; 4M7A E; 3WXL A; 3DAW A; 2KQ2 A; 2HST A; 2RF0 A; 3SWN B; 1PC0 A; 4KH4 A; 4JKE A; 4DUY K; 1FK9 A; 5FWB A; 2P9N B; 1YIT A; 3JSM A; 2D6F A; 4C8M A; 4CF2 A; 3TTC A; 5J2M A; 1UJY A; 4F9O A; 1Z9Z A; 3F2K A; 3L3V A; 3C7N B; 1WHJ A; 1U49 A; 1J9A A; 2RHY A; 1JLQ A; 5FPM A; 2EFI A; 3M3Z A; 2LCS A; 2KTQ A; 1VQ8 A; 3M9Q A; 1NU0 A; 4FW2 A; 4HUF A; 1SAZ A; 2OJ2 A; 3HPH A; 1KD1 O; 5GMK l; 1YJN N; 4QCL A; 1LAV A; 4PWD A; 4RTZ A; 2Z1H A; 2QA4 Q; 3LDO A; 2EX3 A; 4JLI A; 1BI4 B; 3VEW A; 3IAY A; 4H5R A; 2HHU A; 2QKK A; 3QJ6 A; 4DA4 A; 1XOV A; 4K41 A; 3R6N A; 2UUB K; 1Y64 A; 4BAC A; 2VG5 A; 2B5J A; 2F9T A; 5GAN h; 5BRF A; 2Z0W A; 2HBL A; 2E4H A; 5CZ2 A; 2CP6 A; 2KI7 A; 1S6P A; 4F2R A; 4PJJ A; 4CI0 C; 3N4P A; 3J8A A; 1ZHI A; 3CME Q; 4CEF A; 1WQ7 A; 2HKN A; 5C25 A; 3EFS A; 2R58 A; 3SK3 A; 4C8Q B; 1K8A O; 3CG7 A; 5LJ3 f; 2I7N A; 4FM4 B; 4H5V A; 2HSP A; 2DBM A; 3NTI A; 3UAT A; 4C8Q D; 1WSJ A; 4JZ4 A; 2OTJ Q; 1ZBJ A; 4BD3 A; 1M1K R; 4LUD A; 1JXB A; 4CZK A; 3RKX A; 3CPE A; 4F3T A; 1QLY A; 1S72 N; 1L3V A; 4CER A; 1E6G A; 3TR8 A; 2ASO A; 2YHX A; 3K0S A; 1Y7X A; 2HSE B; 1PG5 B; 1S1U A; 4DBR A; 2ASP A; 4BRF A; 2LRQ A; 1IKW A; 4B9S A; 3HOT A; 4GKJ K; 3HTV A; 1FKO A; 5EU7 A; 4JKA A; 2IGI A; 3DLR A; 1QCN A; 1ZYQ A; 2MCN A; 4B9X A; 8AT1 B; 2YUN A; 3AGR A; 1QWE A; 3TTF A; 4CZM A; 5GAE P; 1HXD A; 2UXC K; 1ASV A; 2UXB K; 3EE8 A; 2X4W A; 2D0C A; 2O9S A; 2PYL A; 1JXM A; 3MWT A; 1RBV A; 3MO8 A; 5FU7 C; 3VS1 A; 1W0H A; 5AZI B; 4XU0 A; 3O5Z A; 3LZI A; 3N4Q A; 2WSC E; 2G8U A; 2XHC A; 2E1Y A; 1SKR A; 1TL1 A; 3HPG A; 4ID5 A; 4ID1 A; 3AAQ A; 2NRV A; 1KFZ A; 3HYF A; 3AVL A; 3KLH A; 3MP1 A; 3D6R A; 3OBY A; 1VQO N; 1QVG P; 3CCE Q; 3A8O B; 3CC7 Q; 1G99 A; 2CH5 A; 3L4Q A; 4LF4 K; 1YJ9 A; 3MKD A; 4NYN A; 4RW7 A; 1QQJ A; 1KC8 U; 2HND A; 3CFR A; 2ZPB B; 1Q81 R; 4CJT A; 1C1C A; 2KW4 A; 1NLO C; 3GF5 A; 3O9T A; 2Y1J A; 1VQN A; 4CJ7 A; 3D0P A; 2V52 B; 4F14 A; 1I8F A; 3I56 A; 3M54 A; 5AZJ B; 2A5X A; 4X9D A; 2ENM A; 1BU1 A; 1WB9 A; 3OA6 A; 3VMF B; 2Q2R A; 3U3Y A; 2QW9 A; 2YX6 A; 3EH2 A; 4DR2 K; 3JRZ A; 1JXO A; 2E2O A; 4FWP A; 3V7R A; 4N0A A; 2M9U A; 3ULE A; 1OU8 A; 3EG1 A; 3OSV A; 3THY B; 1I81 A; 2HMI A; 4B9W A; 3UWN A; 4EMH A; 2QI2 A; 4Z94 A; 2BTT A; 1VOM A; 2OTL T; 5GAO p; 1LV5 A; 5LJ5 e; 1UAX A; 2K9G A; 4OML A; 1H3H A; 1U4B A; 3AVB A; 4NXM K; 1GLJ O; 5DM7 R; 1B34 A; 3ZT1 A; 4FT2 A; 1JCF A; 1PSF A; 4FOI A; 5I11 A; 1C0U A; 1WIE A; 1EET A; 1Y96 B; 4R79 A; 3JB9 J; 3VYH B; 2DAQ A; 3OYL A; 2Z1G A; 1Q81 U; 4F16 A; 4E6R A; 2K7A A; 3HL8 A; 1QQC A; 3A5N C; 2QEX A; 1LWC A; 1OKJ A; 1YJN Q; 4RCH A; 3CCM N; 3EE9 A; 3KK2 A; 5AQQ A; 3UIQ A; 3I0R A; 4DTU A; 1BIB A; 1RDB A; 3LWM A; 2LCL A; 4ELZ C; 4C0D B; 4C23 A; 1NGF A; 2KGT A; 4LD0 A; 3CCL A; 4BRQ A; 4OF1 A; 5F0X A; 2FPF A; 3UF4 A; 4J9G A; 3RUX A; 3QET A; 4DB3 A; 1X69 A; 1LW0 A; 4PFZ A; 5FW9 A; 5J2Q A; 1KVB A; 5HLF A; 1NJY A; 3RKY A; 3SWN A; 3AVJ A; 4QQG A; 5E84 A; 1SL0 A; 2QGG A; 5AFU H; 2L8D A; 2HBP A; 1KFD A; 5GAM g; 5GJW B; 2OI3 A; 2RD1 A; 1UGS B; 1W2B S; 3QNO A; 3C7N A; 3HJF A; 2RHI A; 2QQB A; 2DPN A; 3PSI A; 2F4V K; 4MLH A; 1M2V A; 1RL2 A; 5KCH A; 1S72 Q; 3JCM e; 3CPW M; 4LXC A; 4EMK B; 1SRM A; 1B7T A; 1C0T A; 3KWG A; 4LN2 A; 2DA9 A; 5HK0 A; 2E5L K; 4EDU A; 4A54 A; 4YOU A; 3UPU A; 5CVI A; 1RBS A; 4LDW A; 2JMC A; 5C6S A; 1N32 K; 4M78 G; 3B6P A; 3H1V X; 4DKW A; 5AQV A; 2KRS A; 3WEE B; 4M78 A; 4F2S A; 4CGF A; 2MK5 A; 3ZEU B; 4N4H A; 3KJV A; 4D8D A; 3ZLJ A; 3GOI A; 4ZGD B; 4JI7 K; 4C8Q G; 2XHB A; 5J26 A; 2D8J A; 2E1H A; 3I55 T; 2QTD A; 3AVG A; 3P39 A; 4C8Q A; 1VQ4 T; 3NMZ C; 2HVI A; 5CK9 A; 3HSB A; 5D3G A; 5CIU A; 3HXM A; 1VQO Q; 4DS5 A; 2O4I A; 5LJW A; 5IT9 E; 2QWO A; 1ZUY A; 4BR5 A; 3VU3 C; 1R59 O; 1VQP N; 4DTP A; 2COY A; 2NUT B; 4Y28 E; 1BIU A; 3ZX2 A; 3FFK B; 4O5F A; 4CC3 A; 4KTQ A; 2DJQ A; 1I1J A; 4E7L A; 3LP3 A; 4M3U A; 2M0O A; 4M75 G; 2KRO A; 1TUG B; 1E6H A; 3S9H A; 4I9Q A; 1GCQ A; 4CRI A; 4M75 A; 2PVO B; 2AKL A; 3X26 B; 3EZW A; 2ZGW A; 4JAX A; 3EG3 A; 4M77 C; 1X9S A; 4O5V A; 2F2W A; 2RQV A; 5M1Q A; 4A5B A; 4FZZ A; 1X43 A; 2DBK A; 4C5E A; 4W5T A; 3VEN A; 3V6D A; 3JAM E; 1NJX A; 4GVM A; 4M78 F; 4M7D E; 3CJC A; 3SJJ A; 2O4X B; 2EYQ A; 1YI2 A; 1WLJ A; 4DOW A; 2WAC A; 5KYW A; 4CFD A; 3DLL L; 4E19 A; 2RI2 A; 1B9D A; 1LJO A; 3RSE B; 2L3P A; 4AFU C; 2YT6 A; 2V7Z A; 1NNE A; 2EYW A; 3LGF A; 3DYA A; 5CY4 A; 2H3J A; 4OGE A; 2X6N A; 3A8L B; 4JRQ A; 4R0K A; 5L23 A; 3AVI A; 1TK0 A; 5DLQ E; 3E1S A; 