The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
263
|
sequence length |
866
|
structure length |
860
|
Chain Sequence |
LFPWAQIRLPTAVVPLRYELSLHPNLTSMTFRGSVTISVQALQVTWNIILHSTGHNISRVTFMSAVSSQEKQAEILEYAYHGQIAIVAPEALLAGHNYTLKIEYSANISSSYYGFYGFSYTDESNEKKYFAATQFEPLAARSAFPCFDEPAFKATFIIKIIRDEQYTALSNMPKKSSVVLDDGLVQDEFSESVKMSTYLVAFIVGEMKNLSQDVNGTLVSIYAVPEKIGQVHYALETTVKLLEFFQNYFEIQYPLKKLDLVAIPDFEAGAMENWGLLTFREETLLYDSNTSSMADRKLVTKIIAHELAHQWFGNLVTMKWWNDLWLNEGFATFMEYFSLEKIFKELSSYEDFLDARFKTMKKDSLNSSHPISSSVQSSEQIEEMFDSLSYFKGSSLLLMLKTYLSEDVFQHAVVLYLHNHSYASIQSDDLWDSFNEVTNQTLDVKRMMKTWTLQKGFPLVTVQKKGKELFIQQERFFLNMSDTSYLWHIPLSYVTEGRNYSKYQSVSLLDKKSGVINLTEEVLWVKVNINMNGYYIVHYADDDWEALIHQLKINPYVLSDKDRANLINNIFELAGLGKVPLKRAFDLINYLGNENHTAPITEALFQTDLIYNLLEKLGYMDLASRLVTRVFKLLQNQIQQQTWTDEGTPSMRELRSALLEFACTHNLGNCSTTAMKLFDDWMASNGTQSLPTDVMTTVFKVGAKTDKGWSFLLGKYISIGSEAEKNKILEALASSEDVRKLYWLMKSSLNGDNFRTQKLSFIIRTVGRHFPGHLLAWDFVKENWNKLVQKFPLGSYTIQNIVAGSTYLFSTKTHLSEVQAFFENQSEATFRLRCVQEALEVIQLNIQWMEKNLKSLTWWL
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
pubmed doi rcsb |
| molecule keywords |
Leucyl-cystinyl aminopeptidase
|
| molecule tags |
Hydrolase
|
| source organism |
Homo sapiens
|
| total genus |
263
|
| structure length |
860
|
| sequence length |
866
|
| chains with identical sequence |
B
|
| ec nomenclature |
ec
3.4.11.3: Cystinyl aminopeptidase. |
| pdb deposition date | 2015-06-26 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF01433 | Peptidase_M1 | Peptidase family M1 domain |
| A | PF11838 | ERAP1_C | ERAP1-like C-terminal domain |
| A | PF17900 | Peptidase_M1_N | Peptidase M1 N-terminal domain |
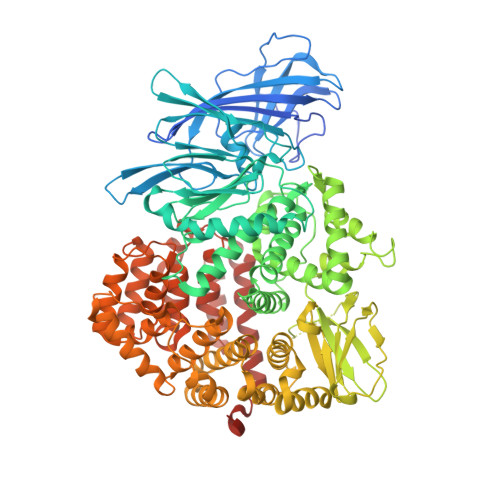
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Alpha | Orthogonal Bundle | Neutral Protease; domain 2 | Neutral Protease; domain 2 | ||
| Mainly Alpha | Alpha Horseshoe | Zincin-like fold | Zincin-like fold | ||
| Mainly Beta | Sandwich | Immunoglobulin-like | tricorn interacting facor f3 domain | ||
| Mainly Beta | Sandwich | Immunoglobulin-like | Immunoglobulin-like |
#chains in the Genus database with same CATH superfamily 3B2P A; 4OU3 A; 4E36 A; 4L2L A; 4QPE A; 5CU5 A; 2XPY A; 3FTV A; 4XMU A; 4FYT A; 4XND A; 3B37 A; 3FTU A; 4K5O A; 4K5N A; 4XO3 A; 4PVB A; 5AEN A; 4JBS A; 3EBI A; 4R5T A; 2XPZ A; 4WZ9 A; 4MKT A; 4XN8 A; 1HS6 A; 3FUI A; 2DQM A; 4XMX A; 5J5E A; 5FWQ A; 3FUK A; 4KXB A; 3Q7J A; 4R5X A; 5AB0 A; 4KX7 A; 4QHP A; 3FUF A; 3CHS A; 4FKH A; 3CIA A; 3FU5 A; 4F5C A; 2ZXG A; 3FTS A; 3FH8 A; 3FUN A; 3PUU A; 4ZW7 A; 4XN5 A; 1Z1W A; 4P8Q A; 3FUD A; 4J3B A; 4ZW6 A; 4XN1 A; 3FUM A; 4Q4E A; 3FTW A; 3FUH A; 4FKE A; 3FUL A; 4QIR A; 4ZX6 A; 4XMT A; 3CHO A; 4DPR A; 4QME A; 3Q44 A; 3EBH A; 4RVB A; 4QUO A; 4KX9 A; 4KXA A; 3B7U X; 4PJ6 A; 3FTX A; 5BPP A; 5DLL A; 4XNA A; 3U9W A; 2HPO A; 3FTY A; 4XMZ A; 3B7T A; 2XQ0 A; 4XMV A; 4XO5 A; 4KX8 A; 3FHE A; 3B2X A; 4KXC A; 4ZX5 A; 3CHR A; 3B7S A; 2GTQ A; 3QNF A; 4MS6 A; 4FYQ A; 2HPT A; 4RSY A; 3B34 A; 4NAQ A; 4Z7I A; 4ZW8 A; 3SE6 A; 3FU6 A; 3Q43 A; 3QJX A; 4XN2 A; 4XMW A; 4NZ8 A; 4PW4 A; 4XNB A; 1H19 A; 3RJO A; 4ZW3 A; 4HOM A; 4X2U A; 2R59 A; 4K5P A; 5C97 A; 5J6S A; 2DQ6 A; 5LG6 A; 4Q4I A; 4ZX4 A; 3B3B A; 4XO4 A; 3FU3 A; 4R7L A; 3FTZ A; 2VJ8 A; 3FH5 A; 4PU2 A; 3FUJ A; 4GAA A; 4FKK A; 4ZQT A; 3KED A; 5AB2 A; 4K5M A; 4KXD A; 5DYF A; 1GW6 A; 4FYS A; 4XN4 A; 4FGM A; 3B7R L; 4XN9 A; 5LHD A; 3MDJ A; 3FH7 A; 4ZX3 A; 5K1V A; 5LDS A; 3FU0 A; 3FUE A; 4ZW5 A; 4FYR A; 4XN7 A; 3EBG A; 1Z5H A; 1SQM A; 3CHP A; 4K5L A; 3T8V A; 2YD0 A; 3CHQ A; 4R5V A; #chains in the Genus database with same CATH topology 3O0L A; 1UOK A; 3I6S A; 3L4W A; 5E4O A; 4X6B A; 3SO3 B; 4OJY A; 5BVP L; 4HFW A; 1PFC A; 1XTM A; 5IHB A; 3PIQ A; 3AS2 A; 4G8G B; 3LQZ B; 4YPJ A; 4G8G E; 3NQY B; 2OJE A; 4OGY L; 1H0D B; 4UNV A; 5JW4 M; 2NW3 B; 5I6X C; 4JO8 A; 3FB0 A; 1C08 B; 3MGO A; 2X4N B; 3HW7 A; 1Q9W A; 4MXV E; 4J3U A; 4IOS D; 2NY1 D; 1ZHK B; 4GG6 B; 3RY5 A; 4MDR A; 1J10 A; 3OKN B; 2WNW A; 3LKS A; 3A21 A; 3DUS A; 3DX9 B; 4NNP L; 2CLR A; 2VC2 L; 4F15 B; 5TDN B; 3A6B H; 4J23 A; 2X4U B; 4XWG H; 2YEZ B; 4X5Q A; 2YA1 A; 2ARJ B; 4U1N B; 2ID5 A; 4BCX A; 1T22 B; 3SDC C; 3T3M F; 4TUK H; 1BG9 A; 5E70 A; 2QVF B; 3VXM A; 1MCE A; 1AS7 A; 3FFC A; 3SE9 H; 2YKL L; 5D1Q C; 5KSA D; 3CZL A; 4QXU L; 4D0C B; 1DDT A; 2C3X A; 1ZSD B; 2NOY A; 1MNU L; 4UB0 H; 3DK0 A; 5KJ4 A; 4N2N A; 2ZOK B; 2E27 L; 4MD0 A; 5DD6 B; 4LKC B; 1G7I A; 3NQX A; 5DR5 H; 3L95 B; 3VD2 A; 1F00 I; 4KY1 H; 1QFW I; 4DQO H; 2F18 A; 5BZD A; 1JFQ L; 1K8I A; 4YDJ A; 4ZFG L; 3LMJ H; 3EF7 A; 4ONF L; 3ZKN C; 4H8W C; 2WAN A; 3RVX D; 1RJL A; 3NIG F; 3K1D A; 2QKS A; 2ZOT A; 3DA0 A; 3NCC B; 1W6W A; 4UT9 L; 4FQC L; 2VER A; 4TQ8 A; 4KPH H; 4HOM A; 2PUQ T; 5C3L D; 3C8K A; 2RIK A; 1I7U A; 5FSJ A; 1RIH H; 4N25 A; 4JO3 L; 5A3I D; 2BOL A; 3QH3 A; 5K9O B; 2IBB A; 3KBE A; 4GRL D; 2ZAH A; 4D8P A; 3VRI B; 4MCM A; 1E30 A; 1PSK L; 2H45 A; 1NAK L; 4YE4 L; 1I1R A; 1NGZ A; 4JO4 L; 1BXV A; 3NAA H; 4M03 A; 3PWL A; 5LJY H; 1FOR H; 2HJK B; 3T6W A; 5EOR H; 5L0Q B; 4JM2 C; 3GBD A; 1OOW A; 2CUM A; 3C60 C; 1D3C A; 5FCU H; 2F08 A; 1NBY B; 1HIM H; 4ZFO B; 2YAE A; 3ELQ A; 4KUC D; 1IAO B; 1GTN A; 2PXY B; 2ATA A; 5K7T A; 4OZI A; 5BW7 C; 5IJD C; 4GAG L; 5DQJ B; 3CXH J; 3CCH A; 5FW7 A; 4LLD B; 3LRS B; 4L8C B; 5BO0 D; 4BZ2 A; 1U93 A; 5KVG E; 2EAD A; 3JWD H; 2Q10 A; 1SBB A; 3QCT H; 1CO1 A; 3BHB B; 3MLR L; 3S2C A; 3V6O C; 2DKS A; 1UB6 B; 4JAM B; 1M42 A; 2IAN D; 3MND A; 4XML H; 5LCV A; 4AK1 A; 3MFF B; 5C0N C; 5DQD L; 3G08 B; 1EGJ L; 4AOF A; 2DQH L; 1HQ4 A; 1AC0 A; 1PX3 A; 1UFU A; 5FUC E; 1WV7 T; 5PCY A; 3KPQ A; 2WLL B; 1CF8 H; 3KPS A; 4MQX C; 4DK3 A; 2HFF B; 4Y1T A; 5BRQ A; 2EC9 T; 1U92 A; 4NLH A; 4Q9B A; 1BOG A; 4XNM A; 4NJ9 H; 1MCH A; 5IHZ A; 4FFW C; 2YQ3 A; 1UA3 A; 1UZ6 E; 1VCA A; 2DLT A; 5F8Q C; 3BSZ A; 3P9L A; 3Q8E A; 3WFD H; 4YK4 B; 4DGV H; 3OGO E; 1XSJ A; 3TWC L; 2C1P B; 1LE5 A; 2O6D A; 2IXQ A; 5E9D B; 3MLT A; 1A4C A; 4C54 A; 4X9Y A; 5E9D E; 2C1O A; 4K71 B; 3FXP A; 4PSR A; 5JHL H; 3D7P A; 1ETJ A; 2RA1 A; 3OWE A; 2MPV A; 5E00 B; 4Z3T A; 1PPI A; 5DFT A; 43C9 A; 4W1T A; 5CIP B; 4XPB H; 1KGY E; 1T0M B; 2DBL L; 3T0E D; 4R26 H; 1JOI A; 2CG9 X; 2NNR A; 3FQX B; 2X42 A; 3EJP A; 3RB5 A; 4FX5 A; 4WW2 A; 1SLX A; 4PJ9 D; 4ES8 A; 2A6D A; 4I9W D; 4JFP A; 1IEB A; 2ADF L; 4DP4 X; 4HZL B; 1F02 I; 1MJU H; 4CGU B; 5AVT B; 4LST L; 4MAU H; 4UTB H; 1ZT4 A; 1H3U A; 3DMM C; 4K2U L; 4PW3 A; 5J11 C; 1LO0 L; 4IKK A; 1D5X A; 5JQH C; 5KTE L; 4YFE A; 1E8Z A; 1E2W A; 4OUU A; 5FGB A; 3KCW A; 4YSR A; 1OAZ L; 1BZ7 B; 2R29 A; 1XHH A; 1Q9O A; 1VB9 A; 3NCM A; 2BX5 A; 2R0W L; 3FSO A; 3WHX B; 4KK5 C; 1KV3 A; 2FWS A; 1ZMY A; 3D4G A; 3ZHL A; 1TRL A; 2GY5 A; 3FT2 B; 3L9B A; 3QJU B; 3QZM A; 3L9R A; 4TUU A; 2GW5 A; 3CZE A; 3T2N L; 3V5D A; 1G6V K; 1XQ4 A; 4KKA C; 1JV2 A; 3ETT A; 5IMM A; 1FZO B; 2CO7 A; 4NO0 B; 5F1S A; 4FLQ A; 5JLZ B; 1XF2 A; 2DQG H; 4HLJ A; 4RRP G; 2VC2 B; 4HFU L; 4HIX L; 3S8W A; 5C9J A; 4W9G C; 5DFA A; 3DRO B; 2ZUW A; 5G5D B; 3VXQ A; 4EXP X; 3RWD B; 4F7T B; 1JHT A; 1CDY A; 3K6S A; 1G9N H; 5DGR A; 3JV5 A; 4XGZ B; 3R4R A; 4OCX L; 4Q97 A; 1VLX A; 3CG8 A; 3B7R L; 2MQG A; 2OL3 A; 3IJS B; 4PB9 H; 3KXF A; 4ERS L; 5IEH B; 4DGY L; 4RFO L; 1SEB A; 3QCU L; 5GGT L; 4Q56 A; 1S4I A; 4YHN A; 3ROL A; 1QHP A; 4EZJ A; 5KBM A; 4NKI H; 4DHL A; 4K94 L; 3IDJ B; 1LDP H; 4JRX B; 3UAJ A; 5KAQ F; 5J6H A; 2P8M A; 3IDN B; 4JRX E; 3IAQ A; 1VGE L; 1TFH A; 4M7K H; 2H2P D; 5E8N B; 4PV9 B; 3WFZ A; 4HD9 A; 4ODU B; 1WDA A; 2VIS A; 1SY6 H; 1GPO H; 1ZTM A; 3STZ B; 3PHO A; 1DUY A; 1UXW B; 4QHL A; 3GRW H; 5LKN A; 3FT4 B; 1EXG A; 4JFD D; 4A38 A; 4DUV A; 1I3R A; 3NFP B; 4RMB A; 3FUI A; 4NPY B; 3OM3 B; 4H26 