3UKU A; 4IIO A; 2CH4 W; 1VSD A; 4JN4 A; 3CCU N; 3SUN A; 3CMA A; 2NLU A; 3OB9 A; 2K6D A; 3CCM Q; 4R56 A; 4WFN L; 1D5A A; 3OVN A; 1ZUK A; 3S3M A; 1A5V A; 1VYV A; 1LAW A; 3HI0 A; 4JJN K; 1W97 L; 5GAO q; 2K3X A; 4UC4 A; 2Y1I A; 1MUZ A; 2F2X A; 1YHQ A; 5KI6 A; 4CJP A; 4M75 F; 5KCO A; 2WSE E; 1QVF S; 1W6X A; 1W2B A; 3CCS A; 3KXT A; 2AA4 A; 3C65 A; 4O0I A; 1ZLM A; 5AQT A; 3ITH A; 4DSE A; 1YWP A; 5GJV B; 1YF5 L; 4BRL A; 1PHT A; 3CPF A; 2RI3 A; 1KLN A; 2IVW A; 3V8K A; 5GAM h; 4FWN A; 3QEX A; 3THW B; 5J1E A; 4DV4 K; 3QWY A; 3RR7 A; 2ZZD A; 4FJX A; 1CKB A; 5KH6 A; 3ZT2 A; 2GFU A; 1S6Q A; 4M63 C; 2L3Q A; 1AWW A; 1M1K O; 4J2B A; 4PJO G; 2VB6 A; 1TXQ A; 1VQL A; 3E01 A; 5IT9 O; 5BPL A; 3T3F A; 4PJO A; 1X9J A; 2EQK A; 2V26 A; 5IHI A; 2OPQ A; 3DRS A; 3RRG A; 4RUG A; 4TVR A; 3A8M B; 3O4W A; 2YUP A; 1IHV A; 3QO9 A; 4AG1 C; 3DM2 A; 3CCJ T; 1VQ7 A; 3HP6 A; 1N8R U; 4JKF A; 5AQI A; 1RAC B; 2A28 A; 4OMM A; 3WQT A; 4PY5 A; 4BRN A; 4OLA A; 3ZET B; 1NEB A; 1XUP O; 3HHT B; 5I21 A; 1TKT A; 3BDL A; 1ATS A; 3KLE A; 2DNU A; 2JTF A; 4J9I A; 1VQ6 A; 3HVT A; 2OA8 A; 2A7Y A; 2XU8 A; 5KE3 A; 4HVJ A; 3QXE B; 4NS5 A; 4XH1 A; 4XU2 A; 3JAM O; 4PL6 A; 4B3T K; 3KLF A; 1GOB A; 3HBT A; 4BRA A; 1JKH A; 2BH1 A; 2DD5 A; 1W2B M; 1VQP Q; 3K5L A; 2G8F A; 5AQS A; 4M78 E; 3SAF A; 1Q8I A; 1WOQ A; 4DQQ A; 4KI6 A; 3CMA T; 3KK7 A; 5GAO m; 4ICL A; 1R0C B; 4LH4 A; 2WFB A; 4B3R K; 4A53 A; 2YHW A; 3ASK A; 3MPU B; 3CCV T; 1D0Z A; 4KV8 A; 1PRL C; 1BKB A; 4JS4 A; 2HMP A; 2QQA A; 4ESR A; 1HLU A; 3QBY A; 4PJM A; 5GAN e; 1BCM A; 1K8K B; 2F8S A; 2DEU A; 1G3T A; 4PJO F; 1NGB A; 1ABQ A; 2D0B A; 3MED A; 3JB9 H; 3VTH A; 2AP1 A; 1N9R A; 1Q9Y A; 4KHS A; 2RJD A; 3JCM J; 4FJ7 A; 5GAO n; 2O8F B; 2OAN A; 4MO5 A; 2EIF A; 4C0Z A; 2QA4 A; 4DTM A; 2V1Q A; 2E12 A; 3PIO L; 1WBB A; 2QXL A; 3MCA B; 3SD4 A; 2DX1 A; 1IV0 A; 5AQF A; 2LNV A; 4XEI A; 1XNI A; 5GAN 8; 2D0Q B; 3C6T A; 1G83 A; 2EKH A; 2JWL A; 1ZSZ C; 2EYX A; 3PV8 A; 4J7I A; 1N9S A; 2KEA A; 4M75 E; 3AA3 A; 4F04 B; 4WQ4 A; 4OX9 K; 5JLF A; 2EAY A; 1BG3 A; 1WGS A; 1EQY A; 3OHR A; 1NKE A; 3CME A; 3CCU Q; 4I6M B; 2DM1 A; 4A4H A; 3V9S A; 2O8B A; 2EJ9 A; 2FF3 B; 3CQX A; 5IHN A; 4N47 A; 2RQT A; 5KRT A; 1VQL T; 1QVF A; 3SQ2 A; 4OMQ A; 4UQG A; 4PNO A; 3U23 A; 3MCP A; 4OPK A; 2L5Q A; 2KKZ A; 2OTJ A; 3IQL A; 3PNW C; 2YHT A; 4PFP A; 4Z02 A; 4CGH A; 3FE1 A; 1IG9 A; 5BRH A; 4B9T A; 2LDM A; 2L89 A; 1MWM A; 1Q7Y U; 2VQE K; 2K5L A; 1GBQ A; 4DWI A; 1YIJ T; 2CGH A; 2X3W D; 4CCH A; 2A41 A; 1NM1 A; 3UT1 A; 2LP5 A; 3KLG A; 1HD3 A; 1VQM N; 3H1Q A; 5AQZ A; 1A5W A; 2QK9 A; 1GLC G; 5F1X A; 2XDD A; 2DER A; 4Z89 A; 3GNG A; 1J3T A; 2A42 A; 4IJN A; 4C8L A; 3NHN A; 3M8S A; 5AQY A; 3P8B B; 2M02 A; 3CF5 L; 3NF7 A; 4DFM A; 3NFA A; 1TSF A; 4IUT A; 4K43 A; 4KAZ A; 2VC8 A; 5JIC A; 3D2F B; 4UY8 O; 3R6M A; 3VQ5 A; 2HD0 A; 4CJL A; 1VRU A; 5EOY A; 1VQ9 T; 1GLB G; 4BGA A; 1BK2 A; 4CI6 A; 1ZUU A; 5LJ5 d; 1EXQ A; 2F8T A; 2HBW A; 2CT3 A; 3VRZ A; 2Y4X A; 3RVC A; 3SQ1 A; 4FYX B; 1KC8 C; 3QLH A; 3A2F A; 4H0V B; 2BUP A; 3WNE A; 3CCE A; 3CC7 A; 1OZ2 A; 3A8H B; 3DLE A; 1VUB A; 5MLC Q; 1K73 O; 1OH6 A; 3CET A; 3ATV A; 3K9J A; 1AZG B; 1IRX A; 4K42 A; 3D7E O; 3VUZ A; 3O3H A; 2DL7 A; 4FWL A; 1RT3 A; 2VG6 A; 2P9U A; 3AVK A; 2CP0 A; 3ME9 A; 1RT4 A; 1QVF M; 1ATN A; 4N56 A; 1BI2 A; 3O9Q A; 2QA4 T; 1QHA A; 1KHC A; 4RMO A; 1XNR K; 1RDC A; 1L5U A; 4CGD A; 1H92 A; 1YBY A; 2RK1 A; 3THW A; 3JCM W; 1Q86 U; 2L2P A; 5KYU A; 1R77 A; 1UUE A; 5D6Y A; 2HF3 A; 4FSX A; 3SMP A; 3KD5 E; 4RTT A; 4JK9 A; 1WHK A; 4JI4 K; 3TSW A; 4KHP K; 2HVH A; 1EWR A; 2CGL A; 4IO9 L; 1WQW A; 4X1V A; 1NGJ A; 2FBT A; 2HHV A; 3P8D A; 4FYY B; 5AQG A; 3V8J A; 1KD1 U; 3T19 A; 2J4R A; 5JS2 A; 4KPY A; 3QO3 A; 2M16 A; 2M1H A; 2Q0U A; 1VSJ A; 4BRG A; 1Q81 C; 4DMN A; 3TAF A; 2FL7 A; 4CJF A; 3KBG A; 4M4Z A; 1VQ5 T; 2OAW A; 4BRO A; 4CBU A; 3ATU A; 4PKH A; 2JM9 A; 1PM3 A; 2YNI A; 2MPX C; 1T6D A; 3BGR A; 1T03 A; 1G15 A; 2VXE A; 2EYZ A; 2RJF A; 4FIB A; 1YJN A; 3QUI A; 5GAN 4; 3RIR A; 3E66 A; 2DTI A; 1MU2 A; 1ACM B; 3C6U A; 2ITM A; 3VQE A; 1QVG M; 4E7I A; 1PWT A; 4E7J A; 1UTI A; 2G8W A; 1BIZ A; 4JLH A; 3FZF A; 4JI5 K; 3KK1 A; 4II1 A; 4JLG A; 4AG2 C; 4Q0B A; 2ECZ A; 5EVZ A; 4EPC A; 2LJ3 A; 4W5R A; 2IS3 A; 3VKI A; 1Q82 C; 2KHO A; 3OS1 A; 3UA7 A; 1NEG A; 1DTQ A; 2CP7 A; 2I2L A; 3F9M A; 3OYB A; 3K5M A; 4IJA A; 4IOA L; 2E6M A; 3CC7 T; 2CYZ B; 4MML A; 3TAG A; 2V8I A; 3M4G A; 1S72 A; 4Y1D A; 2DFF A; 4DFP A; 2XIC A; 3QSU