A; 1UQS B; 5EIQ A; 1EBP A; 5CGY C; 5J06 A; 1FRG H; 2P47 B; 3HKZ A; 2IAN E; 3FBQ A; 4O02 A; 1IQD A; 4L4T D; 3ILQ D; 2GTZ A; 3BGF B; 3EFF A; 3ZUC A; 1CPU A; 1BYO A; 1MWA A; 4LCW D; 4D0B A; 4DTG H; 3PNR B; 3DOI A; 4ADI A; 1BZ9 A; 1Y7V A; 3N21 A; 4P4R B; 4UAO C; 1N4X H; 4GER A; 1BSI A; 2BNQ A; 3ARP A; 2P45 B; 3ESN A; 1NOA A; 3FO2 B; 3VWJ D; 2P5W A; 4I77 H; 2QFP A; 1VAC A; 2QV2 A; 1TZ8 A; 1RUL H; 5B4M D; 4AJW A; 2ENQ A; 5E5U B; 4QY8 A; 4RRP M; 4TLS A; 2BVO B; 4XCN A; 5FKP A; 5JDS A; 3UZV A; 4X6F B; 1X9U A; 3GJF A; 4YDV A; 4WU7 A; 5AVW B; 1FIE A; 3ZIM A; 3PCY A; 4GUP A; 1E5Y A; 3JTB A; 4OM1 H; 1SMO A; 3OQ3 B; 2K9U A; 3ULV C; 1UM5 H; 3Q6F A; 1DE4 A; 1OY3 B; 2BW4 A; 3WLW D; 3OF6 D; 3QEG H; 4WI7 A; 4Z3E A; 5LXV A; 1UUR A; 3HI6 H; 3SSG A; 3LSO B; 3VAE A; 1TLK A; 4NHC H; 3O41 B; 4AY0 A; 1QRN B; 5IMK B; 3UYR H; 2WXL A; 2V7H B; 1C8Q A; 5F72 S; 5EO0 A; 2J1K A; 4ZW3 A; 4Y4H C; 5GS0 C; 5TDO A; 3TO4 D; 4YR6 A; 4XBP B; 3CUP B; 5DQ9 B; 4A2E A; 4DVB A; 5MEN D; 2PR4 H; 1IX2 A; 3TEU A; 5IGX L; 4HAG A; 4Z7U A; 2VDN H; 4AKQ A; 3ZKM C; 3V8C A; 1Y18 A; 3GSX A; 1RUK H; 2BS7 1; 3QWR D; 1QSE D; 1MVU B; 1NCN A; 5DZ9 A; 1OVZ A; 5FPI A; 2J88 H; 3CV5 A; 2APB A; 3HCV A; 3K1K C; 4DGU A; 1R0A H; 2JI3 A; 4XMP L; 4KHX L; 5BZW B; 1BQS A; 3FOL A; 2QSV A; 3BUY B; 5BRZ A; 4CMM B; 4F58 H; 5IND B; 2FEC I; 2EDX A; 4JDT H; 5AAM A; 1RUP H; 4ZUT B; 3HS0 A; 4DK6 A; 3B6S A; 1NGX B; 1VAH A; 3NJL A; 4F57 H; 4JFS A; 4KV5 F; 4IIQ B; 1R19 A; 1K3Z A; 3UC0 H; 4L1D A; 1YNK L; 4Q52 A; 2XBZ A; 4KO7 A; 1C16 B; 5DFV D; 1IM9 D; 1V7N L; 5IVZ A; 4ORG B; 4NI3 A; 4KDT C; 2BP5 M; 1CLY L; 5KSU A; 5G2N A; 3ZYJ A; 1N19 A; 4G8I A; 5FYK H; 3ZO9 A; 1TDQ A; 3OBA A; 4AQ1 B; 3O4L B; 5EBM A; 2ENJ A; 1LEK B; 2R23 A; 2XZC H; 2MQ3 A; 4IS6 A; 3ARB D; 4FQI L; 3RL1 B; 1E4W H; 3TVM B; 3WS3 B; 4CSS A; 1DUZ A; 5EU3 B; 3PDD A; 3WXV H; 5CGM A; 3TL5 A; 1SLW A; 1ZVS A; 2H1X A; 2VLR A; 3JWO C; 3NJM A; 5KN5 A; 3L6F A; 1JHK L; 2EDO A; 3CFJ B; 5FYJ E; 5IIE A; 3QNZ B; 3DOH A; 3QDJ A; 1FO0 H; 4PJD A; 5A6M A; 5T1M B; 3X3F H; 2WYT A; 1UM6 L; 1I4F A; 3QYN A; 1I7R A; 1EAQ A; 1I1A D; 3QHT C; 1DZ0 A; 1AIZ A; 4LCW B; 3MLW H; 2EO9 A; 2OR7 A; 3D2U D; 4LCW E; 4OKV B; 1JYX A; 4Z7W B; 5GIS L; 1KWK A; 2Z35 A; 3EGX B; 5DDH A; 4Z7W E; 3RIA F; 2XN9 A; 2JLP A; 5F1O B; 2HPO A; 5TFS L; 3BKM L; 4KRP D; 2JIX A; 1E9O B; 3O81 A; 1G9M L; 5IBL C; 1M05 A; 3VWJ B; 4U31 A; 4LSQ H; 4E9R A; 2AXF A; 3VWK A; 2Q3X A; 3Q13 A; 2FYV A; 1U42 A; 5E2J A; 2WK2 A; 2APW A; 1X5M A; 2YEQ A; 4G5Z L; 1E9P A; 4NEN A; 4OW3 A; 3I75 B; 5EBW A; 3F28 A; 4FFT A; 4L37 A; 4Y2O B; 1OAX L; 4AQ8 C; 1WUZ A; 2JSF A; 2TLI A; 2VQ1 B; 1KCR L; 4I1N B; 3ZL4 L; 2OSY A; 1JZ8 A; 2DH3 A; 3HMX A; 5GRV L; 5LG6 A; 2XV6 B; 4WFH D; 1PLG H; 1S9Y B; 3D05 A; 5FDY A; 4KGJ A; 3U36 A; 4AIE A; 3L5H A; 5L3U E; 1CE6 A; 5GPR A; 3BAE L; 1NCU A; 2Y7M A; 4MJ5 A; 4WK0 B; 3GML B; 1HE8 A; 3T4A B; 5D72 H; 4F9L C; 4WEM B; 5I2I B; 1HQR B; 3QUY D; 5MEN E; 4N2A A; 2B3R A; 1CFT A; 3Q9P A; 3IJ9 A; 2AK4 A; 1EXH A; 1Z6J T; 3W3B A; 4DP2 X; 2GFB B; 3RU8 H; 2YWY A; 3T4Y B; 1E5A A; 2W1N A; 2NY4 C; 3KWW A; 4MZ2 A; 1AGD A; 1PLA A; 4NT6 A; 1CG9 B; 4MMY A; 3QDH A; 1NMA H; 2OS7 A; 3HE6 D; 3CDF A; 2DL9 A; 3A6O A; 4OQT L; 2PWH A; 2WTR A; 3QZW G; 3G5X A; 2AQS A; 4L1L A; 4WO4 D; 5GS2 D; 4JFF A; 4QOK D; 4LPT A; 4NZT L; 1IL1 B; 1W72 A; 2BSD A; 3KGS A; 5K93 A; 5JPN C; 4QRR D; 2JXM A; 3N2J A; 1ADW A; 2RMN A; 3K2U H; 4X4Y B; 4IKI A; 1BZD A; 2R56 L; 3BH8 A; 4PJF A; 1ZLS H; 4JRF A; 4OZF H; 2R4S L; 2MCM A; 2Y72 A; 2YK1 L; 3W0W B; 3W0W E; 4R4F L; 3O11 B; 2JCC B; 1U8I A; 3CZF A; 2JCC E; 2HH0 H; 2JI2 A; 3NDY E; 5ESA B; 1YEJ H; 1QFO A; 4HH8 A; 4FJ6 A; 4KK8 D; 4WI5 A; 2BY2 A; 4XH2 A; 4K8R C; 4E0R B; 3V5K A; 4G1E B; 4EZK A; 3N9G H; 3LRK A; 3QG6 B; 4YOK A; 4MS8 D; 3WEL A; 4I3S H; 3PV6 B; 3QXW A; 3Q2L A; 2VDQ H; 5EZJ A; 4ZXB B; 1QBL L; 1A3R L; 2WYI A; 4QXG H; 3HN8 A; 1AR1 C; 3DTD A; 2FD6 L; 4BDV B; 2EDW A; 1EXU B; 5F21 B; 5SWS D; 5I1J L; 3FQW B; 1UKT A; 2G30 A; 4N24 A; 1XR8 A; 1IKN C; 2B7D T; 1RK7 A; 5B38 G; 5AVR B; 5I1H H; 4QT5 A; 1F6F B; 5I6M A; 4TS6 A; 4JAN A; 2OMW B; 1XWV A; 1I1A B; 2LOE A; 3VFG H; 2N3J A; 4NZU L; 1FC1 A; 1OAR L; 5GGU A; 4XTR C; 4A6Y H; 1QPX A; 2WOY A; 3AZU A; 4RMR A; 1T6V N; 4FLS A; 4ZAK C; 1ES0 B; 1QLE B; 3PP3 H; 2DQT H; 2HMI D; 5E6I C; 3ZR6 A; 4HXB L; 1S7T A; 3ZTN H; 2OTU B; 2LVC A; 3IVK A; 2H32 B; 1RK1 A; 1OGS A; 3EYO A; 1F42 A; 3D39 A; 4JZO B; 5IB2 B; 5JR1 L; 1I9R L; 3QFD A; 4HH9 B; 3VH8 G; 1Y6N R; 5C8J B; 3FRP A; 3SPV A; 5HYQ A; 2WWM C; 3IK2 A; 1N0L B; 2EKS A; 3WN5 A; 1VGK B; 5DD0 H; 3VXN B; 1S7S B; 1NIA A; 1ZT7 A; 3CJJ A; 4M5Y H; 3IXT B; 2ZUV A; 4PRD B; 1A9E B; 1JV5 B; 4DZB A; 1OPG L; 2P80 D; 3OB0 H; 1GGI L; 3ABL B; 5D6C B; 3TID A; 3L8Q A; 1FFT B; 1N2R A; 5IF0 B; 3ZW3 A; 3GQF A; 1U2Y A; 4TMN E; 1NMB H; 4WFH E; 3QCV H; 1A6T B; 3QRF M; 3TJP A; 3C6L C; 1J9Q A; 4FFV D; 2BFV H; 5L4M A; 5E56 A; 4YLQ T; 1F2X K; 4LAS L; 4AMK H; 1QTP A; 3QUY B; 3X1F A; 2B1H L; 3VRL F; 1MXG A; 5GJT L; 2FJF B; 2VKW A; 2J6E H; 4GRW B; 2XNA B; 4YIQ A; 4MJH A; 4GRW E; 3ESO A; 4M4R B; 3W2D H; 1FFP B; 3VD9 A; 4BH7 A; 4JO2 L; 2J5L C; 1YEI H; 1AXS A; 5J89 A; 3HB3 B; 1TNM A; 3W37 A; 1QO3 A; 4YSQ A; 4H88 H; 2Y25 A; 4WW2 F; 2HCZ X; 3SSB A; 3GBM H; 2F7Q A; 4A39 A; 1IAM A; 5DHY A; 4MLI A; 2W5P A; 4A2F A; 4XOE A; 5E8D H; 3VFW B; 3IKK A; 4TQI A; 4MA3 A; 4YLB A; 3N91 A; 1KIP A; 2X09 A; 3V4P B; 1RMF H; 4M62 H; 4PV8 B; 1I75 A; 4ICW A; 4ALA C; 4TSB L; 2O61 B; 1EHN A; 2CNY A; 3SOD B; 1ZHB B; 3CSP A; 4F5C A; 4BEI A; 2Z0U A; 3CMO H; 3VFU A; 4Q9Q L; 3MBE B; 1HKL H; 5EDS A; 1IBG H; 3MOB H; 4NOB A; 3DXA D; 1U95 A; 4LEX H; 4NQE A; 2RGS A; 3MSA A; 5DTF B; 5CSZ B; 1P7B A; 1K4D B; 1YYM H; 2BC4 B; 4JRW A; 4HET A; 2AJ3 B; 3U79 A; 1U8J B; 2NMS A; 5IRO C; 3FDW A; 4A68 A; 1L0X A; 1T80 A; 3TJE H; 3OAW A; 4NKD B; 4RMV A; 4F33 B; 3PDG A; 1A6A A; 3RHW K; 3HA0 A; 4H57 A; 1A3L H; 5I8C A; 4PJX F; 4NBZ B; 1MH5 B; 3MGA A; 2WP3 O; 3KGU A; 4OF3 A; 1DVQ A; 5SWS E; 1TO4 A; 2ARW A; 3V6Z B; 3PL6 A; 1OCC B; 3P77 B; 1CVR A; 3WEO A; 5DK3 A; 3NIF B; 3OPU A; 5A7B A; 3VE0 B; 4S1R H; 2YN5 A; 3OAU L; 3VD1 A; 4PG2 A; 4OB5 L; 3GBE A; 4KI1 A; 5H4Y A; 1F56 A; 1A02 N; 1CL7 L; 4N87 A; 4MQ7 B; 1LHZ A; 4KXZ H; 4XI5 D; 4IRZ H; 8TLI A; 3PG2 A; 4B27 A; 1BEY H; 4GG8 B; 3PFQ A; 4GT7 A; 5HPO A; 1X9L A; 2GAZ B; 4Z0B H; 5EO1 B; 5IB4 A; 3ZMN A; 3WV0 A; 2EH8 L; 3DYP A; 3FYE B; 1WGO A; 5HJB A; 1UAC L; 1W8E A; 5JUE H; 1WFI A; 2GAB A; 4GAA A; 3L7E A; 5I5J A; 1SHM A; 3VXS D; 3COO A; 15C8 L; 2UUD H; 1SXA A; 1JF6 A; 3MRC B; 4AJY V; 4M61 B; 4XP5 H; 4ZFF B; 2A74 B; 5LWY H; 3S36 L; 4Z1P A; 4OR1 A; 2L3B A; 3MV8 D; 1JXF A; 3QZN A; 5IY5 B; 3ARF A; 4WWN A; 4D0D B; 2J1Y A; 1SYS B; 2AV1 B; 5SX4 H; 2VLK A; 1KIQ A; 2C3V A; 3SGJ A; 3CDY A; 1U8K B; 1QR1 B; 3WGU B; 4LDC A; 2IAL A; 35C8 L; 1EMT H; 2A7B A; 4NRZ A; 1A0Q H; 2F7R A; 3NBN A; 5EN3 A; 1FLT X; 1KJ2 B; 1EEZ B; 4UTB A; 3QS3 A; 5DS1 A; 3UO1 H; 4GQ9 H; 2NT0 A; 3H6J A; 3PS6 A; 4NC1 E; 3TBS A; 1JTO A; 2OQJ B; 4QAM B; 3VGA B; 1OGA B; 1OGA E; 2H47 C; 1ZXK A; 3IE9 A; 4ZG1 A; 4EOW L; 1LDS A; 4KJQ D; 4XPT H; 1JPF A; 4JO1 H; 5EZI H; 5D7S H; 3BP5 A; 2IQA A; 3P7W E; 2GSI A; 3QNY B; 1EZV X; 4ODS L; 5AB0 A; 3KDP B; 3QDG A; 2JKN A; 2Y3A A; 3QQD B; 1A22 B; 3CIA A; 3LS4 H; 1TF4 A; 2VT0 A; 2Z31 B; 3DUR B; 4PI2 A; 3DIF A; 4O5L L; 3UAA A; 1KJP A; 1FAI H; 4Z7A A; 3NCB B; 3KCP A; 5B7E A; 2XTJ C; 2VXU L; 3MA7 A; 4Y7O C; 3VI3 A; 2PPD A; 1ETB 1; 4GQZ A; 4UUJ A; 1G94 A; 3KS0 H; 4XXC C; 1TJX A; 1E90 A; 5FK9 A; 3Q6G H; 3EOB B; 3U2S A; 1MI5 B; 3BP7 B; 4Z7U F; 3MUG A; 1MI5 E; 2J21 A; 5BPP A; 1J1O L; 2PWF A; 3QFJ D; 4IM8 A; 3VDB A; 2HLA A; 1VFM A; 4XVU D; 1T6W A; 2MKW A; 4WNS A; 1F4Y L; 1GIG L; 1Z5L A; 3S88 L; 4P5M B; 3F64 A; 1E3X A; 3MNV A; 1RUM