A; 3TWH A; 3A8Y A; 3NVK A; 2CP5 A; 1D4X A; 1NG2 A; 2ZAE A; 3M0S A; 5M1O A; 3SLC A; 5CE3 A; 2HEQ A; 4XAX B; 3SWN C; 3CD6 T; 1VSM A; 1VQM Q; 1YCS B; 1BOT O; 1RLP C; 2PU9 B; 2FSK A; 3T1H K; 3EFO B; 3DB4 A; 3L2V A; 3K2P A; 4L3Q A; 4CEO A; 2AK5 A; 2D8H A; 3M57 A; 2HHT A; 1TUC A; 3LP0 A; 1PCX A; 1XMQ K; 1U1S A; 3VS3 A; 1KQ1 A; 1K76 A; 4FWQ A; 2RM4 A; 3TAQ A; 3GOX A; 1VSI A; 2KDS A; 1OX9 A; 2HQX A; 2X51 A; 3AO3 A; 4BRI A; 2KY9 A; 1T6C A; 4ELV A; 4DU3 A; 1YNZ A; 3CCQ A; 3OW2 S; 1VQO A; 1O75 A; 3RKW A; 2FZG B; 2Y90 A; 4BY6 B; 2ZPE B; 3PIP A; 4JNE A; 4CHZ A; 4J83 A; 5LL6 Z; 4KBM B; 3ZSX A; 1RAE B; 2VGN A; 3JB9 D; 4KBO A; 1IZ6 A; 5GMK m; 2E10 A; 4YOT A; 1D3B B; 1M90 R; 2RMO A; 4OVL A; 3VF2 A; 4C8Q C; 2CZ6 B; 1LCK A; 3V8L A; 5C10 A; 2VYT A; 1M4Z A; 2P9P A; 2BE9 B; 3EGX A; 4FSS A; 3I8B A; 4Y1C A; 4BR4 A; 4Z4I A; 2BAN A; 4M3T A; 2CZ1 B; 2PY5 A; 1D0X A; 2L3S A; 3KVG A; 1SQK A; 3DOK A; 3LP2 A; 2QAZ A; 1W2B P; 1SV5 A; 1SSF A; 1NJI C; 2BE2 A; 1NE8 A; 1KQS S; 1LNX A; 4CJS A; 2E65 A; 4N41 A; 4HA8 A; 1Z25 A; 2W6K A; 1DLO A; 1UGR B; 4LF7 K; 2PUK B; 4FK0 A; 3ZSZ A; 3VPZ A; 3CC2 N; 3O8V A; 3TU4 K; 1VSF A; 2HOE A; 1S1N A; 3OBW A; 5GAH P; 1WSI A; 1BQN A; 2LT4 A; 4LDX A; 4M7D B; 1N33 K; 1SRL A; 1IGQ A; 1CZB A; 4YOY A; 3CCR N; 1KVA A; 4EHU A; 3H3N O; 4M7D D; 1X3N A; 3SHW A; 3A0I X; 3PMT A; 3VN5 A; 1AWJ A; 2NUZ A; 3CCM A; 3RMQ A; 4FYV B; 4EIA A; 4OB2 B; 4CJU A; 1M90 U; 1K9M U; 3A5Z B; 5EG2 A; 1JEG A; 1DGK N; 1I07 A; 1BU6 O; 4C8N A; 4AF1 A; 1PNJ A; 5DM6 L; 1WHL A; 3M0R A; 3KTQ A; 3SEM A; 5GAM e; 4HUG A; 2HQE A; 2CH6 A; 3LDP A; 3SUQ A; 2E5Q A; 4P5N A; 4X65 K; 4AHV A; 3HPO A; 4M41 A; 2FSN A; 3THX B; 4FJ9 A; 1ITG A; 3M53 A; 5BRE A; 4DQR A; 3C12 A; 1FX7 A; 2PFU A; 2E8A A; 3RNJ A; 4BUG A; 4NXN K; 3RR6 A; 1RDW X; 4CC7 A; 2YXY A; 3LAM A; 3ZEH A; 4RTV A; 3T9N A; 1WZO A; 2HAX A; 2K5I A; 1YJW N; 2FBY A; 1G8X A; 3A8G B; 4C92 B; 3CCQ T; 2EWS A; 3QFP A; 3AA4 A; 4DUZ K; 4RKU E; 5A0G A; 4DFJ A; 2BUD A; 1NK7 A; 2E1Z A; 3PO5 A; 1G2B A; 3AO2 A; 4C92 D; 4CF0 A; 3V81 A; 3TQ7 P; 2ATQ A; 1QW1 A; 1IBL K; 4RW8 A; 1UGQ B; 1MV3 A; 2P9S B; 4JZ3 A; 3OYN A; 4KH5 A; 3WT0 A; 2UXD K; 2R57 A; 2ESW A; 4OB3 B; 5BN9 A; 4N4I A; 2WOM A; 2GBZ A; 2K2V A; 3LWH A; 3IMX A; 4AHU A; 1ZSZ A; 3H45 C; 2BGG A; 3LDL A; 3ZSO A; 1YWO A; 1N8R C; 3FR0 A; 4D8K A; 4HCB A; 4M77 E; 1A5X A; 1T3V A; 3RTV A; 1EIF A; 4JJD A; 4R5P A; 2PZ1 A; 4Q5W A; 3L2Z A; 3G6E T; 5GAN d; 1M3B A; 1U48 A; 2W10 A; 3OW2 A; 4KH6 A; 3U9Z A; 2M0Y A; 4YTL A; 1OUL A; 1LVK A; 2E2N A; 4PKG A; 1V76 A; 1A0N B; 4ORZ A; 4OMP A; 4I9L A; 5AQM A; 1V4T A; 5KPZ A; 2FHD A; 1KXP A; 4Y91 A; 2WBN A; 1S22 A; 3HT3 A; 1RLQ C; 4PJO C; 1NGH A; 1Y97 A; 5HRS A; 2CRE A; 4C0D C; 1I39 A; 2FF6 A; 2V7Y A; 1NMN A; 5GIP A; 4JRK A; 3TRE A; 1CXU A; 2E41 A; 3O4N A; 5I2M A; 2IVO A; 1VQP A; 5FRM A; 2FPD A; 1KVC A; 4X64 K; 3RWU A; 2HHW A; 1IGU A; 2OYQ A; 1TG0 A; 3NH1 A; 4NL2 A; 3VQ9 A; 4PJL A; 4A4E A; 1QVF P; 2RA2 A; 5HK3 A; 2ABL A; 3GBQ A; 1KQS A; 1VQK T; 3QIN A; 4YNQ A; 2A0F B; 4CJR A; 3TCJ A; 3QKJ A; 1KC8 R; 3E9O A; 4JRP A; 2L3R A; 1HPM A; 3BDP A; 2Q0R A; 2AKK A; 4K11 A; 1XQS C; 4GLK A; 5IAY A; 3SUP A; 1MMA A; 4M3W A; 4LF9 K; 1N5Y A; 5AQX A; 1IB8 A; 4EMK C; 3GDQ A; 2DYI A; 4CJ3 A; 4GV6 A; 3WXM B; 1BBZ A; 5HKC A; 1U2V A; 2VWJ A; 4YDU A; 4B9Q A; 4LUE A; 1HYS A; 4Q5V A; 3VQQ A; 4BQZ A; 3CC2 Q; 3D2E B; 3CQY A; 4FLX A; 1UEC A; 1NMD A; 4BGB A; 1X6G A; 5BPM A; 2XDP A; 3P83 D; 1X27 A; 4GVE A; 2DYB A; 4GY2 B; 4FK2 A; 5TKY A; 1NJW A; 2HHX A; 4J7F A; 3L2Q A; 5GAD P; 3CCR Q; 1V4S A; 3EAE A; 5LJ3 j; 3CC4 N; 1OOT A; 3OW2 M; 4B1Z A; 2KMT A; 3L6Q A; 1YN8 A; 3V7C A; 1YIT T; 3CCU A; 5FU6 B; 2OPS A; 3FZL A; 1RTH A; 4MDA A; 4O8U A; 1MMN A; 3OYA A; 4FJM A; 4JKB A; 2NRX A; 2ZD1 A; 2SEM A; 4AT1 B; 1N36 K; 2ZJP L; 4Z4G A; 1Q7Y C; 3PMI A; 5CKB A; 4U67 L; 3TAE A; 1ZA2 B; 1VQ8 T; 1K1Z A; 1TTH B; 4KT0 E; 2DTH A; 4U7B A; 4O4G A; 1TGO A; 4LD9 K; 4A4G A; 1ZBU A; 4GW6 A; 4WQ4 C; 2P51 A; 1VHX A; 3PTA A; 4DCH A; 3SBG A; 2UYT A; 4X62 K; 4JKD A; 4C5G A; 3K5O A; 3OJU A; 2M51 A; 4N5S A; 2HQH A; 4B8A B; 1B07 A; 4EZ6 A; 2V8J A; 3VOV A; 4C1N K; 2VG7 A; 4F7U B; 4B1U B; 1YJW Q; 3BP8 A; 1RTJ A; 4MDB A; 4H5N A; 3C4S A; 3MDQ A; 3TSZ A; 2DTU A; 2FPE A; 3RDV A; 3G71 A; 2ROT A; 2J06 A; 5BR9 A; 2JS2 A; 4HCC A; 3AV9 A; 4KGZ B; 5LJ3 h; 1TYQ B; 2GX9 A; 4IFE A; 5KQ8 A; 5M1N A; 2DXU A; 5BR8 K; 1UB4 A; 3C6A A; 4DQS A; 4CIF A; 5LJV A; 5IXB A; 3O5B A; 3FJ5 A; 2XD0 A; 3QG1 A; 3P38 A; 3EHR A; 1X01 A; 2K5F A; 5GAN 3; 1Q82 R; 2F9W A; 1KSW A; 1Y0M A; 2NRZ A; 2YHA A; 1ARK A; 3A98 A; 3VQA A; 2DXC A; 3VF4 A; 1WUA A; 3ZL7 C; 3V9W A; 3SV4 A; 2EVR A; 2I5J A; 3HOS A; 1TKD A; 1ZBS A; 3BEX A; 4BR0 A; 3CBM A; 1D8Y A; 3RMB A; 3THX A; 3NF9 A; 4QS9 A; 5I2S A; 2G8H A; 1REV A; 1K6Y A; 2P84 A; 2YKN A; 1X6O A; 1TU0 B; 1EP4 A; 5A0D A; 1Y07 A; 1WHM A; 3RMA A; 4K8X A; 4JKG A; 3WQU A; 3MP8 A; 1RAH B; 2D9T A; 4C92 G; 4J9F A; 3MX5 A; 4PL7 A; 1Q86 C; 2LT1 A; 4ELU A; 1I0C A; 2D1K A; 4C92 A; 2VUB A; 1V29 B; 2COZ A; 2RN2 A; 3WXJ A; 1ASU A; 5GIN A; 2QDY B; 4LDY A; 2CUD A; 1YCY A; 2GQI A; 2GEL A; 3JV3 A; 4E1N A; 3Q1J A; 5KGX A; 1JLC A; 1P8Z A; 2P9S A; 2HZY A; 1M1F A; 1KD1 C; 1JLG A; 2P9N A; 3AVH A; 3IKM A; 3GL1 A; 3M6G A; 3LGL A; 2PVD B; 4FWM A; 4K25 A; 1KIK A; 2P5O A; 4IKF A; 4OMF B; 1WHG A; 3S3N A; 1OQK A; 2ED1 A; 3FFI A; 1Q82 U; 3VQ4 A; 1RTD A; 1U45 A; 3NVM A; 1YJ9 T; 1JLA A; 1M1G A; 5FRN A; 4BWM A; 2HNZ A; 1BI3 A; 3FDR A; 2GEM A; 3JS6 A; 4BVP A; 2HW3 A; 4RCB A; 4DTN A; 4O55 A; 1M8V A; 1VQN T; 2HF4 A; 2YCH A; 4O5B A; 1R0A A; 2FB7 A; 3EYZ A; 4FVU A; 2FO0 A; 2MWP A; 1BI4 A; 3HM8 A; 3I56 T; 2BZY A; 4J6Y A; 4DR5 K; 4J8O A; 3UKR B; 4JI3 K; 4I2Q A; 3KHY A; 2O8D B; 2DL5 A; 1X2K A; 2DPP B; 5J3B A; 5CZ1 A; 4OVA A; 5CKF A; 1RAD B; 1RI9 A; 1SKS A; 2O88 A; 3G71 T; 2XI9 A; 4PKC C; 2XO7 A; 4A2A A; 3I8D A; 4I2P A; 4B8C A; 2LSN A; 3CC4 Q; 1NJI R; 4WQ5 A; 3QII A; 4E3S A; 1WSG A; 3HRU A; 4H0O A; 4DU1 A; 3T69 A; 4JK8 A; 3F70 A; 1CZ9 A; 1HUX A; 2XDB A; 1DT9 A; 1S1W A; 3M55 A; 2C06 A; 3CEY A; 1U5S A; 2KYM A; 2IOC A; 4JI2 K; 4E0D A; 2CZ0 B; 3OYG A; 5CCA A; 3O1W A; 3OTO K; 3NTH A; 1QZ5 A; 1T44 A; 2QEX T; 3F9K A; 4IUR A; 4CFC A; 3I0S A; 2E4L A; 1NGA A; 3NF8 A; 5AYF A; 4GBQ A; 2C0I A; 1T3L A; 2X74 A; 4BE0 A; 2G3R A; 1UEB A; 1VQM A; 2DIG A; 3CCL T; 3MK6 A; 4DV6 K; 2QDL A; 3H8Z A; 3E2U A; 4CZF A; 1EZZ B; 2OTL N; 4NOY A; 5AZI A; 3AVM A; 4WTO B; 3NFC A; 2QXF A; 1KRP A; 4NO7 A; 3A5M C; 3G10 A; 4PA1 A; 5E26 A; 1NJI U; 2QWP A; 1D1A A; 4QVZ A; 2OPP A; 1AOJ A; 1K73 U; 1KLM A; 2RHK A; 2FXU A; 3OMG A; 3ALY A; 1SL2 A; 3G4S A; 4YFU A; 4BE2 A; 4C8Q F; 1JJ2 S; 3A1P A; 3E9G A; 5AQJ A; 1IG8 A; 3LYI A; 3O80 A; 3H3O B; 4IWV A; 3DOL A; 1ZBI A; 4BQL A; 3SJH A; 3M1F A; 3ER0 A; 3HRT A; 5L8R E; 3ZT4 A; 2P1J A; 3TEE A; 1X75 C; 4MZP A; 4FWS A; 1WDX A; 2FSJ A; 5LL6 S; 5AZJ A; 5GAO r; 2P0Q A; 4F7U G; 3EKS A; 4A61 A; 2ROL A; 1JRI A; 1OV3 A; 2HBJ A; 1Z26 A; 3MX2 A; 4CVN E; 3CJB A; 2D5R A; 3J80 E; 2BDP A; 4ISE A; 4F7U A; 4JD2 B; 4LY7 A; 3W3D A; 2IAJ A; 2IPO B; 1N8R R; 3VQ6 A; 3THY A; 3X25 B; 1BIA A; 1GLD G; 3CIP A; 2O31 A; 4XTY A; 3SDZ A; 1QKW A; 5GAM d; 4IFV A; 2K4Y A; 2X9H A; 5CKH A; 3EEQ A; 4JV5 K; 5AQH A; 2XRI A; 3ZT3 A; 4J6W A; 2QGF B; 1RT5 A; 4FWO A; 4H0T B; 3NCI A; 4BG8 A; 2D0N A; 3NBP A; 3H6E A; 4ISF A; 3CQT A; 3SUO A; 2GBQ A; 1NZ9 A; 2JLE A; 1MHN A; 3FZK A; 3E19 A; 4AIL C; 1Y96 A; 3PT9 A; 3MN5 A; 3C6H A; 1O9S A; 1QMC A; 3FB9 A; 1NBW A; 1WSF A; 2GWK A; 4PQU A; 5HRR A; 2ATC B; 2E2Q A; 4X9C A; 1QP3 A; 3PSF A; 4WFA L; 3EGD B; 3I56 N; 3QYH B; 1M90 C; 1K9M C; 1G5V A; 2O8C B; 1N27 A; 3P8H A; 1KC8 O; 5BPN A; 4CJ4 A; 2UU9 K; 3ZSW A; 3OA9 A; 4M45 A; 2KK4 A; 4KHY A; 4GKK K; 3FIF A; 5SZT A; 3DI6 A; 4O0J A; 3D7S B; 3L3U A; 4LF8 K; 3VJP A; 1BUP A; 4CJ5 A; 4E7K A; 4F7U F; 3J7A F; 3IG1 A; 2DL4 A; 3I55 N; 1OH7 A; 1SPK A; 3OW2 P; 1VQ4 N; 1KFS A; 1YI2 T; 2ED0 A; 2YKM A; 2E20 A; 3CJ1 A; 3QBW A; 4JI1 K; 2KFZ A; 6AT1 B; 4C8O A; 1KAY A; 2OZS A; 1X6B A; 3G4S T; 4FL9 A; 1D0Y A; 4WCE L; 2XY7 A; 3J7Z O; 2YJE A; 3QWX X; 3VGL A; 4BE1 A; 1J5E K; 2HHQ A; 3TAM A; 4RTY A; 2R5A A; 2GUI A; 1JLF A; 5GAN 6; 3W0L A; 1UGV A; 4EMK A; 4LSN A; 5HCK A; 2LQ8 A; 8ATC B; 1M5Q 1; 2DK3 A; 4CIE A; 1W7A A; 1Z6R A; 1M9S A; 5ADX A; 5C0M A; 1ZC6 A; 3CXC S; 2P9I B; 4FSV A; 3QJY A; 2E3H A; 3VQD A; 3VS6 A; 3KFV A; 4WF2 A; 3BF3 A; 4HCZ A; 2F5T X; 2D1X A; 1WSE A; 3KZB A; 1HQU A; 2PZS A; 3JCM V; 4C8Q E; 1RFQ A; 2MF2 A; 4CJK A; 4CZE A; 1Q81 O; 3MEU A; 2J6F A; 1WFW A; 4M7A B; 3WEE A; 2GUP A; 1G3S A; 2Q3S A; 5HRO A; 1OZ3 A; 4LNP A; 3L8B A; 1YHQ T; 4FW1 A; 2KRM A; 3C0C A; 4OB1 B; 1Q7Y R; 5HY6 A; 4M7A D; 1K4U S; 3ZEU A; 3FGU A; 2OTL Q; 1JBM A; 3CCS T; 1X9W A; 3PIP L; 3MN6 A; 3LLR A; 1TK5 A; 2Y4Y A; 2UUA K; 1RNH A; 2ZPG B; 1IRE B; 3RMD A; 4B08 A; 3J80 O; 2EGC A; 1G3Y A; 2LX9 A; 3LDN A; 1BHL A; 2OPR A; 4M77 B; 5GAG P; 4ISG A; 4EZ9 A; 4M77 D; 5CKE A; 2DRM A; 4B9N A; 3MN9 A; 5KTQ A; 