H; 4FQP A; 1U30 A; 1BFT A; 4HUN B; 2WVS A; 2FAT L; 3N40 F; 2IH3 B; 3MLX H; 5F0M B; 3ZHN A; 3RWJ A; 1OIO A; 2H1P L; 5ECG A; 2QDW A; 4U1M B; 2VIT B; 1AI1 H; 4PGE A; 1P5U A; 5LAX B; 2P5E D; 2CLZ A; 1UWX K; 4OIA A; 4K90 A; 4RQQ A; 5D1Q D; 3WL9 A; 5DJD B; 4HEP G; 2Q0Z X; 3WDH A; 2A9A A; 2FED D; 3PGD B; 1GJI A; 4CU8 A; 2A83 A; 4LFH G; 4RSV A; 2WII A; 4IKL A; 2XWJ B; 1NLB L; 2WBJ A; 3FO1 A; 5EU6 B; 1YEF H; 4D9Q D; 3SGK C; 4E9T A; 4L4V A; 3L81 A; 4NZJ A; 3SDA B; 2NXY D; 2XID A; 2E6J A; 2YFN A; 2DM3 A; 3FQN B; 4LLQ B; 1S7V A; 1MZZ A; 3ARG B; 5DWU L; 3AMK A; 2GIX A; 3BUI A; 2J1W A; 3QJS B; 4L9D A; 1ZLU K; 1CD1 B; 2IQ9 L; 4NQX A; 4IBZ A; 5GHW L; 3BVT A; 5EPM B; 2AJZ B; 1RZP A; 1BFS A; 2E59 A; 3MCK B; 3N42 F; 3MME B; 1TIU A; 2H1L M; 4QRP A; 3NFS L; 1UT2 A; 3DGC R; 3VFM A; 5DRZ A; 4Y5V A; 3VXS E; 1HYS D; 4MJI A; 5KYU A; 3IM1 A; 4GB9 A; 1MDA A; 3DAR A; 1FJV A; 4K9J A; 4ZFZ B; 4CU6 A; 3B2U C; 4HA1 A; 5FYK U; 5F7Y C; 1YYC A; 2I7O A; 3QBT B; 1BFV H; 1T5W B; 1MCO H; 4X6D A; 1IRA Y; 5IML B; 4CC4 C; 2D73 A; 4NQU A; 3N43 B; 4OCN C; 1T04 A; 3LIZ H; 3C6S B; 3IFL H; 4UQD A; 1CJY A; 1JWS B; 5TGB A; 4B0M M; 4P5T A; 4N1C A; 3QUX D; 5C0C B; 12E8 L; 1X5K A; 5C0C E; 1V3J A; 4ZZK A; 3UTQ A; 4FQH A; 2C9S A; 1EV2 E; 3MRE B; 4TTD D; 3U9P L; 2PW2 B; 4JM4 L; 4HUV A; 1U3Y A; 1IUZ A; 4HWZ B; 4UM8 A; 1ET8 A; 2XAC C; 4EDW H; 1K4C B; 5TJW K; 3NA9 H; 4J6R L; 1ZAG A; 3P0V H; 1TYE B; 3P4N A; 1HEZ A; 2GPZ A; 2P48 B; 1L0Q A; 2P8L A; 2V17 H; 1VDG A; 3QH5 A; 1PG7 H; 2PKD A; 2E86 A; 1JVL A; 3D25 A; 1FIG L; 1BMZ A; 1U3H C; 4KXZ I; 3WNK A; 4OII H; 4HJG A; 3P11 H; 3C8D A; 3CVH L; 2HRG A; 4WEU D; 5JOR B; 1TLI A; 5ADO L; 5JXA H; 2EQD A; 2VUK A; 3Q2C A; 4LCI H; 4M7J L; 1OSZ B; 4U6Y A; 4U1H B; 3LKN B; 1AKJ B; 2NX5 D; 2OJZ L; 4W70 A; 5FOB A; 3B2X A; 1KB3 A; 3TCL B; 5F7H A; 5ALB H; 3DET C; 1VF5 C; 1NCB L; 2NY0 B; 4U0R B; 4Y4F A; 3H55 A; 4LKX A; 4HJ0 D; 3QFJ E; 4PJE F; 3WFH A; 5JHD D; 2NSM A; 5I9Q B; 2MNU A; 3QJX A; 2FB4 L; 1ZJB A; 3SJV A; 1I7T A; 4HX1 B; 4QF1 A; 4YX2 L; 4IHB A; 4CDH A; 2DMG A; 3NJV A; 2VXV H; 4P2R B; 2YWZ A; 1BZQ K; 2DWS A; 5DZN A; 3DVF A; 2VXT H; 3LEX A; 5SX4 I; 4JN2 B; 1KG0 A; 3CN3 A; 3MRQ B; 3H3K A; 4CNI A; 4ZBJ A; 5IV2 A; 3RVW D; 2CIK A; 4HWE H; 5K9J H; 3D12 B; 5T3X L; 3L9E A; 3WNP A; 4BH8 A; 2P5E E; 5TPW H; 4PJF F; 4ZSO A; 1FE8 L; 1KEN H; 3BZ5 A; 3OWR A; 5DHV H; 1RZI A; 3EH5 B; 3D0L A; 4GRM B; 3IET A; 2ATP B; 4OI5 E; 1UYW L; 2FO8 A; 1MFD L; 4WW2 C; 4K5M A; 1FO0 A; 1WA0 X; 2GT9 B; 1KPR B; 4EE2 A; 4KKB D; 1B4T A; 4D9Q E; 3WFC L; 1NLD H; 2YA2 A; 1TMQ A; 3MSF A; 4N8U A; 3T09 A; 2OPC A; 4MX7 A; 3RVV D; 4DN3 H; 1T3F B; 5E1G A; 1NKG A; 1SJE B; 4A1R A; 4LL1 A; 3AWG A; 1IC7 L; 3G6D L; 3O2D L; 1EAJ A; 4D3C H; 5C71 A; 3BAX A; 3QXD B; 3WY2 A; 5E6U B; 4HS8 H; 3H42 H; 1HX0 A; 3RNQ A; 2V0A A; 1XR9 A; 1MHP H; 1MAJ A; 3MJ9 L; 3B9D A; 4NRX B; 1REI A; 5T3X D; 5FEB A; 2X4T A; 1DCL A; 5D1X A; 4XCC L; 4P3C H; 3GHB L; 5IOP A; 5IB5 A; 1XH0 A; 1XO8 A; 4R90 H; 1AZR A; 3BWS A; 3E6H B; 3GSO B; 2JV6 A; 2W07 B; 3OKD B; 5D55 A; 3TV8 A; 1AHM A; 1AS8 A; 3UPA A; 1YQ2 A; 2V9U A; 1G6R H; 3FSW A; 3LH8 A; 3DGG A; 1O6S B; 4RBP H; 3OKL B; 1S9K C; 1I8M B; 4GKN A; 1JPG B; 4GSU A; 4K7F B; 3MRG A; 5F93 C; 5KBD A; 2WBX A; 1DBA L; 3RFZ A; 2FNW A; 4JFZ L; 4NZD A; 3NOI A; 5B39 B; 1UJ3 A; 1OP9 A; 5EEY A; 3NQK A; 1UWE L; 4MNH B; 3GNM H; 4LSR H; 4P57 B; 1DQT A; 2XII A; 2BHY A; 4MA1 C; 4DCQ B; 3RWE B; 4N6R B; 4DP9 X; 1ZE3 H; 4AM0 B; 5T3Z H; 1CD0 A; 1DQV A; 4R9H A; 5I8E A; 5FSS A; 4ONG H; 4QUO A; 1YNL L; 2BWD A; 1A7B A; 2VM3 A; 4N2B A; 1EO8 L; 3B7U X; 3HR5 B; 5KSB A; 1LEI B; 2CBM A; 5GRU L; 4M7Z B; 5DZA A; 3S3J A; 1FAK T; 2IDS A; 2Z9T A; 4WJ5 A; 4BFB D; 2DQD H; 4U2Y A; 3GSW B; 3QIW C; 2IEP A; 1BVL A; 2OSL B; 4XPF L; 1J8R A; 1Q94 B; 4QEG A; 1U6A H; 2PCF B; 1D9K B; 4CST A; 4Z9K B; 1WH0 A; 1KXT B; 3DX8 B; 5J1A B; 3IJE A; 3GMO A; 4M9P A; 4YHM L; 3M8O L; 4S11 A; 1RJZ B; 3H54 A; 4PM1 A; 5DJ8 B; 3SE6 A; 2VN6 A; 4XN2 A; 2AEP L; 5ESV B; 4Y16 A; 3WO3 B; 2BSR A; 3MNW A; 4L0Y A; 2NY7 L; 4N5E C; 3UZQ B; 3LD8 B; 3WY4 A; 5HJ3 B; 5JHB A; 3VXR B; 3PV7 A; 1IIL E; 3VXR E; 5C6P B; 3BWA A; 2FK9 A; 5DJ6 B; 1EHL L; 5IW6 A; 3X1B A; 2HX8 A; 3FQT A; 1JI1 A; 1GOH A; 2GIT A; 3QRG L; 5JHD E; 1YSO A; 3H0T B; 4U3C A; 4DEU A; 2OK0 L; 1PKO A; 2DLG A; 4QHU A; 3CDG A; 2VDO L; 1A1N A; 1KJM B; 2Z64 C; 3CVH B; 4RGM B; 2C3G A; 1FVD B; 4IRJ D; 1KNO A; 3JYB A; 2Q86 A; 2WSD A; 3KLQ A; 4ISV A; 2X4R A; 1ORS A; 4MMZ C; 4UYP A; 3SDX A; 1J5O H; 1EBA A; 5T1L A; 2BLF A; 2VSK B; 3CH1 A; 3T8D A; 2VW6 A; 4IML A; 5AEA A; 3RWC B; 3SY0 B; 3CLF H; 2ZIC A; 2LR4 A; 3NP3 A; 1CK0 H; 5ITF B; 3RZC B; 2CBO A; 1EAN A; 1NWO A; 1VIW A; 4HP5 A; 1IBZ A; 4OZ4 A; 4RAU A; 4XPG L; 1NEZ A; 4J3O D; 4G1M A; 2XA8 L; 4J6I A; 5F88 A; 3LNH A; 3MRF A; 2UZI L; 4YO0 A; 5JNY A; 4MAS A; 1OAQ H; 2OFQ A; 4NZR L; 2JQM A; 3EYF A; 4R2G D; 4X50 A; 1CIC D; 3UG4 A; 5EOT B; 1EDY A; 2AVF A; 4HGW A; 4P5R A; 1GAF H; 2ZQK A; 3L5X H; 4IMK C; 5FO9 A; 2UWE B; 5BVJ A; 2K3T A; 1QWH A; 2UWE E; 4OLE A; 3TPU B; 4WWK D; 1Z3R A; 2CKB A; 5DHX B; 4XMX A; 3DBS A; 3ALP A; 1QAW A; 4XE5 B; 2BWQ A; 2AJU L; 5ICY B; 1NCW L; 4E9X A; 1F8T H; 2GHZ A; 3SPI A; 2LU3 A; 3JWN E; 1I8K B; 5CZR A; 3V10 A; 2DYS B; 5I1G L; 4PUB H; 4Y2D B; 1B3I A; 4BXV A; 3L7F A; 3S35 H; 3JV6 B; 4OCY L; 3H1Z A; 1SLU A; 2W0L A; 5AAM C; 5ID0 B; 4YBQ C; 1NGQ L; 2GJZ B; 2QPS A; 1IKF H; 3FU7 B; 4MCZ A; 4PJG F; 3TV3 H; 3LZF L; 3MLS L; 3QK0 A; 2DWY A; 4FZ8 H; 2X2H A; 1U4G A; 2F5A L; 1J8H D; 1AP2 B; 5IW1 A; 5AYQ A; 5KVG H; 4FJY A; 3S96 A; 4P2O A; 5IG7 B; 3J2P A; 2XSK A; 1ZRU A; 1ICT A; 1K6Q H; 1GYW A; 1OTS C; 3N85 L; 3PAB B; 3PUC A; 4X8C A; 4LHU B; 3E3Q E; 5J12 B; 5DJY A; 2AJS H; 2WVV A; 3JVG A; 4HU8 A; 4YL4 A; 4J5V A; 4O4Y L; 2I27 N; 5F9J B; 2J6E A; 4FYQ A; 3A4D A; 1EZU A; 4N5P E; 1MRE H; 3ZT5 A; 1H3X A; 3PWV A; 1NDG B; 1RPU A; 3UYP B; 1XH3 A; 4O2C A; 3FQU A; 4KRO D; 3O6M L; 1HTY A; 4YBL C; 3ZR5 A; 1LVE A; 4AW7 A; 1DQL L; 4ETQ A; 2ITE A; 1HH9 A; 3QUZ C; 2BRR L; 4MJ6 B; 1ADQ A; 1TO5 A; 4YHL L; 3GXP A; 4I3G A; 5D93 B; 4D7B A; 4A2G A; 2FJH B; 1P7K A; 1PZD A; 3B3B A; 4ENE C; 1I31 A; 3PJS A; 3QHF H; 1L6Z A; 1T66 D; 3SQR A; 4B50 A; 1EFQ A; 1SDD B; 2NW2 B; 2C4X A; 3LZO A; 1HDM A; 2BSE D; 4B9J A; 2VMJ A; 2VR3 A; 3Q8F A; 4BQ9 A; 5I6K A; 1G4R A; 4PTU A; 4N9G B; 1MIE L; 2BST A; 4LAR L; 4Y7M A; 4ZI8 A; 1E3F A; 4CRP A; 2P52 A; 1R3H B; 2GCY A; 1NUG A; 4KMT L; 4ZPT A; 5HI5 D; 1JAE A; 1O9V A; 3QJH A; 4N2O A; 1ZV9 A; 3FH7 A; 4M8V A; 1GZQ A; 5B1K A; 4X80 L; 5BVP H; 3VI3 F; 4JEG B; 4JLR B; 3BZ4 A; 2OW6 A; 5GZN A; 2EDE A; 1JNL L; 2RB8 A; 5DRW B; 1IAI M; 3ZN6 A; 2QR0 B; 1DN0 A; 4HKZ B; 4F7E B; 4Y9G A; 4Z7O F; 5M95 B; 4OTX L; 3L45 A; 3Q3G A; 5MI0 B; 3QI9 B; 4PWF A; 4OGY H; 1DQJ A; 3PJN A; 4X0K A; 4XND A; 1KBV A; 4M29 A; 1LP9 A; 2X9Z A; 4EO5 A; 2UY6 A; 3SGD I; 3T3P F; 3ZHF A; 1NCJ A; 1G7P B; 5EU5 A; 4KXB A; 2VER N; 4NNP H; 3VCL B; 2VC2 H; 2OP4 L; 4EDX B; 3FU5 A; 4N66 E; 2HTL D; 1E9Q B; 1G7L A; 2O93 L; 2B3I A; 3AAZ A; 5KOV C; 1GRW A; 1YED A; 4KWU A; 1GYC A; 4P23 D; 3U4E B; 4CJ1 A; 3B2V L; 5D7I A; 2FO4 A; 1I8M A; 4X5X B; 5HVW A; 3QZW A; 5I71 C; 3C09 C; 4XC1 L; 1WIO A; 2YKL H; 3ZPD A; 3ARE B; 3V4L A; 4QXU H; 1H8N A; 1Z9L A; 3GMM B; 5L7X L; 2PTV A; 1MNU H; 5C0E A; 3CHO A; 2DH2 A; 2F74 B; 4HV8 A; 3B3I B; 2E27 H; 3AG1 B; 1TQB B; 4AYE C; 5LQB L; 1IFH L; 1YMH A; 1R34 A; 1X7Q A; 1JFQ H; 1HEH C; 2XX1 A; 4KKC D; 4ZFG H; 4G9D A; 4AIZ B; 1SJH A; 5L8P E; 1FN4 A; 4ONF H; 3V0V A; 5ABZ A; 5L2J B; 1DVF A; 1JZE A; 5FGR A; 4KVC L; 2F68 X; 1AC6 A; 1SN2 A; 4FQC H; 4NQC D; 1J8H B; 2K6Y A; 2VCO A; 4RMQ A; 2IH9 A; 4DZ8 A; 5GXB B; 1J8H E; 2C5D C; 3C8H A; 4JO3 H; 4S1D C; 3FU6 A; 4WI2 A; 3V6F B; 1JD7 A; 1L4I A; 5GGS B; 4EN2 A; 5KVE E; 1EAP A; 2AEI T; 2XWX A; 4LHQ B; 1DO6 A; 1AIF A; 1NAM B; 2XCM C; 4R96 A; 1AKP A; 1PSK H; 1ZCR A; 3PRE A; 5CFA A; 1NAK H; 5DJZ B; 3O6F B; 4JO4 H; 4W9I C; 4UDU B; 3SE3 A; 2C9R A; 4Z7Q B; 3KHO A; 5B38 A; 4X6D F; 4ONH A; 3QIB A; 1I8L C; 1U8H B; 4LSD A; 4Z7Q E; 4KRO B; 3RWH B; 5I1L A; 3TN0 B; 4JRL A; 1XB3 A; 5KYY A; 3SYA A; 4EQ3 A; 1J0Z A; 1QZN A; 3CN4 A; 3ISR A; 1TQC B; 1UD2 A; 3U6X S; 2J25 A; 4GAG H; 5LNG A; 4LRI A; 3P40 A; 4HOX A; 1WGV A; 4NO5 B; 5CZV L; 4ZD3 H; 2NPL X; 4IBU A; 2NY6 B; 1FL6 A; 2X89 A; 2C4F T; 3G6A A; 3LVE A; 2QKI A; 4BQB A; 4QRU A; 3V79 C; 3MLR H; 4YXK L; 1WBX B; 1PJW A; 3ASO B; 2ED8 A; 5LHD A; 1R75 A; 5BUA A; 5DQD H; 4DET A; 5ALC H; 5B7F A; 3SDY L; 3VH8 A; 1XBD A; 2DQH H; 3J8F 7; 2M83 A; 1A6V L; 5IES L; 2ALA A; 3PNW A; 4G8F B; 2G75 B; 5B1J C; 1DEE B; 3HKF A; 3V2A R; 3WUW B; 5DJ2 A; 3IU3 B; 2IPK A; 5A2B A; 1OSP L; 5MP6 L; 1ZT1 B; 3HN3 A; 3NES A; 4XPA L; 1BII A; 3ZBM A; 4DKD C; 3MO1 A; 1SXN A; 3QHZ L; 5EFI B; 3LYR A; 4P4K B; 5EZL B; 5CJX A; 3NVQ A; 4KAQ L; 1I8A A; 1RHH B; 4I5B A; 1B0R B; 4UZS A; 4UDT B; 1KR6 A; 1JCV A; 4Y5Y C; 3L4V A; 1JWM A; 3ZDY F; 1L6P A; 1SUJ A; 4BXS V; 4JZJ C; 3P7Q E; 2Z65 C; 4R4B A; 1JER A; 1P4L B; 3BQU A; 1V3M A; 5JPM A; 1Q0E A; 4N0U A; 1MZY A; 4WKZ B; 4Z78 A; 2K45 A; 1EFX D; 2P7T B; 2ADF H; 3NEE A; 3QQ3 B; 3X1N A; 3KS0 J; 4F9P C; 2Y7Q B; 1HJC A; 4Z7N E; 2OMY A; 1JVK A; 1TFP A; 2H2S C; 4K2U H; 2OXG A; 1LO0 H; 1N59 B; 1XIW C; 1XZ0 B; 2ICE C; 3LAF A; 3IJY A; 5KTE H; 2XQ0 A; 5HYJ A; 3BAY A; 1ML0 A; 4XO5 A; 1P7Q D; 4WK2 B; 1OAZ H; 4H25 B; 5IQ9 A; 2CBP A; 2WXI A; 1CBJ A; 3RPI A; 1CMO A; 1EHK B; 3DDG A; 3EYU L; 2HFE B; 5EUO A; 2DDQ L; 2F54 A; 2FZ3 A; 4LU5 L; 2YD9 A; 1T03 L; 1G4M A; 2I07 B; 4IBT A; 3TXA A; 4H13 C; 4LSV L; 3T2N H; 5IBT L; 1KIR B; 1HI6 A; 1UD4 A; 3TJH D; 1ZHN B; 4GUO A; 5FYL E; 3CSY B; 5EOQ L; 3HRZ B; 3Q1E A; 3OZL A; 1SN5 A; 2YFO A; 1KLU A; 2Y9R T; 5AG8 A; 5EV7 A; 1WD9 A; 2BNU A; 4KOB A; 1AQM A; 3LIZ A; 4WV1 C; 4CVZ B; 3N0A A; 5D6D C; 5GGR Y; 3SIK A; 4HFU H; 1S5H B; 1I8I B; 4HIX H; 5E2T L; 4C4B A; 4NQC E; 4GKS A; 2EDN A; 4PRP A; 1HYR C; 3FBO A; 3T1W A; 3MAW A; 5IJV A; 4BUH A; 4HUU B; 1GC1 L; 4A2L A; 1ERN A; 1PAZ A; 2WV3 A; 4Z95 H; 1U8M A; 2X7L A; 3KH4 A; 4M93 C; 3LTV A; 2AJY L; 4B60 A; 4OCX H; 5E1A A; 2MHA A; 4UIN L; 3LKR A; 1JYV A; 1SXS A; 4DGY H; 1NUF A; 2D3J A; 4IU3 B; 4XXD A; 3RVU C; 1IVL A; 4RFO H; 3SCM A; 3QCU H; 1SUH A; 2EDJ A; 5GGT H; 1GYA A; 4LCC B; 4ANX A; 4NXK A; 4H1L G; 4LQ1 A; 2Z3F A; 3FC8 A; 2PCP B; 4K94 H; 5KAN G; 1YPZ E; 3KPN A; 5F1N B; 4GAJ L; 4U1J B; 4DO6 A; 1VGE H; 4KU1 A; 5TDP B; 1Y1D A; 4WU5 B; 1VFB B; 4YHY C; 3JWD C; 4CC0 A; 2IF7 A; 1MCI A; 1ZHL A; 2JKL A; 4Z76 A; 1H15 A; 4R84 A; 3IXA B; 4W9J C; 5CO1 A; 5AVY B; 2CII B; 2ALW A; 3X11 B; 1QUN B; 3SQB B; 1U9K A; 1ETZ B; 5KSB F; 3FTU A; 1DJG A; 2QPD B; 4C4U A; 5KYN A; 1QSF B; 2IBG A; 4AYG A; 2QLV B; 3WEA A; 1QSF E; 1XCW A; 4JDV B; 3QR1 A; 4CKQ A; 4E2O A; 1K3I A; 2YGN A; 1XGR B; 4PJA B; 3W9E B; 5BUP A; 5CEX A; 2GTW B; 3B9K D; 4PJA E; 1RJY B; 4QRB A; 4R7V A; 4ANK A; 1PVH A; 2MOQ A; 3CSG A; 4G42 B; 5AKT A; 2AAV A; 5JOF A; 1BJ1 J; 5DA4 A; 2R2B B; 3CN0 A; 4L1S A; 1D7D A; 4N2E A; 1AZN A; 2G9H A; 3GTN A; 3QR0 A; 5AW3 B; 5CJ5 A; 3K6O A; 4XVT L; 3FSZ A; 3ZYZ A; 4X6C F; 2GEQ A; 2WW8 A; 3BPO C; 4BM7 A; 4AQO A; 4KJW D; 1TZI B; 2BZ7 A; 3V0D A; 1UVQ B; 1UAT A; 4ZTP L; 4PWK A; 3RP1 A; 4WDI B; 4U1I A; 1P69 B; 2BHF A; 1KXV A; 3OR6 A; 4AH2 A; 1EZM A; 4UNT A; 2VYR E; 3DX0 A; 1T5X B; 4NSK A; 3VX0 A; 1LWH A; 3Q2N A; 3CDZ B; 3Q01 A; 2CU9 A; 3NZS A; 4JQV A; 1U58 A; 5C09 B; 3QEQ D; 4KJP C; 2D31 B; 5IGX H; 3MV7 D; 1FYT B; 3L1E A; 5C09 E; 3SDX F; 1MAM H; 1FYT E; 2FRG P; 3K7U A; 3ARD C; 1NCA L; 5E6Y A; 3FF7 A; 5IB1 B; 4BZ1 A; 5HDB B; 4KHT L; 4EKJ A; 1P53 A; 4XB3 A; 2YRB A; 1PZ2 A; 2LQR A; 2VWE E; 43CA B; 4YDN A; 3D0A A; 3CFB B; 4HAL A; 1CGW A; 3HG3 A; 3BPL C; 5HHM B; 5F89 A; 4KZ0 A; 4XNX L; 5HHM E; 1GGC L; 4F93 B; 3WJL A; 4UQ2 B; 4XO4 A; 4XP9 H; 4XMP H; 4KHX H; 5GJS L; 2CLV A; 3GB7 A; 2E9B A; 1ZV2 A; 2V3D A; 4KML B; 4ZYK B; 1ZPL A; 3F6L A; 4TYU A; 3N06 B; 5DO2 D; 2Q76 A; 2AEQ L; 4N1E A; 2APT A; 4NC0 B; 4HEM A; 2NY3 D; 2PO6 D; 1KBG L; 1OB1 B; 4OI9 A; 1G9N C; 1IAI I; 2BP0 B; 5HHP B; 3FU8 A; 5DEJ A; 1R1C A; 1YNK H; 2NY2 B; 2KLS A; 2HP4 A; 1KJV B; 1YC7 A; 4OVV A; 4V1D B; 2ZX9 A; 4FCA A; 1A6Z A; 1FV1 B; 1QEW A; 2RJM A; 3FZU D; 4N28 A; 5F7W C; 1V7N H; 4JB9 L; 3K72 A; 1YJD L; 1CLY H; 2RTT A; 4UIK H; 4M1G L; 3K5S A; 4NS0 A; 2E7D A; 4I0P B; 4YAQ B; 2YC1 A; 3VXM D; 5A16 A; 3FFC D; 2JKT M; 4FQI H; 3UCQ A; 4OU3 A; 1MN4 A; 4HHA B; 1IND H; 1IZJ A; 1J48 A; 4HMC A; 3NUK A; 4PY7 B; 2KER A; 3L4X A; 3CBR A; 1JZ3 A; 4AUJ A; 1W9X A; 2XFX B; 1I85 C; 3NSF A; 5DC9 B; 2VZ1 A; 3MOQ A; 1M7D B; 1NPN A; 1DJI A; 4G8G A; 1M9W A; 2VRK A; 3LQZ A; 2OYP A; 3RJD A; 2IOM A; 3ILP B; 3MKG A; 2N0K A; 4JFY B; 4QWW C; 2X88 A; 1MF2 L; 1UM6 H; 1MHH B; 2NW3 A; 3CHS A; 1C08 A; 3I74 A; 2CUH A; 3RTQ C; 4XNU L; 2WKO A; 1A4K B; 2KZ1 B; 3E80 A; 4MAY B; 3U48 A; 1S9V B; 2X4N A; 4M1C C; 4IJ3 B; 4BZ2 L; 5BMF L; 2NR6 C; 1ZHK A; 2IGF L; 4U1G B; 5GIS H; 3ZKQ D; 4GG6 A; 3CH8 A; 3OKN A; 4U2Z A; 1ULV A; 3DX9 A; 2OM5 A; 4FFY A; 4WEE A; 5TFS H; 3BKM H; 3ECU A; 4QRS B; 1IEA B; 5LCV L; 5TDN A; 3JYC A; 4NUQ A; 2X4U A; 5I74 C; 2YEZ A; 3KPO B; 4ZX6 A; 1G9M H; 3N44 B; 4Y5V C; 1TCM A; 2ARJ A; 4U1N A; 1T22 A; 1BXA A; 3BEW A; 4N2I A; 3KR3 L; 4GRW H; 4G5Z H; 3ZZQ A; 1T83 A; 1TKW B; 4P48 A; 2OCC B; 4M6O L; 4ESK A; 1AHW C; 3QPX L; 4JR9 L; 5E94 B; 4D0C A; 3VG9 B; 4IOI B; 2MMX A; 2DLI A; 3BSZ L; 1ZSD A; 4G6A C; 3TBB A; 3ZQX A; 3CQP A; 3DLQ R; 1OAX H; 2ZOK A; 1TTC A; 2OMX B; 1NIE A; 2B59 B; 5DD6 A; 4LKC A; 4IK6 A; 1KCR H; 3L95 A; 2NYY C; 3BW9 B; 2EP6 A; 5GS2 H; 1ASQ A; 4HHW A; 3U9U B; 3BI9 X; 4YVN A; 3TTS A; 3IYL W; 3N1M C; 3CAF A; 3MBX L; 1HIL B; 1UT1 A; 4ZTE A; 5I75 C; 2DWD B; 4M8R A; 2L8A A; 2G2P A; 4H1L H; 1WA2 X; 1WZM A; 3BAE H; 5K9O A; 4B9P A; 4KK9 D; 3KPS D; 3HUJ B; 3QEQ E; 3A4A A; 5JIK A; 3HUJ E; 4GS7 B; 3PHS A; 2W81 C; 2P43 B; 1U8O B; 5F9A C; 4IBQ A; 3IF1 B; 2HJK A; 5F97 E; 1UH3 A; 4PLK C; 1ZA3 A; 3K8K A; 3QQ4 B; 4OQT H; 1OAK L; 1MFM A; 2E8Y A; 2R29 L; 3OLI A; 4N3T A; 4OF8 A; 2HRP L; 1IAO A; 2PXY A; 6FAB L; 3EUU A; 1MLC A; 3QRB A; 4NZT H; 5A6L A; 5DQJ A; 3S3S A; 2R56 H; 4HPO L; 2JJV A; 3LRS A; 5D7I F; 1RIU L; 2R4S H; 1AQK L; 2P4A B; 2PO6 B; 2YK1 H; 1EIB A; 5FM8 A; 4R4F H; 2BHZ A; 3BHB A; 1XXF C; 4FHJ A; 1WAP A; 3AWF A; 1UB6 A; 2Y3N A; 3MFF A; 1LTX A; 3BFW A; 1UEM A; 2X44 D; 1CLO L; 8FAB A; 3BSH A; 2XX0 A; 5E2W H; 2WLL A; 5LN8 A; 1IM3 D; 2HFF A; 9CGT A; 1ACY L; 4JHV A; 1FVC A; 4AG4 L; 3LEV L; 4X98 B; 3H9Z A; 3VXM E; 1MRC L; 1A3R H; 1QBL H; 3HMW L; 1HE7 A; 5FM5 M; 5BNX D; 3AIT A; 2FD6 H; 1PZC A; 2ESV B; 3EIF A; 2OL3 L; 4BE6 A; 5I1J H; 2ESV E; 2C1P A; 5HPM A; 2KLR A; 1RKR A; 4N8C H; 2PPC A; 5E9D A; 2EDL A; 4D9R D; 3Q0H A; 3TLI A; 4TUL L; 3RWG B; 3V4V B; 1OD9 A; 2FWO A; 4RWH A; 1Y0L B; 1NZR A; 5E00 A; 1EHX A; 3IDY C; 4P07 A; 2BMK B; 1KB5 B; 2Y06 L; 5HD8 D; 5CIP A; 4WQO A; 1RJL B; 4NN5 C; 4G3Y L; 1T0M A; 3KBF A; 2D7O A; 2EUZ A; 1A65 A; 2TAA A; 2UVF A; 5EU4 B; 3FQX A; 4NZU H; 1IC4 L; 1OAR H; 4Z7V B; 3CWG A; 4ZUW B; 3QH3 B; 4Z7V E; 2BW5 A; 3MNZ B; 3PPE A; 3NCE B; 2LTQ B; 3QUM B; 4HZL A; 4CU7 A; 4HXB H; 4O02 L; 2EIB A; 4PY8 I; 3IGA A; 5D1X C; 5D2N B; 4G7Q B; 1PY0 A; 1MWA L; 4P2Q B; 5D2N E; 4FI6 A; 3QJQ B; 4HDI B; 1I9R H; 4XP6 L; 5JR1 H; 4P2Q E; 1JRH I; 4JHA H; 3KXF D; 1GYV A; 3V52 L; 4POZ D; 2Z91 A; 4X4M E; 1IGC L; 1CDC A; 1BZ7 A; 3P3T A; 4RIR B; 1ZKA A; 5CLW A; 2KYI A; 5E1Q A; 1QF0 A; 4I13 B; 3WHX A; 2IAA C; 4BA0 A; 3BAJ A; 3UAJ D; 3L2N A; 2F7P A; 1OPG H; 3FT2 A; 1AFV L; 3GSU B; 1GGI H; 5IVX F; 4UTA A; 3LHP L; 4C47 A; 1WBY B; 4LGS B; 5T1D L; 4AM0 Q; 1YQV L; 4FXL A; 