1NYF A; 2VV5 A; 4GY5 A; 4KHQ A; 4RTW A; 1KAZ A; 3CHW A; 1QQO A; 1TAQ A; 1UA0 A; 2NUT A; 1IJJ A; 2JJ9 A; 2JZ2 A; 2ZJR L; 2VAS A; 3QNN A; 4PLI A; 4JYA K; 4MDX A; 2FEI A; 2YSQ A; 2AJQ A; 2I7P A; 3NH0 A; 4XU3 A; 3O4Q A; 3EH1 A; 3NGP A; 4LH5 A; 1XC9 A; 4OKK A; 3PX0 A; 5LJ5 f; 2OCE A; 3LAN A; 3SFM A; 2GV9 A; 3O9S A; 3C95 A; 5J2P A; 4J5Y A; 1NJZ A; 4L8R B; 3VS0 A; 1Q95 G; 4R55 A; 1VQ7 T; 3ZX3 A; 4Z4C A; 5FPD A; 2VQF K; 3NH2 A; 2W40 A; 3H46 O; 2NZT A; 3R8E A; 4HTU A; 3TPQ A; 2X4X A; 1S1V A; 2O4X A; 5DK5 A; 4B3M K; 4QS7 A; 3TTD A; 3UE5 A; 4CES A; 4DTR A; 1VQ6 T; 3AHU A; 3VER A; 4LDV A; 3QDR B; 2ZPH B; 4QQD A; 3RSE A; 1JCG A; 2JVV A; 2FRY A; 4ANJ A; 1NK0 A; 2YI1 A; 2FYK A; 1Q86 R; 3CC2 A; 4DEX A; 2RQR A; 1UFF A; 1NK8 A; 1QSL A; 1D9D A; 3DXK B; 2F96 A; 2DD4 A; 1DKG D; 1ESV A; 3AAP A; 1WNS A; 2F7T A; 4BC4 A; 1NGI A; 4DTJ A; 1R0B G; 2CUB A; 2PBD A; 4CC4 B; 4FWR A; 1WJR A; 5CQX A; 3CCR A; 1RT7 A; 1NKC A; 2KXC A; 1BO5 O; 2GCX A; 2HCK A; 4A63 B; 5LJ5 j; 1KD1 R; 4DOV A; 4Y0W A; 1AZE A; 1WA7 A; 1MV0 B; 4YDU C; 4KI4 A; 1FWZ A; 1BI0 A; 4Z0U A; 4OJR A; 4KB0 A; 4JD2 A; 1LWF A; 3WNG A; 3CCJ N; 1TS9 A; 1EX4 A; 2X6S A; 3WNH A; 1TKX A; 2VWF A; 5M1F A; 5LJ3 e; 3RES A; 2YJF A; 2E6N A; 4A59 A; 3THV A; 3I55 Q; 3JCM f; 4H0P A; 3I5S A; 5A2Q E; 3MEC A; 1GCP A; 4BRE A; 4FYW B; 1VQ4 Q; 4B1Y B; 4EAH D; 1RDU A; 1YXQ A; 2ITG A; 1UGP B; 1CSK A; 1BGX T; 1GBR A; 2RK2 A; 1EFN A; 3R9P A; 3M5A A; 4XIU A; 1HNZ K; 3H0I A; 1C0G A; 4LFC K; 1SEM A; 4A4F A; 3CXC A; 1XNQ K; 2MOX A; 1YJW A; 4PJO E; 2YV0 X; 1S3X A; 2RI5 A; 4RTX A; 3FLC O; 4CGJ A; 3U4L A; 3HP9 A; 3CME T; 3CMA N; 2KFN A; 2KNB B; 4DLG A; 2DET A; 2UUC K; 2EWN A; 2DTO A; 3EGD A; 2NNW A; 4MLE A; 3CCV N; 1NGC A; 2O8C A; 1T0J A; 5I42 A; 1DTT A; 1ZA1 B; 2EGE A; 4GMJ B; 1NK5 A; 1W9H A; 1YT3 A; 2ASM A; 4DR4 K; 5TKW A; 4CF9 A; 2KZM A; 2BIV A; 3O8M A; 2OTJ T; 1AD5 A; 4B9M A; 2VWK A; 5C0Y A; 3LPT A; 1WHH A; 1F21 A; 2J7I A; 3DXM B; 2NYS A; 3JSC A; 1U04 A; 3J7Y P; 3WUH A; 1S5J A; 1BQM A; 3GG4 A; 1OPK A; 2A6A A; 3M59 A; 3QHS A; 4JDS A; 3ZET A; 2WSF E; 1S9G A; 4M7D C; 1ZBL A; 4KH0 B; 5BRD A; 1NLP C; 5D6X A; 2RO0 A; 4CZL A; 4AM6 A; 3LH5 A; 4J8F A; 3AT1 B; 4OMO A; 1HSQ A; 2DEQ A; 2XMF A; 3PFS A; 3EFR A; 1KHI A; 2D4W A; 2P9K B; 3AA5 X; 3MEE A; 4CGG A; 1K8A U; 2PQW A; 4BRM A; 2ZGY A; 4KRF A; 1BWF O; 1HKC A; 2KVQ G; 3MEV A; 4XTZ A; 1NOY A; 2R5M A; 1IKY A; 3P31 A; 4PKI A; 3ZX0 A; 3AO5 A; 3UB5 A; 4OPA A; 3ZSY A; 4A57 A; 1LOJ A; 2RJC A; 3LH0 A; 3U3G A; 5HP1 A; 4FJ8 A; 1VQL N; 4IUV A; 3V9Z A; 4K0K K; 2FLO A; 3BW1 A; 4ZGE B; 2P9I A; 4GNI A; 4IDH A; 3H6Z A; 3HFO A; 4BRH A; 5CKD A; 2HBK A; 3CXC M; 3I33 A; 1CLQ A; 3CCE T; 1K8K A; 3TAR A; 4YOR A; 2E3I A; 2EJG A; 2P5G A; 4BC2 A; 1MA9 B; 1TVR A; 4HWI A; 2EYY A; 3JCM h; 3SPY A; 4MDW A; 4C92 C; 1YIJ N; 1Z9Q A; 4XQ3 A; 1NOZ A; 3TSU A; 4NCA A; 2O8F A; 3M58 A; 2QG9 B; 4D6W A; 4NL3 A; 2PUO B; 4IEE A; 1AT1 B; 1RAB B; 2BKD N; 3ZCM A; 4C5I A; 3OYM A; 1IKV A; 2J05 A; 3VTI A; 3MXM A; 3LL3 A; 3REB B; 3AVF A; 1ILY A; 1GXI E; 3ZFJ A; 3CFP A; 3Q7B A; 2AHJ B; 2XKO C; 2QA4 N; 1B92 A; 1UJ0 A; 3L42 A; 2GFA A; 1MMG A; 3WVD B; 3JCM b; 4Z4F A; 1PD1 A; 1XJW B; 3EGU A; 4EIK A; 4OKJ A; 4DBH A; 2RPI A; 2ZPF B; 4F3O A; 2CGK A; 2EBP A; 3RMS A; 3QZ5 B; 4KH1 B; 4I6M A; 3CYM A; 1VQ9 N; 4B6M A; 2QQS A; 3CC4 A; 1PD0 A; 4JUV A; 1BIS A; 1Y57 A; 1AWO A; 1UE9 A; 2AZV A; 2YRV A; 3REA B; 1VAZ A; 4CIG A; 4YOW A; 4CEC A; 1N34 K; 4J2D A; 1TAU A; 2CT4 A; 2B6A A; 2KSS A; 4LE9 A; 2OZM A; 1HRH A; 4N76 A; 1OVQ A; 5ECA A; 1TK8 A; 2C0O A; 2Q36 A; 3CER A; 1JQQ A; 4ELT A; 3CCJ Q; 1WLP B; 1I5O B; 2J6K A; 5GAN 2; 2A08 A; 2FBV A; 5FPE A; 1IO2 A; 4DV2 K; 2A36 A; 4WE1 A; 2DIL A; 3NE6 A; 4FO0 A; 3DLH A; 3HM9 A; 1YJN T; 1E7O A; 1N5Z A; 3ID6 A; 2NWM A; 1I4K 1; 1OU9 A; 2RF2 A; 2QYZ A; 4ELY C; 1K9M R; 4O8K A; 2IIR A; 2B4J A; 4WQ5 C; 4CJW A; 3H0F A; 1Z05 A; 1JU5 C; 2ZQB A; 4QS8 A; 3J7A P; 2LJ1 A; 4FXD A; 3NPF A; 3G25 A; 3SMS A; 2BTF A; 4CHY A; 4C1N I; 3CMA Q; 4OB0 B; 4FVM A; 1L3U A; 1K73 C; 2DKG A; 3VS5 A; 1C0M A; 1HYV A; 1VSK A; 3D2F A; 3O08 A; 5JLH A; 3FZM A; 1UA1 A; 4IBN A; 1MNE A; 3CCV Q; 4DR3 K; 5KRS A; 5KGW A; 4BC5 A; 3X28 B; 3VQC A; 1U3O A; 1K0S A; 1RT1 A; 2IVN A; 4CBW A; 1GLL O; 2LMJ A; 1OI1 A; 2H3E B; 3KD1 E; 1T0H A; 3ZSR A; 3NGI A; 1VQ5 N; 4E89 A; 1S72 T; 5FRO A; 3JCM Q; 1QCF A; 2DXB A; 1MND A; 5EXW A; 2JTM A; 4LF5 K; 2MYS A; 4KGV B; 4KGX B; 2E6Z A; 1GFD A; 3R1X A; 4HHT A; 3RBB B; 2NRR A; 3VEY A; 3HRS A; 4NCB A; 1WYX A; 4HCK A; 4KRT A; 1RT6 A; 3AA2 A; 2YDL A; 3D45 A; 3IFR A; 2B86 A; 1CXQ A; 1SZ2 A; 2WC9 A; 2CDT A; 4CK3 A; 4CED A; 1YTA A; 3OYE A; 1QWF A; 1FMK A; 1QQM A; 3QER A; 4DTO A; 4XTV A; 1T05 A; 3VUB A; 5GAN f; 3HKS A; 3QDK A; 3HVR A; 2LC4 A; 3M8R A; 4N4G A; 1HJR A; 4FPB A; 4VUB A; 3ULD A; 2SRC A; 4KO0 A; 5C24 A; 4IUP A; 1VQL Q; 2P9L B; 4YOX A; 2GTO A; 2E2P A; 1ABO A; 1TOV A; 3M0T A; 4EFH A; 1B34 B; 1IKX A; 4J6X A; 5CAI A; 1VYT A; 4QQ6 A; 1ZBX A; 5LJ3 g; 3FZH A; 1U1T A; 3CC7 N; 3VEX A; 1VQO T; 3BFM A; 2O9V A; 1D09 B; 4FJ5 A; 3EN9 A; 2XY5 A; 4DHY A; 3KKR A; 5AQN A; 4CHN A; 3PX4 A; 1YIJ Q; 4AH9 A; 4DFK A; 4JRI A; 3M56 A; 1W1F A; 4HT8 A; 4CZH A; 4B9L A; 5JS1 A; 3CD6 N; 1RBR A; 1YVN A; 4B3S K; 2X78 A; 2DRK A; 2WON A; 3SPZ A; 5ADX H; 4PLL A; 4XTW A; 4B1V A; 3FLG A; 2V1R A; 4DV3 K; 4J9E A; 4G9Z A; 3H08 A; 4NYF A; 3VET A; 3HST B; 1JLE A; 4J9H A; 1M3C A; 1QKX A; 2LX7 A; 4BRP A; 3B6O A; 5GAN j; 3AVC A; 4JHD B; 2Q97 A; 3QEI A; 3M0P A; 3G1J A; 3SQ0 A; 5FDL A; 1SSH A; 4H4M A; 1FYN A; 2MMP A; 1KG1 A; 3KLI A; 5HOO A; 4DG1 A; 1VQ9 Q; 1NLV A; 4A2B A; 1SG5 A; 1UG1 A; 1XTD A; 2CSQ A; 4HVU A; 1Q86 O; 3G7Z A; 3GLX A; 3JXU A; 1VIF A; 4PFO A; 2W6L A; 4CHQ A; 3HK2 A; 4F4K A; 3VQ8 A; 3CPW S; 4CD1 A; 2RQW A; 4J2E A; 1FXX A; 1M2V B; 2KAF A; 4KUL A; 2X4Y A; 4B3O A; 1VYU A; 2QKB A; 5CR2 A; 2HNI A; 1GLE G; 4BRD A; 3TAB A; 1HNI A; 4RG8 A; 1NKB A; 3L0Q A; 1SF9 A; 4Y92 A; 4I7F A; 2HMA A; 4FLT A; 1D9F A; 3CCM T; 4H5W A; 4A4K A; 3O6W A; 2LNH B; 5JMV A; 1MGQ A; 2QK0 A; 3EFO A; 4IUQ A; 1KCF A; 3THK A; 3M8Q A; 3BZK A; 4H0Y B; 1IXD A; 5I3U A; 4M78 B; 2CP3 A; 2DY4 A; 2QTV A; 1JB0 E; 4X34 A; 2F1L A; 4M78 D; 4HXJ A; 1XC3 A; 2HHH K; 2KZZ A; 2OTL A; 4QW2 A; 2X35 A; 2O01 E; 2LCC A; 1E4F T; 2DZC A; 2K2M A; 1WN7 A; 2XG8 D; 2E64 A; 3HFN A; 2EHG A; 4E47 A; 2D0O A; 2YHY A; 4IDK A; 4CE9 A; 4FJH A; 5T7B A; 2QAS A; 2GTJ A; 2DE0 X; 3JB9 G; 4ZGJ B; 4FJJ A; 1VQ5 Q; 3ZSQ A; 1X3M A; 2PYJ A; 4J9C A; 3PGG A; 3TSP A; 5C42 A; 4W5N A; 4IG0 A; 1HKG A; 2YNF A; 1TV6 A; 2ZF5 O; 1HX1 A; 4BI3 A; 1GLA G; 2P4T A; 5E41 A; 1D3B A; 1VSH A; 4CJE A; 4X66 K; 3P1G A; 4M75 B; 3UA6 A; 3VS4 A; 1WXU A; 1R8O B; 4GLM A; 3K9K A; 3M0Q A; 3MEW A; 4L59 A; 4M75 D; 4FM2 A; 1D1C A; 4FLU A; 1RUW A; 3HPW A; 1XI1 A; 1HNX K; 4CZG A; 2OX7 A; 3U9D A; 3I35 A; 4EP4 A; 3EO3 A; 4ZTF A; 2NUB A; 5LJ3 d; 2EQU A; 5KYW B; 2P0P A; 2CH4 A; 1KQS M; 2R7Y A; 4F8R A; 3OYK A; 2RKI A; 3DMJ A; 3AVN A; 3GBT A; 1ZBH A; 1YAG A; 3CCQ N; 1W4S A; 1W70 A; 1Q18 A; 2RB6 A; 4M7D G; 4PHT X; 3IRX A; 5J39 A; 3CXC P; 4ILH A; 1M1K U; 1L3S A; 1TKZ A; 4M7D A; 3WNF A; 4FT4 A; 4B1W B; 1J54 A; 1VSE A; 3SQ4 A; 2EJF A; 1VQP T; 1B9F A; 3UKU B; 1VIE A; 3K58 A; 2GQV A; 4DQP A; 1BB9 A; 2RN8 A; 3JB9 F; 3S41 A; 1XQH A; 3CD6 Q; 5EC7 A; 3T1Y K; 1NGE A; 1BI1 A; 1V1C A; 1H1V A; 3L2U A; 4BVO A; 4FZY A; 3AO1 A; 1GOA A; 1FMV A; 1YTU A; 1JLB A; 2XY6 A; 3AO4 A; 3DRP A; 1WBD A; 4LC9 B; 1KSP A; 3HSC A; 3DJC A; 3G6E N; 5I22 A; 4JPB W; 2CSI A; 3NGY A; 2EQJ A; 4FLY A; 3CPW A; 3L4I A; 5AQL A; 3UDC A; 3OYI A; 2ZWH A; 4EP5 A; 2PAV A; 1WXT A; 5HRN A; 5GIO A; 2PVG B; 2VKN A; 3I55 A; 3CM6 A; 3JCM c; 1UOC A; 2VWB A; 1LW2 A; 3O1B A; 1VQ4 A; 2P9L A; 4A5A A; 1S1X A; 3QML A; 3OMC A; 3OSS C; 3AAR A; 3O3F A; 2JN0 A; 2Z1J A; 1SKW A; 3OY9 A; 1M3A A; 1GRI A; 4CGI A; 1IBM K; 1ZQ1 A; 3MN7 A; 1RAF B; 3MET A; 2RQU A; 5JVG L; 3K5N A; 4DF4 A; 4GV3 A; 2K3Y A; 2VVK A; 4AHR A; 4DSF A; 3AY9 A; 5F2R A; 3CCU T; 4M7D F; 1E4G T; 4DTX A; 2E6L A; 5DLO A; 3OYC A; 3EG9 B; 4CF1 A; 5FLX E; 1SL1 A; 1VQK N; 1LCU A; 4V19 S; 2JT4 A; 3BDU A; 3S3O A; 4PJO B; 4JJB A; 3GIB A; 3SCX A; 3J81 E; 1PSE A; 4PJO D; 2CGJ A; 2ZM6 K; 3URG A; 2O4G A; 3LPU A; 4AM7 A; 1LWE A; 3BY5 A; 3FRN A; 4IFY A; 4DU4 A; 1NPP A; 4BDY A; 4GV9 A; 1KTQ A; 4IO8 A; 1M90 O; 1K9M O; 4JHD A; 2DZ9 A; 2K3B A; 4ILG B; 4H4O A; 4FJL A; 5GMK j; 4M3X A; 4XK8 E; 1NBE B; 2EQI A; 3SZ2 A; 3CBP A; 3ZSV A; 3WXK A; 1JO8 A; 3LAL A; #chains in the Genus database with same CATH homology 1J9A A; 5HBM A; 1JLQ A; 4Q0B A; 2FBX A; 3VQD A; 2KTQ A; 2PZS A; 4FW2 A; 1WSE A; 4HUF A; 3HPH A; 1HQU A; 3UIQ A; 4CJK A; 4W5R A; 2IS3 A; 1Q9X A; 4QCL A; 1LAV A; 5HRP A; 4PWD A; 2Q3S A; 2Z1H A; 5HRO A; 2EX3 A; 3OS1 A; 1BI4 B; 1DTQ A; 3L8B A; 3IAY A; 2HHU A; 4FW1 A; 3OYB A; 3K5M A; 3V1R A; 2E6M A; 1X9W A; 4W5O A; 3TAG A; 2VG5 A; 4BAC A; 1TK5 A; 2B5J A; 1RNH A; 4Y1D A; 2DFF A; 5CZ2 A; 1S6P A; 4DFP A; 3RMD A; 4B08 A; 4F2R A; 