4KZC A; 1A5F L; 1FZO A; 1GYU A; 4F7M B; 4S2S B; 2G5R A; 4NO0 A; 5F7B A; 5JLZ A; 4HUW B; 5I66 B; 5J7C C; 3FBK A; 1TTG A; 3GKW L; 2DLE A; 3AT9 A; 3QIU B; 3ZDX F; 5CM1 A; 2Z4Q B; 4FER A; 4Z23 A; 2OAA A; 3DRO A; 4LAS H; 5G5D A; 2OUL B; 4CSZ A; 3RWD A; 4JUT A; 4F7T A; 2B1H H; 4BFI B; 1U92 B; 3GM0 A; 3FOR A; 5GJT H; 1GW6 A; 2R15 A; 1BOY A; 3F65 A; 2AZA A; 2HSI A; 4XVJ L; 4Y9B A; 4XGZ A; 3NSY A; 2BNQ D; 5B39 G; 4JO2 H; 4FAB L; 5EZO L; 1PIG A; 5IEH A; 1MCW M; 2P5W D; 2B1A L; 1GXE A; 2IAM C; 4DP5 X; 3CVB A; 2TLX A; 1NC2 B; 2EZ0 C; 3IDJ A; 3VXU B; 1UD6 A; 4OZG B; 5FIE A; 4JRX A; 5A2J H; 3VXU E; 3IDN A; 4OZG E; 4W9F C; 1P84 K; 4NO3 B; 3CZJ A; 2YPK A; 4PV9 A; 5E8N A; 4YFL I; 3VD3 A; 1YEW A; 2UZX B; 3EOA B; 2HB0 A; 4HUM B; 4K5O A; 1MFE L; 1TVB A; 4TSB H; 3STZ A; 4H8U A; 5CEZ L; 1UXW A; 2IG2 L; 4I3R H; 3FT4 A; 1FL3 B; 2NXZ C; 3BO8 B; 3NFP A; 4NPY A; 3WS6 A; 4HJS A; 4BA8 A; 1UQS A; 3TX4 A; 5BV7 L; 3P7V E; 2VMF A; 3C08 L; 4OBS A; 5C7X L; 1MWO A; 1FFN B; 2Z93 B; 2GC4 C; 1XSO A; 1U33 A; 2X3B A; 1QTY T; 3GIZ L; 3GSR B; 3OXS B; 4LVO C; 4X8J B; 2WD0 A; 1ZT4 B; 3NCW A; 4ZMQ A; 4RLD A; 4YNY B; 3O3D B; 3GRG A; 1IM3 B; 1XX9 C; 4UM9 B; 4P46 B; 3JUZ A; 2OI9 C; 4P4R A; 3RIK A; 1FA4 A; 3KLD A; 3B9A A; 3BEV B; 1Q9O B; 2Z7Z A; 5JW5 B; 2B39 A; 1VSC A; 1KTO A; 1D7F A; 3FO2 A; 4UOZ A; 1CXK A; 1W0Y T; 4HS3 B; 5JW4 N; 4D9R E; 4JPW H; 3LRL A; 1OAU L; 3NG5 A; 3V5D B; 1ZK9 A; 3AU1 B; 5CEZ D; 2BVO A; 4MA7 H; 3VI4 B; 5LBG A; 3OAU H; 4Q9C A; 4X6F A; 3VI4 E; 4OB5 H; 2CO7 B; 3KL0 A; 1XF2 B; 5BRZ D; 2BNR B; 2WHZ A; 4KXC A; 4XC3 L; 1CL7 H; 2BNR E; 2EIL B; 1MIM L; 5DPF E; 4CAG A; 4H40 A; 1E0O B; 1IC0 A; 4NBY B; 2C3W A; 3V0I A; 3WTV A; 2K6W A; 4M48 L; 3LSO A; 4XNY L; 4N3U A; 5DEZ A; 3O41 A; 3ZE0 B; 5HI3 C; 2CZN A; 5E0F A; 1DDH B; 5MEP B; 1MHE A; 1QRN A; 3AXI A; 5IMK A; 2EH8 H; 3ZE0 E; 5JHM A; 1ACZ A; 1UWX H; 2V7H A; 4F7P B; 2H5U A; 3OL2 A; 1UAC H; 1QFW L; 4XBP A; 3CUP A; 3DX1 A; 1E2V A; 5DQ9 A; 4BIN A; 4W81 A; 4ZDH B; 1DQD L; 4ENZ A; 4OY9 A; 3OG6 B; 4QHN B; 1KJ3 L; 4J3T A; 3S36 H; 1KRO A; 1MVU A; 5CXP A; 3SKN B; 1MG2 C; 2ZKH L; 3E38 A; 5E99 L; 2VIS B; 3FHA A; 2YSS A; 1ZLU H; 2FJG B; 4PAZ A; 5BZW A; 1I1C A; 2PR9 A; 1MCW W; 2VLL B; 3EOY G; 3HE7 C; 35C8 H; 3N1P C; 3BUY A; 3QDJ D; 3QPK A; 4CMM A; 1PZU B; 3VFS B; 3OKM A; 3JWN G; 1J11 A; 2L7Q A; 1TJI L; 2QTV A; 1IQD B; 4ZUT A; 2GTZ B; 3HHM A; 3UBX B; 4IIQ A; 2D0H A; 1MFV A; 1T1Y B; 1M4K A; 3HMX L; 4RX4 A; 2GJ6 B; 2KPW A; 3ZDZ B; 2GJ6 E; 4UV4 L; 1KYU A; 2EVH A; 2MWC A; 1AZC A; 4X9F A; 3ZDZ E; 2XN9 D; 1C16 A; 5C0A B; 4EOW H; 2X1X R; 2NTF A; 5C0R L; 4ORG A; 4ANV A; 4H44 C; 2JIX D; 2BNQ E; 5JVI E; 2BY0 A; 5F7N C; 2DQE L; 3NJX A; 5I73 B; 3VWK D; 4AQ1 A; 3O4L A; 4JL2 A; 3H9Y A; 4KQ4 L; 1V05 A; 4KJY A; 4LHU G; 4ZS7 L; 3RL1 A; 4F8O A; 5HTS A; 4XCN B; 3TVM A; 3WS3 A; 3QWO B; 2Z1P A; 5EU3 A; 3UGU A; 1YVL A; 1IZK A; 2VXU H; 2BVY A; 4E9S A; 5CZK A; 2RD0 A; 2GY7 B; 3D2T A; 5I8K L; 1P1V A; 1LLA A; 3CFJ A; 3QNZ A; 1L5G A; 4XO8 A; 1G7M B; 3NID B; 5LG1 A; 1PEW A; 1XED A; 2XBT A; 1J1O H; 4K0O A; 1W72 L; 4MWF B; 5T1M A; 5AAW C; 4EAE A; 2FQE A; 5GZO A; 2QQL L; 1F4Y H; 1GIG H; 5EO0 B; 3S88 H; 2E9J A; 4L8B B; 4OKV A; 1QI4 A; 2YXF A; 2FAT H; 3K6F A; 2HKF L; 3EGX A; 2VW4 A; 2H1P H; 2CO4 A; 2AK4 D; 1E4X H; 1E9O A; 2F8I A; 3BKJ L; 3FQY A; 5CCG E; 1NTD A; 5CLT A; 5DJC A; 4JJH A; 4MS6 A; 1LQB C; 3IDG B; 3GO1 L; 2GKI A; 2FH8 A; 1T0N B; 3FQ1 A; 3I75 A; 4JFF D; 4Y2O A; 1NLB H; 5J56 B; 2MEJ B; 1X5I A; 3FOL B; 3F04 A; 5JXE D; 2GSM B; 1XZW A; 5JOP D; 5BRZ E; 4FAD A; 4LQF L; 4IOF D; 3DO7 B; 3JUY A; 3QZP A; 1BQK A; 5I6L A; 3OKY A; 5D7J B; 4WCY L; 5DWU H; 1WBZ B; 4LVH C; 4APQ B; 4K55 A; 1CYW A; 1E6J H; 2E6P A; 4JE4 B; 1S9Y A; 5C0I B; 1S78 D; 1MRF L; 5GHW H; 1HHG B; 5FM4 A; 2DW5 A; 2NDF A; 4RDQ F; 1JHL L; 3QAZ B; 4W4N A; 1OXY A; 3GML A; 2A6J B; 2FT7 A; 3T4A A; 5I2I A; 2G0K A; 2WSP A; 1VES A; 3CSF A; 4KKP A; 4BV1 A; 1ZAN L; 4J3O G; 1QPK A; 1UXS B; 2R23 B; 3TIE B; 1OCR B; 1UZG A; 2CO3 A; 1VEP A; 2GFB A; 2KHA A; 3T4Y A; 4YN3 A; 1SNR A; 4WGV B; 2CHW A; 1YA5 A; 2WKO F; 4RET B; 2O6E A; 3NGB C; 4LPC A; 4MXV F; 5SWQ B; 5I28 A; 2I7S A; 1KC5 L; 2I9L B; 4LOU D; 4U4I A; 1GY2 A; 3VD7 A; 4BYH A; 4EN3 A; 5FO8 A; 4ZW5 A; 3IMU A; 3H7L A; 4PWE A; 3QDJ B; 4PJD B; 2XN0 A; 2YD0 A; 12E8 H; 1IL1 A; 1JSY A; 3QDJ E; 4PJD E; 3O8X D; 1IJ9 A; 4X4Y A; 1Y1U A; 4P0D A; 4WTZ A; 2DQJ H; 4BZY A; 3U9P H; 4JM4 H; 4RNR B; 2FSE B; 4U3H A; 3QYC A; 3W0W A; 4ZMX A; 5GS3 A; 1NCC L; 1SXC A; 5D5M B; 1TOX A; 4PCT A; 2JCC A; 1YLB B; 2HZH A; 4J6R H; 5AD0 B; 2AK1 L; 2H6U A; 3LZN A; 1RXL A; 1WSS T; 5ESA A; 3NRP A; 3M07 A; 3Q06 A; 2O6F A; 3CTT A; 4LLV B; 3ARY A; 1X7S A; 4E0R A; 3THM L; 3QG6 A; 4WY7 L; 3D39 D; 1X5G A; 2ZXG A; 1FIG H; 3PV6 A; 3M6D A; 2ZPK H; 1FNF A; 3CVH H; 4D9L L; 4ZXB A; 3CZS A; 5ADO H; 2NZI A; 2YPL B; 4WEN B; 4N0Y H; 4M7J H; 1MVF A; 4BDV A; 1EXU A; 2YPL E; 3FXI C; 4L37 B; 2WXR A; 3FUH A; 2W59 A; 3FQW A; 2PYF B; 2LLN A; 1UM4 H; 2O30 A; 2YEV B; 2OJZ H; 4G7R B; 5IJB C; 3GHE H; 4MJ4 A; 4ICX A; 3AS0 A; 4AX3 A; 1BJM A; 1NCB H; 3X13 B; 4HPH A; 4FA6 A; 3O3B B; 3IS0 X; 4M4P A; 2EID A; 1XD1 A; 4HGM A; 4XMV A; 1ID2 A; 5KBK A; 3TO2 B; 2OMW A; 3SDD C; 1I1A A; 3Q8C A; 4XWO C; 2BZD A; 2FB4 H; 3ECW A; 2GBA A; 4YX2 H; 5AZU A; 4R97 C; 1U0Q A; 1CD8 A; 4JG5 A; 1ES0 A; 5EO9 B; 3IDX H; 1WF5 A; 4X9H A; 2HT3 D; 5K69 A; 3Q43 A; 2OTU A; 3BUB A; 2H32 A; 3L3G B; 5T3X H; 4YFD B; 5IB2 A; 4JZO A; 4K89 A; 3BPC B; 2XQY A; 4HH9 A; 3U25 A; 1FE8 H; 3ZE1 B; 5C8J A; 4KRL B; 3ZE1 E; 3NPS C; 3JWN H; 1HL4 A; 1VGK A; 1RUQ L; 4G80 B; 5FCU L; 3VXN A; 1S7S A; 2XWC A; 1ED3 A; 4NN6 C; 1UYW H; 3IXT A; 1MFD H; 3REV B; 4OFY D; 4PRD A; 1A9E A; 1JV5 A; 3MLU L; 3NYK A; 2ZNW A; 1GGU A; 3WFC H; 3FUJ A; 4PY8 J; 5D6C A; 1ZXQ A; 3GHP A; 2DIE A; 3MV9 B; 2OSX A; 5IF0 A; 3MV9 E; 1SN0 A; 1IC7 H; 1UZ6 F; 2MH4 A; 3G6D H; 1XGZ A; 3O2D H; 3WII L; 1VAD B; 1XGU B; 1A6T A; 4LXF A; 3PP4 L; 4NQE D; 2B4C L; 3RWF B; 3VM7 A; 5AAL A; 4KIT B; 4EI6 A; 3EJQ A; 3MRI A; 4H4N A; 4H0G A; 5D4J A; 2DAV A; 3MJ9 H; 1A1O B; 4BWU A; 3V5K B; 2MFN A; 1AGC B; 3CBJ B; 3QUY A; 3K2K A; 1SHX A; 1CF8 L; 2L04 A; 1BQU A; 3GHB H; 1ESP A; 3LDB B; 4XCC H; 1W0W B; 2KZB A; 3QQ9 D; 3KPP B; 4NUM A; 2XNA A; 5E23 A; 5EZJ B; 1CXI A; 4FQQ B; 4OI7 A; 4M4R A; 3RVU D; 3SZ6 A; 1G87 A; 1FFP A; 1KGC D; 2CDG B; 2EDR A; 4L3E B; 3S3P A; 4DGV L; 2P26 A; 2YAM A; 4L3E E; 2DIB A; 4V11 A; 3VD4 A; 5INC B; 3FTV A; 2OZ4 L; 2JJT A; 2QHL A; 3PL6 D; 4JCW A; 2IMM A; 4JFZ H; 5C0G B; 3KDM B; 5CP3 A; 1HNY A; 2E7L C; 4CKV X; 5KJR L; 2EYS B; 1UWE H; 3VFW A; 5HYI A; 3TYF B; 3O8X B; 4L9L C; 5B3J E; 1E4X I; 4KX7 A; 2X9Y A; 3IN0 A; 4PV8 A; 1AVA A; 3N3E A; 3L2L A; 3V4P A; 3SDC B; 1IC5 L; 2O61 A; 1DVU A; 2XA3 A; 4OM0 H; 3T9W A; 1E67 A; 1JE6 A; 1ZHB A; 3ELA T; 3U6R B; 1Z9N A; 3FTS A; 3MBE A; 2ED9 A; 2YO3 A; 3SCM C; 4AC4 A; 3CC1 A; 1EO8 H; 5CSZ A; 5DTF A; 5AVV B; 2UZY B; 1K4D A; 5GRU H; 2KPN A; 4UTB L; 4LSP L; 3FTW A; 1JPS L; 2Q20 A; 5A6I A; 2XBD A; 5IVN A; 3DRQ B; 2FAU A; 2Y7N A; 1D6E B; 2ZD0 A; 1U8Q A; 2ZP0 T; 4XPF H; 2ZUU A; 4W9L C; 4RVB A; 4NKD A; 3SPV B; 1MCN A; 1IGM L; 3X3G L; 4Z0X A; 5FF6 B; 3UMN A; 1MH5 A; 4YHM H; 2E7H A; 3M8O H; 1EJO L; 3RY6 A; 1JTP A; 2AEP H; 3ARF D; 3P77 A; 1NKO A; 3DO2 A; 2ZWK A; 3DSF L; 2VLK D; 2ZUQ C; 3VE0 A; 4IGB D; 1HHK B; 2X9X A; 1TQU A; 3LZL A; 3UJT L; 1W15 A; 4I4W B; 1EHL H; 4J3O H; 3HOG A; 4MQ7 A; 3QRG H; 2X38 A; 1OB0 A; 4MMX C; 2OK0 H; 4MVB C; 1GZH A; 3EJS A; 2AQQ A; 1NCO A; 1BEX A; 4GG8 A; 3W56 A; 3C60 B; 2EE0 A; 2VDO H; 4KUC F; 4R0L C; 4QYA A; 3L1G A; 3MCL L; 3NRF A; 2XI9 A; 2KI6 A; 4N2L A; 3UDW A; 2G5U A; 1V10 A; 5GGV L; 4L8S B; 1CGY A; 3QDG D; 1N6V A; 2JVU A; 5I19 L; 3O6L L; 3RGV A; 1PS3 A; 2FHF A; 4HIQ A; 1NGW B; 4DK5 A; 4M61 A; 