4CEF A; 3TWH A; 3U6F A; 1BHL A; 1RTI A; 2OPR A; 4DQI A; 5C25 A; 1VSM A; 3CFO A; 4EZ9 A; 3CG7 A; 2IDO A; 4B9N A; 5KTQ A; 4KHQ A; 3K2P A; 3L2V A; 3EY1 A; 1TAQ A; 4CEO A; 1WSJ A; 1UA0 A; 2HHT A; 3T1A A; 1JXB A; 3QNN A; 4LSL A; 3SV3 A; 4F3T A; 3LP0 A; 2AJQ A; 2G8I A; 1S1T A; 3NH0 A; 3O4Q A; 1L3V A; 4CER A; 3TR8 A; 4LH5 A; 1XC9 A; 4OKK A; 1HPZ A; 1S1U A; 3PX0 A; 4B9S A; 1IKW A; 3LAN A; 2GV9 A; 3C95 A; 3TAQ A; 5J2P A; 1VSI A; 1FKO A; 4KTW A; 5EU7 A; 3OYH A; 2IGI A; 3DLR A; 1ZYQ A; 1NJZ A; 4L8R B; 3AO3 A; 4Z4C A; 4ELV A; 4DU3 A; 3NH2 A; 1VK0 A; 1ASV A; 4HTU A; 1S1V A; 2D0C A; 5DK5 A; 4CHZ A; 4KHW A; 3ZSX A; 2PYL A; 4CES A; 4DTR A; 3NGZ A; 1RBV A; 1W0H A; 1HMV A; 4DF8 A; 1GOC A; 3LZI A; 5SWM A; 3OYD A; 1NK0 A; 4FZY A; 1QSL A; 4OVL A; 3ZT0 A; 2G8U A; 5J2N A; 1NK8 A; 3VQ7 A; 1SKR A; 1D9D A; 3HO1 A; 2F96 A; 1TL1 A; 3HPG A; 4Y1C A; 3OYF A; 4ID5 A; 1WNS A; 4ID1 A; 3HYF A; 4Z4I A; 3VQP A; 2BAN A; 3AVL A; 3KLH A; 4BDZ A; 4DTJ A; 2PY5 A; 4CFB A; 4M3T A; 1NKC A; 1RT7 A; 3LP1 A; 3DOK A; 4OPJ A; 3LP2 A; 2DFH A; 1SV5 A; 2BE2 A; 4NYN A; 4RW7 A; 5CYQ A; 2HND A; 4CJS A; 4N41 A; 3CFR A; 1Z25 A; 1DLO A; 1C1C A; 4CJT A; 2KW4 A; 3V4I A; 2Y1J A; 3ZSZ A; 3NAE A; 4FK0 A; 4KI4 A; 4Z0U A; 4OJR A; 3D0P A; 5D7U A; 4KB0 A; 1LWF A; 3WNG A; 1VSF A; 1EX4 A; 1J53 A; 2X6S A; 3WNH A; 1TKX A; 1XWL A; 2JGU A; 1WSI A; 3U3Y A; 2YNG A; 3THV A; 1BQN A; 3L2R A; 4B9U A; 3QEP A; 1CZB A; 3MEC A; 1QTM A; 4FYD A; 1KVA A; 2HMI A; 3LWL A; 4M3Y A; 3VN5 A; 2ITG A; 1BGX T; 3LDS A; 4XIU A; 1LV5 A; 1HQE A; 4CJU A; 1UAX A; 2YV0 X; 5CYM A; 1U4B A; 3AVB A; 4C8N A; 3C94 A; 3KTQ A; 4QOZ B; 4CGJ A; 3HP9 A; 3ZT1 A; 4HUG A; 4RW4 A; 3SUQ A; 4DLG A; 1JXE A; 1C0U A; 1RIL A; 4AHV A; 4CFA A; 4M41 A; 1EET A; 3DLG A; 3HPO A; 4FJ9 A; 1ITG A; 4DQR A; 5I42 A; 1DTT A; 4GMJ B; 1NK5 A; 1W9H A; 1YT3 A; 2Z1G A; 3OYL A; 4CF9 A; 2KZM A; 1QQC A; 3HL8 A; 1LWC A; 3LAM A; 4B9M A; 2VWK A; 2A1R A; 3LPT A; 1U47 A; 3KK2 A; 3SI6 A; 1F21 A; 2A1S A; 4RW9 A; 2FBY A; 1U04 A; 3I0R A; 4DTU A; 4DFJ A; 1S5J A; 1BQM A; 1VRT A; 3AA4 A; 1RDB A; 1C1A A; 3LWM A; 4ZTJ A; 1NK7 A; 3PO5 A; 1RBT A; 1S9G A; 3AO2 A; 4CF0 A; 3V81 A; 2YHB A; 1ZBL A; 4LD0 A; 2ATQ A; 4MQ3 A; 4RW8 A; 3TAP A; 1NK9 A; 3QET A; 3OYN A; 3PX6 A; 2DFE A; 1LW0 A; 4CHP A; 5J2Q A; 2WOM A; 2GBZ A; 3AA5 X; 3QEV A; 1KVB A; 5HLF A; 1NJY A; 3MEE A; 3AVJ A; 4CGG A; 4AHU A; 1SL0 A; 4KRF A; 3MAQ A; 2BGG A; 1JL2 A; 1KFD A; 3ZSO A; 4CK1 A; 3K59 A; 1NOY A; 1A5X A; 1IKY A; 3RTV A; 4HCB A; 3AO5 A; 3ZSY A; 4R5P A; 3QNO A; 3HJF A; 5HP1 A; 4FZX C; 3U3G A; 4FJ8 A; 3V9Z A; 1U48 A; 3V1O A; 4CF8 A; 1C0T A; 4K8Z A; 3CQ8 A; 1CLQ A; 4I9L A; 3TAR A; 1NK6 A; 1IHW A; 2P5G A; 4KB1 A; 1TVR A; 3QIO A; 1RBS A; 3SPY A; 3HT3 A; 1NOZ A; 1T7P A; 4NCA A; 3B6P A; 1Y97 A; 5HRS A; 1I39 A; 1NJW A; 1HYZ A; 3ZCM A; 4E1M A; 1CXU A; 1IKV A; 3O4N A; 3OYM A; 4F2S A; 3MXM A; 4M42 A; 4OLB A; 4CGF A; 3AVF A; 5FRM A; 3RWU A; 1KVC A; 2D0A A; 3KJV A; 2HHW A; 1ASW A; 3CFP A; 2OYQ A; 1JL1 A; 1C6V A; 3NH1 A; 1B92 A; 2XHB A; 3VQ9 A; 3JYT A; 4FJK A; 4CK2 A; 4QAG A; 3AVG A; 2G8K A; 4Z4F A; 3QIN A; 2HVI A; 4YNQ A; 4OKJ A; 3L2W A; 4CJR A; 2RPI A; 1XHZ A; 4F3O A; 5D3G A; 3NF6 A; 4JRP A; 3HXM A; 4DS5 A; 3BDP A; 2O4I A; 3CYM A; 4BDP A; 3SUP A; 4M3W A; 1BIS A; 3O3G A; 4CIG A; 1N5Y A; 4J2D A; 4DTP A; 4FJG A; 4CEC A; 1TAU A; 4CJ3 A; 1BIU A; 2B6A A; 2OZM A; 1RDA A; 2VWJ A; 1HRH A; 4N76 A; 4KTQ A; 2IC3 A; 1HYS A; 1TK8 A; 3LP3 A; 4E7L A; 4Q5V A; 3VQQ A; 4OGC A; 4ELT A; 4M3U A; 4FJN A; 4NCG A; 4FLX A; 3NHG A; 3TAN A; 2FBV A; 3S9H A; 3P83 D; 1IO2 A; 2ZE2 A; 4I9Q A; 2HHX A; 4FK2 A; 4WE1 A; 3NE6 A; 3L2Q A; 3SYZ A; 3HM9 A; 1X9S A; 4M3Z A; 3ZSR A; 1RT2 A; 4FZZ A; 2RF2 A; 2B4J A; 4CJW A; 4W5T A; 3V6D A; 1NJX A; 2ZQB A; 4GVM A; 5K14 A; 2OPS A; 3SJJ A; 1RTH A; 1S9E A; 4HUE A; 4CJQ A; 4FXD A; 1WLJ A; 4O44 A; 4CFD A; 3OYA A; 4FJM A; 2ZD1 A; 4E19 A; 1B9D A; 4Z4E A; 4Z4G A; 4CHY A; 3TAE A; 3RMC A; 4RW6 A; 4FVM A; 1L3U A; 1HYV A; 1C1B A; 1C0M A; 1VSK A; 3DYA A; 5CY4 A; 4CEA A; 4O4G A; 4OGE A; 1UA1 A; 2X6N A; 4IBN A; 3AVI A; 1TGO A; 1ZBU A; 4GW6 A; 5KRS A; 5KGW A; 4JRQ A; 2P51 A; 1YVU A; 3VQC A; 1TK0 A; 3RR8 A; 1RT1 A; 3K5O A; 3OJU A; 1VSD A; 1UWB A; 3SUN A; 4N5S A; 4B8A B; 3KD1 E; 4EZ6 A; 1BCO A; 2VG7 A; 3NGI A; 4DSI A; 4E89 A; 1D5A A; 1RTJ A; 3OVN A; 5FRO A; 3S3M A; 2DTU A; 1A5V A; 3F73 A; 1LAW A; 1NK4 A; 3AV9 A; 4JS5 A; 1QSY A; 4HCC A; 4FK4 A; 2Y1I A; 5KI6 A; 1O1W A; 4CJP A; 1FKP A; 4CEQ A; 4HHT