4J39 A; 3R0M A; 2K7P A; 2A74 A; 4LT7 A; 3V6O E; 4QNP E; 2W65 B; 2EDB A; 1E40 A; 2OBG A; 2KZK A; 2GBU A; 4M7K L; 1HZH L; 3DVG B; 4XPG H; 3TC9 A; 1YEC L; 5HDQ H; 2XA8 H; 4D0D A; 1H5B C; 1SYS A; 2AV1 A; 4N4Y B; 2UZI H; 4AYB A; 3T5W A; 4NAJ A; 4NZR H; 4AYF B; 1QR1 A; 1U8K A; 1JPT L; 4F5C E; 4HLZ G; 3EH3 B; 2PND A; 1BQR A; 2YMO A; 2VZO A; 2D03 H; 1FNE B; 3BUQ A; 3LQA L; 4PS7 A; 1KJ2 A; 5IW3 A; 3FL7 A; 2CRI A; 1EEZ A; 5ETU A; 2AQ1 A; 2NT1 A; 3QOR C; 4IBY A; 1MQK L; 4DTG L; 2AJU H; 2OQJ A; 1NCW H; 2BHU A; 3H9S B; 1OGA A; 5B1B B; 1NAM H; 3H9S E; 2DZE A; 2WO4 A; 5FHA L; 1A6A B; 4MDJ B; 1PD0 A; 3RDT D; 5FFL A; 2WAH A; 5I1G H; 2YAO A; 1KGC E; 3QNY A; 4AEN B; 5F8R C; 1R3L B; 3RBG A; 1MQ8 A; 3QQD A; 4OCY H; 1NGQ H; 2IH8 A; 3TH3 T; 2DYP D; 2Z31 A; 3DUR A; 2A4Z A; 5H4Z A; 3LZF H; 3MLS H; 1CGS L; 2NY5 L; 1ZE3 C; 2WBJ D; 3CSB A; 2L7J A; 2WXM A; 1W91 A; 3Q2W A; 4GQP H; 3N85 H; 1U2H A; 3DMM B; 4FQL H; 4TUJ B; 1IQW L; 4WWP A; 4PJ7 B; 1BF2 A; 1MI5 A; 3BP7 A; 4TRP L; 3U46 B; 4O4Y H; 4PJ7 E; 3OJ2 C; 4PRN B; 4XMZ A; 3D34 A; 3TCX A; 3CBK B; 2DMK A; 4P5M A; 3O6M H; 1YY9 D; 3GWJ A; 4QRP D; 1DQL H; 4RAV A; 2CDF B; 3AG3 B; 4AL8 L; 2IH3 A; 2K8Q A; 4MJI D; 3ARU A; 3IFP B; 4AEI L; 4RSY A; 2VDN L; 2BRR H; 1I3U A; 2XWT B; 1RUK L; 2VIT A; 4YHL H; 3MRM B; 1C1E L; 5LAX A; 3LQM A; 4FRW A; 4TSC L; 4XMW A; 1SXZ A; 4NCC 2; 2CD0 A; 1AGB B; 2WLK A; 5DJD A; 1OWR M; 1KBK A; 5IQM A; 4ZOA A; 3PGD A; 4F2M B; 4IGB B; 3D08 A; 5LAP A; 4F2M E; 4FNL L; 2VLK E; 2AXJ A; 4P5T D; 5E2U L; 5EU6 A; 5H35 A; 1KT2 B; 3C5Z C; 4GAF B; 2SOD B; 2XWZ A; 3NH7 L; 1MIE H; 1KIU B; 3AUV A; 4HAK A; 1RUP L; 3SDA A; 3EDE A; 1CDB A; 2V5S A; 4MHH I; 3MTR A; 3FQN A; 4LLQ A; 3ARG A; 4KMT H; 2DLF L; 4AJ0 A; 1V3K A; 2OJ1 A; 1CD1 A; 5E4E B; 3TPK A; 1OCZ B; 2H6P B; 3B1T A; 4X80 H; 5HJG A; 5EPM A; 3KVP A; 2X05 A; 1B2Y A; 3MCK A; 4Q14 A; 3MME A; 2GI0 A; 1JNL H; 1QAB A; 5F0J B; 4G6J L; 3QDG B; 1T20 B; 1MW0 A; 2AQP A; 4C4K O; 3QDG E; 1RZ7 L; 4OTX H; 4Y8A A; 1KEL L; 2VQ4 A; 4K07 A; 1V8H A; 1OP5 K; 3UAD A; 2D2C C; 4JFE B; 1T4K B; 5FYL H; 4JFE E; 3SOT A; 3CHQ A; 1T5W A; 3UOA B; 1WCB A; 3KLA B; 4E36 A; 5IML A; 3EJY C; 2VME A; 4Z7W F; 3UTS D; 3HVD A; 4GMT L; 3C6S A; 1N9P A; 5FHB L; 1JWS A; 3U2S B; 3FKU S; 4UAO B; 5C0C A; 1III A; 2WLN A; 3H2P A; 3MUG B; 3RVT C; 2LE9 A; 3FNK A; 4Y4F D; 3KLL A; 3B2V H; 3MRE A; 3GMR B; 4PJ5 B; 5AL0 A; 3Q05 A; 2PW2 A; 4XC1 H; 4PJ5 E; 3SJV D; 4HWZ A; 4AQE B; 5L7X H; 4AT6 B; 3SGE H; 4XRC B; 1K4C A; 5JA8 B; 1ZIB A; 1F97 A; 3EZJ B; 3FN3 A; 3M17 B; 3AM8 C; 3D48 R; 5L0Q C; 4XVO A; 1UKS A; 4PI0 A; 1AGF B; 3OZK A; 5D70 H; 3RDT B; 3G9A B; 5FB8 B; 5LQB H; 3NQZ B; 1ECZ A; 3SQO L; 3KM2 A; 5LGJ B; 3MZT A; 3BLP X; 1BQL L; 2VM4 A; 1A8Z A; 4OFD A; 2IBZ Y; 1S3K L; 4FQX C; 2DYP B; 5JOR A; 1JZ7 A; 5DO8 A; 5DMY A; 4KVC H; 2Z34 A; 3U9W A; 2YAP A; 2WBJ B; 4U1H A; 3TFK D; 4UI2 A; 3LKN A; 1AKJ A; 4YXL L; 1QAS A; 2Y6S C; 4G82 A; 3TCL A; 2NRD A; 5FUC C; 4I48 B; 3NZH L; 1RUR L; 2VR6 A; 3T73 A; 2YD1 A; 4H8V A; 4PJC B; 1N12 A; 2VIR B; 4ZS6 D; 4BD4 A; 4PJC E; 1N67 A; 1RFD L; 4FF9 A; 2OMT B; 4C57 C; 4AI7 A; 4HX1 A; 5H37 A; 3F7K A; 1CT8 B; 4P2R A; 4E5X B; 1X5H A; 2PP7 A; 2ODH A; 2I9T B; 4KKR A; 4LIQ L; 4MJI B; 4JN2 A; 4FAA B; 3MRQ A; 5EEU A; 4MJI E; 3MXV L; 2I85 A; 2WJ7 A; 3CFQ A; 3ARW A; 4HU2 A; 5D4H A; 3JZ7 A; 5CZV H; 3IEA A; 1JDC A; 3U0P A; 1CFQ B; 2V5T A; 5AQ0 A; 5GRX G; 1E7U A; 4I1R A; 1BX2 B; 3F8U B; 1PZ5 B; 2BOC A; 3ARR A; 4IK7 A; 5DC2 A; 4GRM A; 1F1A A; 1MAK A; 2AMG A; 2VMC A; 4YXK H; 1O6T A; 2ATP A; 1BRE A; 2JCC F; 2QMK A; 4EMZ A; 2NY1 C; 5BO1 L; 2GT9 A; 3SDY H; 1T2Q L; 1KPR A; 4FD0 A; 5CIL L; 1A6V H; 5IES H; 4K7P H; 1BIH A; 5AVZ B; 1IMH C; 1LO4 L; 1F3J B; 4N9O B; 1T3F A; 2VR7 A; 1OSP H; 1SJE A; 5MP6 H; 1R3J B; 2VDL L; 4XPA H; 3ZL1 A; 1QPP A; 3ULU E; 1TQT A; 1ZDP E; 1PZ3 A; 3QHZ H; 3FU0 A; 5KSA C; 2EIM B; 3QXD A; 3P4N B; 4UQA A; 5D1Z D; 4KAQ H; 2X53 A; 4W9K C; 4H0L C; 1HJB C; 4NRX A; 4A66 A; 4WI0 B; 3GLZ A; 4GSQ A; 3EDD A; 1MW2 A; 4GFT B; 4I3S L; 3E6H A; 1SFD A; 3GSO A; 3OKD A; 3RY4 A; 2W07 A; 3WNM A; 1KPV B; 4JKP L; 3S98 A; 3RVX C; 1RAM A; 1O6S A; 4Y2N A; 4IDJ L; 1JPG A; 4POY A; 3NTC H; 4K7F A; 4U6Y B; 3DX6 A; 4YST A; 3T6V A; 4Y16 D; 1JMZ A; 3UTS B; 3P1V A; 3F3H A; 1XAK A; 3UTS E; 4M7L T; 5A3I C; 3TA4 D; 5B39 A; 4UOJ A; 5EF3 A; 5I1H L; 4MNH A; 2YQG A; 2IFG A; 2WAQ A; 1DQQ B; 4JPV L; 4ZO6 A; 1EH9 A; 1FGV L; 2VSD A; 3MBW B; 4GRL C; 4IRS A; 3RWE A; 1E42 A; 2NXZ D; 4N6R A; 2CR0 A; 1T1X A; 5AVS B; 4DCQ A; 4E9Y A; 5K9K B; 3GI9 L; 2H9G A; 4G43 B; 1JJU A; 3EYU H; 2H26 B; 3LEX B; 3V4U L; 2DDQ H; 1LEI A; 4ZOC A; 3HR5 A; 4JFQ B; 3L5W A; 4LU5 H; 4Y8D C; 4CNI B; 5IV2 B; 2Z4T A; 5CSG A; 2CKB L; 4LSV H; 3T1F B; 5IBT H; 4RMS A; 3GSW A; 2VZT A; 3HFM L; 5EOQ H; 4PRH B; 1Q94 A; 5DSC B; 2PCF A; 4FCH A; 4PRH E; 1SOQ A; 1D9K A; 3IDU A; 2TRH A; 3OJD A; 1P4I L; 5T1L D; 1CYX A; 1NSN L; 4QRQ A; 1MJJ B; 1KXT A; 3DX8 A; 5J1A A; 5EOC L; 2FQD A; 3K4D A; 4UNU A; 5E2T H; 2Q3A A; 1RJZ A; 2HFT A; 4GQR A; 4YFL F; 1NCI A; 3ZRS A; 1N10 A; 5ESV A; 3FO0 L; 5ESQ A; 4IBV A; 2YAR A; 4WVR A; 3K46 A; 2YD8 A; 3UZQ A; 4MX7 B; 3AXL B; 2AMU A; 2OYK A; 1RPQ A; 4GSR A; 5I8O L; 4GRW F; 3VXR A; 2DQ6 A; 1G1Y A; 1I1F B; 2AJY H; 1WFN A; 2N1T E; 2NQC A; 4UIN H; 3GD1 C; 4ZMY A; 3BIB X; 4DEP C; 1WAA E; 4A67 A; 3H0T A; 4ELD A; 2APV A; 1NC4 B; 2ADY A; 4URK A; 3ZI6 A; 2Y2W A; 3UTT B; 3M7O A; 4OSU H; 3VJ6 A; 4YDL B; 2CWR A; 4GAJ H; 4GH7 B; 3CVH A; 2O26 U; 2AVG A; 1FVD A; 4L3F A; 1XF3 B; 3QP3 A; 3U4B L; 1X9Q A; 2FX8 L; 4L48 A; 3EHB D; 4C8X A; 4A55 A; 3NN7 A; 2OTP A; 2QHR L; 3T0E C; 3F83 A; 4OAW B; 3RWC A; 3SY0 A; 3RFZ B; 1XF1 A; 3ZLQ C; 2EIC A; 5FR3 A; 1ADQ L; 3GBM L; 3HZM B; 4OGA D; 2V33 A; 3RZC A; 1BM3 L; 5ITF A; 1SM3 L; 4PJ9 C; 5DK0 B; 1Z7J A; 3TT1 L; 4LLU B; 1DJW A; 3CFI C; 4GBX D; 4I87 A; 4P2O D; 3V0F A; 3ZTJ G; 2DPK A; 1Y6M R; 5E24 E; 4IYK A; 5EOT A; 3LNG A; 1LXM A; 4LRN L; 4B96 A; 1UWG L; 1HKL L; 5DK2 B; 3EAK A; 1AR1 B; 5D1Z B; 4LIQ E; 3TPU A; 4YZW A; 4TNV K; 1NH6 A; 3P2D A; 5F7K C; 3LXU X; 4NM4 L; 4WJ5 B; 5ICY A; 2Z7U A; 3LN5 B; 2FR4 B; 3FQ2 A; 3I9I A; 3U0W L; 3LRG A; 2DJS A; 4AZU A; 7PAZ A; 1Z9P A; 4XVT H; 3JSS A; 3PUU A; 1I8K A; 5E6Z A; 3V0E A; 4BWW A; 4GO6 B; 4MZR A; 3IJE B; 4EF3 A; 1OAY L; 4FFZ A; 4ZTP H; 4Y2D A; 4Q6I A; 4AE1 A; 4QHK I; 2HX9 A; 3JV6 A; 5HYS B; 4C58 B; 3D69 A; 4LZK A; 5ID0 A; 1FNS L; 1RSF A; 4FP8 L; 3BWU C; 3FU7 A; 5DLM L; 3VSQ A; 5ICX B; 2UYL B; 2GJR A; 2VDL B; 2VQT A; 1AP2 A; 2MNJ B; 5E6I B; 5DZJ A; 3CPU A; 3KG5 A; 4DPB X; 2PWG A; 3B7T A; 2VXQ A; 1CFV H; 1Q8M A; 3PAB A; 4LHU A; 3FHE A; 1UEN A; 4ODB D; 4QHU B; 1XEU A; 3GZP A; 4FXK B; 1NCA H; 1FWX A; 5EU7 E; 4KHT H; 5F9J A; 4ODW A; 5E9N A; 1NID A; 3TBT B; 5DXW A; 1EAO A; 2BJM L; 3UH8 A; 1NDG A; 3KD4 A; 2RBL A; 3UYP A; 1J5D A; 1GGC H; 4PS8 A; 2JMR A; 3ATD A; 5GZN D; 1GHF L; 1U36 A; 5JUE L; 2P46 B; 5J57 B; 2X4R B; 1CTM A; 3UBX G; 3ZYI A; 3BGM B; 5GJS H; 3G5V B; 5T1L B; 1FJO A; 4IML B; 5FDW B; 2OTW B; 4MJ6 A; 3RHW F; 5DMG D; 1BV8 A; 4M6A A; 2PWE A; 2BIH A; 3US1 A; 2AEQ H; 2XG4 B; 5E7F A; 2FJH A; 1Z9S B; 4OZ4 B; 1AD0 A; 2BIP A; 1KBG H; 1NEZ B; 2P49 B; 1Z9O A; 3ILQ C; 3KYN B; 1KB5 H; 1SDD A; 3A4E A; 4FQJ L; 2NW2 A; 5A2I H; 1UH2 A; 4CAD B; 5IEK B; 2LFE A; 2XY1 A; 5AB2 A; 4JB9 H; 1IGY B; 5AW4 B; 2EC8 A; 1YJD H; 1IBH A; 5FUZ L; 2PCY A; 3WNJ A; 2WLI B; 4M1G H; 4DT2 A; 3I84 A; 2BII A; 3FK3 A; 4K94 C; 4TNW K; 4HIE B; 4DUX A; 3VFT B; 3VWJ C; 4MZN E; 3AGV A; 1R3H A; 2Y36 L; 1HIN L; 2WNK A; 4UV7 L; 2NNA B; 1FZK A; 4BWT A; 5B4M C; 1BA9 A; 3LPG A; 2ZKX A; 1E4H A; 2ZP8 A; 3LIU A; 5D7S L; 3RWI B; 3CLE L; 3EOT H; 1RVF L; 5I5R A; 3U7Y L; 4HG4 J; 1H8O A; 5DRW A; 5HYE A; 1S6N A; 2QR0 A; 2GC7 C; 4HKZ A; 5FA3 B; 3BIS A; 3QI9 A; 2R83 A; 1MF2 H; 3L3J B; 1DVF