A; 4DQS A; 4CIF A; 4NCB A; 4C8K A; 3AA2 A; 1RT6 A; 3D45 A; 4DSK A; 1CXQ A; 4CK3 A; 4O0I A; 4CED A; 1YTA A; 3OYE A; 4DSE A; 3ITH A; 2YHA A; 3QER A; 4DTO A; 3VQA A; 1T05 A; 1RCH A; 3V9W A; 3SV4 A; 4J2A A; 2I5J A; 3HVR A; 1TKD A; 3M8R A; 1HJR A; 1KLN A; 3ULD A; 3QEX A; 4KO0 A; 5C24 A; 1D8Y A; 5J1E A; 3RMB A; 3RR7 A; 3MXJ A; 1SUQ A; 3NF9 A; 4FCY A; 3TI0 A; 4FJX A; 2FC0 A; 2G8H A; 1REV A; 1IKX A; 3OJS A; 3ZT2 A; 4FLZ A; 1S6Q A; 1K6Y A; 2YKN A; 1EP4 A; 3RMA A; 3DLB A; 4K8X A; 4PUO A; 3PY8 A; 4AHT A; 4J2B A; 4FJ5 A; 2XY5 A; 3E01 A; 4ELU A; 3KKR A; 4CHN A; 1QE1 A; 4AHC A; 4AH9 A; 3T3F A; 4DFK A; 2OPQ A; 3PX4 A; 3DRS A; 3RRG A; 4FM1 A; 2RN2 A; 3ISN C; 3G0Z A; 4B9L A; 1ASU A; 3EZ5 A; 5JS1 A; 3KIO A; 1RBR A; 1IHV A; 2X78 A; 3QO9 A; 2WON A; 4E1N A; 3DM2 A; 3SPZ A; 3HP6 A; 4FJI A; 1I3A A; 5KGX A; 1JLC A; 3AVH A; 1HNV A; 3MEG A; 2KFN A; 1JLG A; 4PY5 A; 4OLA A; 4B9V A; 3H08 A; 1WAJ A; 4NYF A; 2P5O A; 3HST B; 1JLE A; 4IKF A; 3S3N A; 1RTD A; 3FFI A; 3VQ4 A; 1TKT A; 3B6O A; 1U45 A; 1JLA A; 3KLE A; 3AVC A; 3QEI A; 5FRN A; 4BWM A; 2HNZ A; 3V1Q A; 3HVT A; 1IH7 A; 3SQ0 A; 5FDL A; 3SNN A; 2OA8 A; 4H4M A; 3DLK A; 3KLI A; 4HVJ A; 4KFB A; 4DTN A; 1T8E A; 4DG1 A; 4O55 A; 4DSL A; 2HW3 A; 4CEB A; 4O5B A; 3KLF A; 1R0A A; 1GOB A; 3EYZ A; 1JKH A; 4CHQ A; 3KK3 A; 1BI4 A; 3K5L A; 2G8F A; 3HK2 A; 4F4K A; 1Q8I A; 3VQ8 A; 4DQQ A; 4KI6 A; 3OS0 A; 4I2Q A; 4TSX A; 4J2E A; 4ICL A; 1FXX A; 4DLE A; 5CZ1 A; 4KHU A; 4LH4 A; 4B3O A; 3PUF A; 3OS2 A; 3K57 A; 1SKS A; 2QKB A; 3MXI A; 4KV8 A; 3TAB A; 4JS4 A; 1HNI A; 4RG8 A; 1NKB A; 4I7F A; 2XO7 A; 1RDD A; 4FLT A; 1BCM A; 1QS4 A; 3I8D A; 1ZBF A; 1D9F A; 2F8S A; 4I2P A; 4B8C A; 2LSN A; 2W42 A; 2D0B A; 1L3T A; 3MED A; 1Q9Y A; 4E3S A; 4KHS A; 1WSG A; 4FJ7 A; 1KCF A; 4CEE A; 4DU1 A; 3M8Q A; 5I3U A; 4DSJ A; 2DY4 A; 4ZHR A; 1CZ9 A; 4W5Q A; 1S1W A; 4DTM A; 2IOC A; 3VA0 A; 4E0D A; 4Z4H A; 4KTZ A; 3OYG A; 2KZZ A; 3C6T A; 1WN7 A; 3IS9 A; 3PV8 A; 2EHG A; 3F9K A; 3AA3 A; 4IDK A; 4CE9 A; 4FJH A; 5T7B A; 1NKE A; 4CFC A; 3I0S A; 2E4L A; 3NF8 A; 3V9S A; 2X74 A; 4BE0 A; 4FJJ A; 4N47 A; 5KRT A; 3NDK A; 4G1Q A; 3SQ2 A; 3ZSQ A; 2PYJ A; 4UQG A; 4W5N A; 5C42 A; 4IG0 A; 2YNF A; 1TV6 A; 4OPK A; 3V9U A; 3AVM A; 2QXF A; 4CGH A; 5E41 A; 1KRP A; 1IG9 A; 2HNY A; 3G10 A; 3OYJ A; 1VSH A; 4PA1 A; 4CJE A; 4FM0 A; 1EKE A; 3P1G A; 4B9T A; 4DWI A; 2OPP A; 1X1P A; 3CM5 A; 1KLM A; 4CCH A; 1SL2 A; 3ALY A; 4IG3 A; 1TL3 A; 3KLG A; 4YFU A; 4H8K A; 4FM2 A; 4BE2 A; 4FLU A; 1XI1 A; 1N6Q A; 3QES A; 4KHN A; 1QHT A; 1A5W A; 4CEZ A; 2QK9 A; 1WSH A; 3DOL A; 1ZBI A; 4EP4 A; 1X9M A; 4ZTF A; 4FLW A; 2NUB A; 3LAK A; 4C8L A; 3M8S A; 3QIP A; 4F8R A; 2R7Y A; 3VA3 A; 2RKI A; 3DMJ A; 3OYK A; 3DLH A; 3AVN A; 3NF7 A; 3ZT4 A; 2P1J A; 4AHS A; 3NFA A; 4DFM A; 2QKK A; 1BL3 A; 1ZBH A; 4KAZ A; 1L3S A; 3IRX A; 3KKS A; 4CJL A; 3VQ5 A; 1TKZ A; 1VRU A; 1Z26 A; 2ETJ A; 3ASM A; 3M8P A; 2D5R A; 3DRR A; 3WNF A; 5DMR A; 2BDP A; 2HHS A; 1EXQ A; 4LY7 A; 1J54 A; 2F8T A; 1VSE A; 3SQ4 A; 2IAJ A; 3VQ6 A; 1B9F A; 3K58 A; 4BWJ A; 4KRE A; 3SQ1 A; 3QLH A; 4DQP A; 3A2F A; 2XY8 A; 3WNE A; 4IFV A; 3DLE A; 3V9X A; 1J5O A; 4DS4 A; 4CHO A; 2XRI A; 3O3H A; 1RBU A; 3ZT3 A; 2YNH A; 1RT3 A; 1GOA A; 1VSL A; 1RT5 A; 3AO1 A; 3L2U A; 2VG6 A; 1JLB A; 2XY6 A; 3AO4 A; 3AVK A; 3DRP A; 3NCI A; 1YTU A; 3NBP A; 1KSP A; 1RT4 A; 1XHX A; 4N56 A; 2HB5 A; 2Z1I A; 3SUO A; 2JLE A; 4AIL C; 3NGY A; 4FLY A; 1RDC A; 4EP5 A; 3OYI A; 3QEW A; 4CGD A; 1QMC A; 1WAF A; 1L5U A; 1WSF A; 5HRN A; 4PQU A; 5HRR A; 3CM6 A; 3PO4 A; 4M3R A; 1QSS A; 1UOC A; 3P5J A; 2G8V A; 1LW2 A; 1S1X A; 3KD5 E; 4MFB A; 3O3F A; 2HVH A; 2Z1J A; 4CJ4 A; 3OY9 A; 3ZSW A; 1SKW A; 4M45 A; 4CGI A; 4KHY A; 2FBT A; 2HHV A; 3AVA A; 4CJV A; 5SZT A; 3DI6 A; 4O0J A; 3T19 A; 3K5N A; 4DF4 A; 3VQB A; 3L3U A; 5JS2 A; 4AHR A; 3RRH A; 4CJ5 A; 4OKE A; 4DSF A; 4KPY A; 1VSJ A; 4DMN A; 2E6L A; 4E7H A; 3IG1 A; 4E7K A; 4DTX A; 3TAF A; 3OYC A; 4CJF A; 4CF1 A; 1SL1 A; 4DTS A; 1KFS A; 2YKM A; 3S3O A; 4KKO A; 4FLV A; 2YNI A; 3LZJ A; 1C6V X; 2KFZ A; 3SCX A; 3BGR A; 1T03 A; 1G15 A; 4C8O A; 2OZS A; 2KQ2 A; 3LPU A; 2O4G A; 4DU4 A; 1LWE A; 4IFY A; 2XY7 A; 4BDY A; 4BE1 A; 1FK9 A; 1KTQ A; 1MU2 A; 3C6U A; 3TAM A; 3VQE A; 2HHQ A; 4E7I A; 4E7J A; 2GUI A; 1JLF A; 4C8M A; 3JSM A; 4H4O A; 4CF2 A; 4FJL A; 4M3X A; 5J2M A; 2G8W A; 4LSN A; 1BIZ A; 4JLH A; 3SZ2 A; 3ZSV A; 3L3V A; 4CIE A; 3KK1 A; 1U49 A; 3LAL A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...