D; 2B16 A; 4GMS L; 3WLW C; 1SYV B; 2R2H A; 4XNU H; 1CYG A; 3SE4 A; 1GJU A; 3Q2V A; 5GSR B; 1F6L L; 3OSV A; 5BMF H; 4GBX B; 1HNG A; 2IGF H; 1A66 A; 2R1W B; 2LV4 A; 3EXL A; 3VCL A; 3W38 A; 4TLU A; 4K23 L; 3BYT A; 4EDX A; 1NFI B; 2QVI A; 2XWD A; 2WVV B; 5LCV H; 3TO4 C; 1E9Q A; 4MYP A; 5ACM A; 2JK5 B; 3TLR A; 3DO1 A; 2AJX L; 4JPS A; 2EKJ A; 1YEK H; 3U4E A; 4TVU A; 4X5X A; 5AW5 B; 4DGI L; 3GZO A; 3KR3 H; 2I32 A; 4HJT A; 3QIB D; 1WIC A; 1OGP A; 3FOM B; 3ARE A; 1MFC L; 1UKQ A; 1RUM L; 1PU0 A; 3ETB J; 3NFN B; 4JR9 H; 4M6O H; 2K78 A; 3BSZ H; 2F74 A; 3VYN A; 4X7C C; 3B3I A; 2C9U A; 3RZW A; 1OF0 A; 4KXA A; 3D1M C; 1CDG A; 2M26 A; 3QXU A; 1VZG A; 4KRM B; 1NAN H; 1P7K B; 2VDK B; 4AIZ A; 1FQ9 C; 2ETW A; 2F19 L; 2X89 D; 4O00 A; 3MBX H; 5L2J A; 3BAK A; 4LBA A; 5K59 E; 3D09 A; 1J8H A; 4FG6 D; 1USQ A; 1AAJ A; 5DFV C; 5DUR D; 3C9N B; 2BST B; 4OVU A; 4RGN D; 3V6F A; 5CR1 A; 3OLE A; 5GGS A; 1K2D A; 1NAM A; 1JBJ A; 5DJZ A; 1E8Y A; 3O6F A; 1H6T A; 3GKZ A; 4UDU A; 4CW1 B; 3ARB C; 4HT1 L; 4DP8 X; 3QEU B; 1U8H A; 2EXW D; 1UGN A; 3BZ4 B; 1L9N A; 2YA0 A; 3L54 A; 4YHP B; 3N43 F; 2K3H A; 3VMO A; 3WFB L; 2R29 H; 3TN0 A; 1TR7 A; 3A3Y B; 2HRP H; 4NYL B; 4WK4 B; 1UCT A; 4XNZ C; 2IV8 A; 3ZTJ H; 3P7P E; 4RMT A; 4HCI A; 1X9R A; 3QA3 B; 4HPO H; 4NO5 A; 4UT7 L; 1RIU H; 1YN6 B; 1BFV L; 1AQK H; 1JZ6 A; 3BQU D; 4DW2 L; 3WY1 A; 4ABV A; 1XF4 B; 2FBO J; 4GXU M; 1I1A C; 3IGK A; 4N2G A; 4DEW A; 5E8P B; 3DWT A; 2HYM A; 3LIZ L; 4PGZ A; 2R1Y B; 5CZX D; 5IQN A; 5LBV A; 1S7Q B; 5EF0 A; 1M3I A; 2BVU A; 2WXH A; 4UQE A; 5TK4 A; 2EO1 A; 1CLO H; 3NAC L; 4G8F A; 3C6E A; 2G75 A; 4KRP C; 6TMN E; 1JF1 A; 3RTL A; 1ACY H; 3HG4 A; 4AG4 H; 3WUW A; 2YUW A; 3LEV H; 3IU3 A; 4JQX C; 1MRC H; 3HMW H; 1ZT1 A; 4I18 C; 1AQ8 A; 3EJJ X; 4FUL A; 3WXW L; 2CCL A; 2OL3 H; 5EFI A; 4P4K A; 5EZL A; 4WWI D; 2F54 D; 4R5T A; 1X5Z A; 2WCS A; 1F4A A; 1RHH A; 1B0R A; 4TUL H; 1L6X A; 4WFG D; 1EEU A; 3B83 A; 5A2A A; 4UDT A; 3LKQ B; 4MJJ A; 2XPG B; 5D9S B; 2O5X L; 3PF2 A; 1URL A; 2Y06 H; 4PQX A; 4G3Y H; 3FUN A; 1P1Z B; 1RHF A; 3ARS A; 3NJI A; 4DZ8 B; 5AG9 A; 3KLH D; 4YGA B; 3UIZ A; 1P4L A; 2C34 A; 5JS3 E; 1WZ1 L; 3G7T A; 1IC4 H; 1D5I H; 1MFU A; 2YN3 A; 4AGO A; 2FHB A; 4PRP D; 4WKZ A; 1KWG A; 1NMC B; 5K39 A; 2P7T A; 3FON B; 1G9M C; 4QIR A; 3QQ3 A; 4O02 H; 2JJW A; 3QUY C; 2Y7Q A; 1A7N L; 1AIF B; 1QSV A; 1PZB A; 4G8E B; 4Z77 B; 1MWA H; 3BD3 A; 2ZYR A; 3LVT A; 3WNN A; 4XP6 H; 3GXT A; 1N59 A; 3VD0 A; 1XZ0 A; 2V9T A; 3V52 H; 1IGC H; 3SCM D; 1QFW M; 2VN3 A; 4NQS B; 4H25 A; 4NBX B; 5CLY A; 4PRE B; 5JT9 E; 5HDR A; 2HFE A; 4J4P A; 2D3N A; 3HE6 C; 1E4J A; 4U1S B; 2I07 A; 1AFV H; 3HG5 A; 4JDZ B; 1NL0 L; 4Y0G A; 3QJO A; 3LHP H; 1HH6 B; 4PME A; 1KIR A; 5BOZ G; 3VXP A; 4DO4 A; 5T1D H; 3ZKX C; 3CSY A; 4DQA A; 3RGB A; 4KZE H; 1UGK A; 1YQV H; 3HRZ A; 1A5F H; 2DY2 A; 2RAC A; 4UCF A; 3RJO A; 4QRU B; 2D2O A; 3RX7 A; 4CVZ A; 4F8Q A; 2OCJ A; 5D7L B; 3U30 C; 3VRJ B; 3GKW H; 1I8I A; 4WUU B; 5F7A A; 3AG4 B; 3IFO B; 4X9B A; 5B21 A; 1LIL A; 1JGV L; 1WIU A; 2X6C A; 3H3B C; 4R7X A; 2J8H A; 2KQN A; 5A43 C; 4XCE B; 4KK8 C; 2QQN L; 2JJU A; 2ZX8 A; 3QWQ B; 2A73 B; 4HUU A; 3DNZ A; 1OPO A; 5DJ2 B; 2IPK B; 4MS8 C; 1U8C B; 2QDV A; 4QDG A; 4XMK L; 4FQ1 L; 4MVB D; 4OIF A; 3P3S A; 3VA6 A; 5EZO H; 4D3C L; 4FAB H; 2PPA A; 2CPU A; 2JNZ A; 4IU3 A; 1N3N B; 2LGN A; 4PHX A; 2B1A H; 5EH1 A; 3OR7 B; 5CJB A; 3HZV A; 3PJL A; 4I5B B; 3KNB B; 4LCC A; 1BEC A; 2PCP A; 2R7E B; 4BZ1 L; 5I17 B; 2F53 B; 4P3D B; 1MNN A; 5F1N A; 2F53 E; 2P5P A; 5TDP A; 1NPJ A; 3FB8 B; 2MOG A; 2ZUS A; 1MFE H; 3VTT A; 3RA7 L; 2DM7 A; 5CEZ H; 2ZF9 A; 4X4X B; 2IG2 H; 4LPW A; 1X3D A; 1AE6 L; 3DHU A; 3IXA A; 4MDI B; 3DO4 A; 4IKJ A; 1IE4 A; 4B9F A; 2CII A; 2Y2Y A; 3EG9 B; 4J3V A; 5BV7 H; 3X11 A; 1QUN A; 3C08 H; 5C7X H; 3SQB A; 3DJS A; 1YG9 A; 3GIZ H; 1ETZ A; 2FDB P; 4GKZ B; 5C86 A; 4BQC A; 1K85 A; 4XHJ C; 1QSF A; 3MC0 A; 4W4O C; 4WZ9 A; 5T1K D; 2HMI C; 1WZK A; 3R18 A; 4JG9 A; 3GDB A; 2LFG A; 5F44 A; 3SBW C; 4JDV A; 4ADG A; 4KRN A; 3MZG B; 5BRP A; 1U3H B; 1H3P H; 1OP3 K; 1SMA A; 1IB5 A; 1XGR A; 2R27 A; 1OW0 C; 4PJA A; 3W9E A; 2GTW A; 3O4O C; 3RPI B; 1RJY A; 1OOA A; 5EUO B; 4NCC 1; 5EUO E; 3W11 D; 4X6E B; 2PYE A; 1Y6K R; 4G42 A; 4P9H L; 1OAU H; 2XKN B; 4JCL A; 2K1M A; 4WFG E; 2R2B A; 4JL1 A; 1WCQ A; 1TJH L; 2Q8A L; 5DZO A; 1KS7 A; 3ZME A; 1HHH B; 4Y5X C; 1TZI A; 1UVQ A; 5B7M A; 1AMY A; 1MIM H; 4WDI A; 3TBW B; 3GT6 A; 3S97 C; 1SZN A; 4PRP E; 4FFV C; 4M48 H; 3AY2 A; 4XNY H; 4Z6A T; 3BP6 B; 4NC1 C; 2OZN A; 1U8M B; 1T5X A; 2Q6W B; 3CDZ A; 3OHX C; 3LKR B; 2NYZ A; 1QFW H; 2ZXD A; 3O45 B; 4MLS A; 5C09 A; 2C26 A; 2D31 A; 4OFV A; 1FYT A; 3GTT A; 3AQY A; 4V33 A; 1FLR L; 3NEO A; 1S7R A; 3QAQ A; 4FFY L; 2WUB H; 4LFH D; 4QHK J; 2VDP B; 2B9A A; 1CBV L; 1L0Y A; 5IB1 A; 5FLV A; 4WGW B; 1KJ3 H; 1OSY A; 3DUH A; 2ICE S; 4R83 A; 2ZKH H; 43CA A; 2EH7 H; 5E99 H; 3CFB A; 5CMA B; 3II2 A; 4CN6 A; 3QE5 A; 3ZE2 B; 1DJX A; 3AS1 A; 4X8G A; 4EUP B; 3ZE2 E; 4UQ2 A; 2D4D A; 4Z76 B; 2P8P B; 5I0V A; 5DEG B; 1J5O L; 5A3Y A; 3ERX A; 4G8G D; 3RAJ L; 4HBC L; 1N68 A; 4ZYK A; 2UWE F; 1XH2 A; 1TJI H; 2RAM A; 1N6U A; 4JZN B; 1QHO A; 2WXG A; 1E4K C; 2J2Z B; 1F4X L; 2OMB A; 3BFQ G; 4WI3 A; 3EAD A; 4E41 D; 3HMX H; 1OB1 A; 3F58 L; 2BP0 A; 5IBU B; 1NCD H; 1KJV A; 4V1D A; 1OM9 A; 2V3E A; 3QPQ D; 3WU1 A; 3D3V B; 1ZVY A; 5C0R H; 5CEX B; 3D3V E; 1CC3 A; 4EBQ L; 3L2M A; 5BQD A; 3QJF A; 4I0P A; 4YAQ A; 1KUM A; 2AEO A; 2CH8 A; 2NLJ B; 4KQ4 H; 5EZP A; 5G4M A; 2CA3 A; 4FA8 A; 4ZS7 H; 4HHA A; 4PWH A; 3S5Y A; 4XZ4 A; 3NN8 B; 3FN0 L; 1O7S A; 5ERW B; 1M6U A; 5IR1 B; 2C3H A; 3JTS B; 2XFX A; 3ROO B; 2EXT A; 3HIZ A; 4MA1 B; 4PUB L; 3UBX I; 1M7D A; 3RX9 A; 5I8K H; 4N2C A; 4U1I B; 3OGC B; 1M53 A; 5IT2 L; 4F3F B; 4KZD L; 2JJS A; 1DJH A; 4JFY A; 1W72 H; 4XI5 C; 3BZD A; 1MHH A; 4O9H L; 2FL5 B; 1OAJ A; 2BK8 A; 3HNS L; 1J0I A; 2CBQ A; 1A4K A; 4F8P A; 4MAY A; 1E6O L; 2EUW A; 1S9V A; 4DFH A; 3FH8 A; 5KVG L; 5T1K B; 2JLL A; 4XCY B; 3BH9 B; 3L08 A; 1U58 B; 2ZSV B; 2NYK A; 1G43 A; 2HKF H; 2R9H D; 3LH2 H; 5HXM A; 4DAG L; 3QIW B; 5L88 L; 4HKX A; 2BVP B; 4QRS A; 3LNF A; 4DP0 X; 4HLE A; 3KPO A; 3BKJ H; 1XF5 B; 3O0R H; 3WIH A; 2IMN A; 4M43 L; 3BKC L; 3LS5 L; 1Q4N X; 3EBH A; 5D71 L; 2XKS A; 3RPB A; 5LGH L; 1QW8 A; 4Y19 A; 2IWK A; 3S48 A; 3ARO A; 2X1O A; 1TKW A; 2X1W L; 3TF4 A; 3A6C L; 4IOI A; 3HN5 A; 3AY4 A; 5DMJ B; 4POU B; 3UGE A; 1O9W A; 3ZK0 A; 2PXY D; 2YJQ A; 2X0W A; 3D0V B; 4ZX5 A; 2OMX A; 3KWV A; 2B59 A; 3WYR A; 1J9T A; 4F2E A; 3BCD A; 4LQF H; 1E5C A; 3B34 A; 3U9U A; 4WCY H; 3FT9 A; 3GSQ A; 1JGE B; 5F7D B; 1AGE B; 1T1Z B; 5J13 C; 1HIL A; 1SLV A; 2DWD A; 1MRF H; 4FTV B; 1YMM A; 2Z8W C; 5CJQ H; 1FDL L; 4FTV E; 4OD2 B; 4Z18 A; 3HFF A; 4NUG L; 2CR6 A; 4J27 A; 4PJI A; 3SBQ A; 1GXW A; 1XT5 A; 2Z7Y A; 4KJQ C; 5T33 H; 5IUS A; 4YGV B; 3HUJ A; 1ZAN H; 4MMZ B; 2JEL L; 4CVX B; 1U8O A; 2IOO A; 4MNG B; 4YEF A; 2IV9 A; 4HWB A; 5F0P B; 4MNG E; 3IF1 A; 3MPC A; 1YDP B; 5E9D D; 2DWE B; 4J3O F; 1J5C A; 1KC5 H; 3QQ4 A; 5LVE A; 3EYQ D; 5CKG A; 5IB3 B; 5A2L H; 3HI5 L; 1KZY A; 4ZMT A; 4M6N L; 4J3S A; 3B5T A; 2VO8 A; 3M7F B; 5I18 L; 4G7S B; 3NEX A; 4CC1 A; 2ZE0 A; 4UTA L; 4TQE L; 2PO6 A; 4E41 B; 1TZG L; 1OTU D; 1QSZ A; 4E41 E; 4XVU C; 1NCC H; 4LMQ I; 4GG6 E; 4BUQ A; 2AK1 H; 2WIN B; 4ANW A; 4X9G A; 2WLZ A; 2GHW B; 1C5D B; 4JVP A; 3THM H; 4WY7 H; 3BEW B; 2ZKW A; 2YMX L; 3H4F A; 1Q9K B; 3BZF B; 4M6M H; 4HIJ B; 4KY2 A; 4X98 A; 4D9L H; 3F84 A; 5FO7 B; 3WN6 A; 2R39 A; 5KYW B; 1TVH B; 2ESV A; 3CU1 A; 1TYR A; 2FED C; 2VLM D; 4ZW7 A; 3EYV B; 3RWG A; 3V4V A; 1BGX L; 2D3L A; 1TG8 A; 3QAR A; 1Y0L A; 2NXY C; 4E5X G; 2BMK A; 1KB5 A; 2NUT B; 1R33 A; 2N0M A; 5HBB A; 3DX2 A; 4NO0 D; 5ANM B; 5CF3 A; 5ANM E; 4DPR A; 2R8S L; 5EU4 A; 3U6X A; 4Z7Q F; 2GVY A; 3RGH A; 3C75 A; 4PS3 A; 4Z7V A; 4ZUW A; 4YBL B; 3ZGH A; 4P3W A; 1TLM A; 3MNZ A; 3N42 B; 4EI5 A; 2IO5 A; 5IMO B; 3QUM A; 1UJT A; 3QUZ B; 4R7N B; 5SV4 A; 2WQW A; 5D2N A; 1A1M B; 5JW4 R; 1HYS C; 3T2I E; 4P2Q A; 5GGR A; 4UZU A; 4HDI A; 4LVE A; 4PN6 A; 4WTX A; 2Z7W A; 4X4M A; 3X17 A; 2PTT A; 1PCS A; 4BFK A; 1RUQ H; 1ZA3 B; 1B3J A; 1TEN A; 4RIR A; 4U3X A; 4W6W B; 4HSQ A; 4JRX D; 3MLU H; 1MLC B; 3GSU A; 1A3Q A; 3RLS A; 3J27 A; 2APX A; 1Z0M A; 5I5I A; 2WLH A; 1WBY A; 3QUX C; 4Z8F L; 3D4C A; 4ZD3 L; 4PVM A; 4EKX A; 1J05 B; 1E5U I; 4RMW A; 4F7M A; 4F20 A; 4S2S A; 1U8P B; 1M9S A; 2VZV A; 3BT2 L; 4TTD C; 3WII H; 3PP4 H; 4HUW A; 5F3B A; 2B4C H; 1TU2 A; 5I66 A; 1ASP A; 2EVJ A; 3QIU A; 4OCW H; 1A21 A; 5ALC L; 2Z4Q A; 1WJ3 A; 3OAZ K; 1VZI A; 3LZR A; 1M7I B; 2A99 A; 4Q0X H; 4LCY B; 5HI4 C; 4BFI A; 5KUK A; 3BWZ A; 4M7Z C; 1CGU A; 1FVC B; 1UWF A; 2WMP B; 4KVN L; 2ZX6 A; 4TLK A; 4PSP A; 5CCH E; 3P3R A; 5HPM B; 4H2C A; 1IAI L; 1M2O A; 2OZ4 H; 5K5Y A; 3AWE A; 1BG1 A; 3UTS A; 3EBG A; 3OLG A; 1NC2 A; 3HAE B; 3VFP B; 3VXU A; 4YQX L; 4OZG A; 2FWO B; 3K2I A; 3SKM B; 5KJR H; 3TBV A; 4JFH D; 4X4Z A; 1FFO B; 1RUA L; 4NO3 A; 4Z7N F; 5FA4 B; 3TA3 B; 4LSS L; 3EOA A; 2UZX A; 4ZUU A; 5L4J A; 4XOB A; 1IC5 H; 1O75 A; 4TVP L; 1SOX A; 4MRB A; 2MNW A; 3U7A A; 3D06 A; 5I71 B; 4D7P A; 3C09 B; 1FL3 A; 4HJ0 C; 3BO8 A; 4NKM B; 3SHS A; 1LYQ A; 2FIK A; 3IJH A; 3GIV B; 1CGT A; 3MLV L; 1FFN A; 2Z93 A; 4JM0 A; 4OUO A; 3GSR A; 3OXS A; 4X8J A; 2HVB A; 1QRN D; 2F8X C; 1II4 E; 3IGA B; 4LSP H; 1JPS H; 1LNC E; 4YNY A; 1IT9 L; 3O3D A; 1IM3 A; 2WYH A; 3MUH L; 2XTS A; 4UM9 A; 4P46 A; 3RVW C; 3L3I B; 2Z91 B; 1BZO A; 3EF4 A; 5C9K A; 3FUD A; 3ALW A; 1IGM H; 1XGP B; 2LAG B; 3X3G H; 1JK8 B; 5TH2 A; 3BEV A; 5JW5 A; 1CTO A; 1ZWI B; 3F00 A; 4TQP A; 1TET L; 3OKE B; 1EJO H; 32C2 B; 3VGG A; 4HS3 A; 2VLM E; 3X12 B; 3P02 A; 3DSF H; 3AU1 A; 4KKB C; 4TVP D; 5F6H I; 4OJA A; 3VI4 A; 1KGU A; 2VSC A; 1UWY A; 1KV7 A; 3UJT H; 3RVV C; 2Y1Z A; 2BNR A; 4WJK B; 5C0B B; 3NPZ B; 2AW2 A; 3GSN B; 2FEE I; 5FYJ L; 4JFW A; 1JZH A; 1PE5 A; 3BMV A; 3FFD B; 3M1O A; 4X7E C; 5DUP H; 4RX4 D; 1YPZ F; 4NN7 C; 4PGD B; 4B3V A; 5GGQ L; 2R69 H; 1AAC A; 2J4W H; 4PCS A; 1DDH A; 5MEP A; 4Z95 L; 2YXV A; 4F7P A; 2ZCL L; 4J28 A; 1HC1 A; 4DIX A; 3MCL H; 2R59 A; 1ZS2 A; 2COQ A; 3JWN C; 5EF1 A; 5GGV H; 4XHQ A; 2DQF B; 2EIJ B; 2HJL B; 2WYZ A; 5D7H A; 5I19 H; 4TNE A; 3O6L H; 4ZDH A; 2ZTB A; 3O4L D; 4NHU B; 3UP1 A; 3S5L B; 3RPK A; 3CK0 H; 5GRZ A; 4MNQ B; 1U8N B; 3MOA L; 4QHN A; 7TAA A; 4MNQ E; 4BGD A; 3TVM D; 2VMD A; 3SKN A; 3ABG A; 1HZH H; 4PJB B; 1XTL A; 2YPK B; 1YEC H; 2IDT A; 4PJB E; 2ZXB A; 4P2C G; 1WAE A; 1JWU B; 2E7B A; 1A64 A; 1TVB B; 3ZZL A; 5FYJ D; 3TEZ A; 2FJG A; 3UZE A; 4OLZ L; 4Y4K D; 1JPT H; 2VLL A; 4PJA F; 5DS2 A; 5KVM C; 4Q6K A; 4S10 A; 3VFS A; 2N59 A; 5J12 C; 2FHC A; 3LQA H; 3H2Q A; 3ATA A; 1H8S A; 1MHC B; 3ZST A; 5GZO D; 3UBX A; 1LWJ A; 1MQK H; 1T1Y A; 3A24 A; 2GJ6 A; 1SVB A; 2PW1 B; 1XXD C; 4X7T B; 3F5W B; 5FHA H; 1OP5 H; 2BOB B; 3T8V A; 5BUG A; 5C0A A; 5HYS G; 4Y5Y B; 3SD2 A; 4AYM C; 2WLY A; 4JFX B; 4PBD A; 2J56 A; 4JZJ B; 4AZ8 B; 5DC4 B; 1P84 J; 3KPL B; 2ZP9 A; 2IOI A; 2XYC A; 4WI8 A; 5K1N A; 2E76 C; 1CGS H; 4U6H B; 1U41 A; 5C08 A; 2NY5 H; 3WJJ A; 1KSR A; 3QWO A; 2QVK A; 2D7P A; 4JFH E; 1MEX L; 3EGS A; 2AIT A; 4GI8 A; 2XXM B; 3UAB A; 3I50 H; 4HJJ L; 1A7Q L; 2ICE B; 1IQW H; 1XIW B; 3JTT A; 4MB3 A; 1ASO A; 2CQV A; 3KPR D; 4TRP H; 1G7M A; 2K7Q A; 4MWF A; 4Q4E A; 1M71 B; 4NQD B; 3BYY A; 2W8C A; 1MHE B; 2D0G A; 5GS1 C; 4NQD E; 3EJT A; 3L3D B; 3OQR A; 5AW1 B; 3SE8 L; 4B9K C; 1QRN E; 1UAD C; 3AGW A; 2X28 A; 1OLZ A; 1WHP A; 1WO2 A; 4L8B A; 3OMN B; 2WXQ A; 3T0A A; 3ULU H; 3VGB A; 1MJ7 L; 3FB5 B; 4IRJ C; 4AL8 H; 2XJL A; 4KNS A; 1F86 A; 1NEZ G; 2F8V A; 1MAM L; 4AEI H; 5M7Q A; 3HG1 B; 4I1P A; 1RZG B; 1C1E H; 4WV1 B; 1ZK5 A; 4TSC H; 5F7E L; 2ZKY A; 4C83 A; 4CVU A; 3T0D A; 4A2D A; 2WWK O; 4GKL A; 4P49 A; 4TUO B; 1W80 A; 1IBF A; 3IDG A; 4X7F C; 4Y1A B; 3WEN A; 4TVP E; 4EN3 D; 2YSS B; 1T0N A; 3FSA A; 4FNL H; 4Y1A E; 4J3O C; 4XP9 L; 3MLZ L; 5E2U H; 4M93 B; 4LLW B; 4GJT B; 1ZV5 A; 2WZ5 A; 3NH7 H; 4UT6 A; 3OKM B; 4R2G C; 1CIC C; 3S37 X; 5JOE A; 1JRH L; 3DO7 A; 3HNT L; 5L9D H; 2CBT A; 4P9M L; 5D7J A; 1BVZ A; 1WBZ A; 3W0W D; 4WWK C; 2DLF H; 4QR7 A; 4JY4 B; 4APQ A; 4RFE B; 4PEE A; 4QXV A; 3TAS A; 5C0I A; 1J0Y A; 4JY6 B; 1DBK L; 1HHG A; 5DD3 L; 1INQ B; 1KKK A; 2NTF B; 5L4F A; 3FO9 B; 3FU9 A; 3II3 A; 5SX5 K; 2A6J A; 3NCY P; 4LP5 A; 2G2R B; 3IE4 A; 4UIK L; 4G6J H; 3UEQ A; 4YHY B; 4QYO B; 1P7H L; 1RZ7 H; 3VTM A; 5T1J A; 1UXS A; 3KVD D; 1QLE L; 3TIE A; 3O4L E; 4DNL A; 1KEL H; 4UQ9 A; 4ZXB D; 4KJY B; 4FLH A; 1IND L; 5K9Q G; 2H8P A; 1CUR A; 5SWQ A; 5CJ8 A; 1U3N A; 5IFJ B; 2A1W L; 2HXA A; 1EDH A; 2I9L A; 2VUO A; 5LBS A; 5EEZ A; 4GMT H; 4M1D L; 4AVH A; 5FHB H; 4Y4K B; 3L13 A; 1L5G B; 4K5L A; 1HHI B; 4FS0 A; 5D4Q A; 3TWY A; 1PX8 A; 4L5F E; 1DHK A; 5IJC C; 2X4O B; 2XPY A; 2AI0 L; 4TMP A; 3HBZ A; 2ZXC A; 3K2M C; 4RNR A; 5DLH A; 2FSE A; 2IDU A; 2XTL A; 4RZC B; 3I02 A; 5AD0 A; 5D5M A; 1KCB A; 3BZE B; 1ZOX A; 2FUT A; 4KRO C; 3UEZ A; 4W6Y B; 4E42 B; 1KBW A; 4LLV A; 3BPO B; 3SQO H; 4QRA A; 4IHQ A; 1P6W A; 3NJH A; 1TQC C; 2YPV H; 5DJC B; 1BQL H; 2WQR A; 1SOS A; 1S3K H; 3APD A; 2CRZ A; 1QAT A; 1PZS A; 2VR5 A; 4FXG C; 3GTV A; 2YPL A; 1T66 C; 5AW6 B; 1FVE B; 5B8C B; 2PYF A; 3WY3 A; 1RZ8 A; 1TTB A; 4AQZ A; 1MCL A; 4YXL H; 4EZM A; 3QNX B; 3HUJ F; 1RH8 A; 2GB2 A; 1HSB B; 2WP3 T; 3KPR B; 3B4N A; 3NZH H; 1OO2 A; 1FF5 A; 3KPR E; 3X13 A; 1RUR H; 3O2W L; 3O3B A; 1HCV A; 4O2E B; 1QW9 A; 3TO2 A; 1YNT B; 2DOC A; 4XNQ A; 1RFD H; 5HI5 C; 3V4P L; 1X5F A; 1COB A; 1PQZ B; 4YDI L; 1GTF A; 4MJ2 A; 3HPL B; 3BPL B; 2YD4 A; 5FFO A; 2HPT A; 4U1L B; 4LIQ H; 3CN1 A; 5EO9 A; 4AMW A; 3DTU B; 1GOG A; 3MXV H; 1D5Z A; 5BZZ A; 3L3G A; 5CGT A; 3PU7 A; 4JVW A; 4Z7O B; 2DM2 A; 5T5N G; 3BPC A; 4PVL A; 1TNN A; 1OCW L; 1I7Z B; 3HB3 D; 2B5I C; 2HVJ B; 3E8V A; 4G80 A; 1HES A; 4MYW B; 4EN3 B; 5FO8 B; 3W39 A; 1NM4 A; 3REV A; 1SQ2 N; 5BO1 H; 1BXX A; 1T2Q H; 4JKL A; 4N8V B; 4OGZ A; 5JXE A; 4ONO A; 5CIL H; 3TZV D; 1HFU A; 3SPH A; 1X6L A; 1IC1 A; 2GJJ A; 2HTL C; 4D91 A; 1LO4 H; 2XQY K; 1QA9 B; 3MV9 A; 4P23 C; 2YLK A; 4M56 A; 1WS8 A; 2VDL H; 2L7U A; 4X5L A; 3MBE D; 1VAD A; 1XGU A; 3EO9 L; 1QZ1 A; 3PAU A; 3RWF A; 4KKQ A; 4XBE L; 4RZK A; 4G8A C; 2ENY A; 2DMH A; 5HHV L; 4Z5R B; 1A1O A; 3GLA A; 3UYO D; 1AGC A; 5CSB A; 5E2W L; 1W0W A; 4OCR L; 3KPP A; 5C70 A; 4PRI A; 4FQQ A; 1TJM A; 5TMN E; 4KKC C; 4JKP H; 2OYL A; 2CDG A; 1T89 C; 4KTE L; 2D7M A; 4L3E A; 4YHK L; 2PPE A; 1W6L A; 4IDJ H; 4N8C L; 4NFD A; 5INC A; 4TSA L; 1XAU A; 4Z7V F; 2QBW A; 1O7D E; 3CFC L; 2WJ5 A; 1MW3 A; 3D52 A; 3P7U E; 3KDM A; 5C0G A; 1DLF L; 1D5B A; 1ND0 B; 3OHM B; 2EYS A; 4NQT A; 3T0W A; 5IQO A; 1AQD B; 1PL3 A; 3BGA A; 4JPV H; 3TU6 A; 3TYF A; 3O8X A; 2D9C A; 3PGF L; 4FMF A; 5I74 B; 1FGV H; 3OMA B; 1G1K A; 2QLY A; 3AC0 A; 3QQX A; 5FGS A; 4ZNE E; 2LW3 A; 1K5N B; 3A22 A; 3QGV A; 3C2A L; 3SDC A; 1WWB X; 3UL4 A; 1HEJ C; 5C92 A; 1BLI A; 4UMG A; 3JT0 A; 3T2O A; 3GI9 H; 2ILL A; 3U6R A; 3DOS A; 4KKL D; 5L8L B; 5I16 B; 2QT6 A; 4M7L L; 2CCW A; 4NQQ A; 4XHJ D; 5F9W C; 1AHW B; 1X4X A; 3V4U H; 1MFA L; 1L9T A; 1IAR B; 1SRD A; 2J20 A; 2UZY A; 4JKW A; 3V55 A; 1KJ2 L; 4M8Q B; 2CKB H; 4OZI F; 1BD2 B; 5C0F B; 4NJA H; 1S9X B; 5E2V L; 3DRQ A; 1BD2 E; 1NGY A; 3R06 B; 3N41 F; 3HFM H; 1D6E A; 1VPO L; 4JHA L; 3EZ8 A; 3BT0 A; 4JGN A; 1P4I H; 3RGV D; 1NSN H; 5FF6 A; 1ED3 B; 1JIB A; 5EOC H; 5I75 B; 1PTZ A; 2OCF D; 1V7M L; 2W9D L; 1T6B X; 1GJW A; 3FO0 H; 2FEJ A; 2XOT A; 3MX0 B; 2XLL A; 4JRY B; 4JRY E; 3D9A L; 1HHK A; 5I8O H; 4U3S B; 3GJE A; 2CAL A; 2NUP B; 3T2Q A; 4I4W A; 3QIS A; 1Z9G E; 4EI6 B; 3MRI B; 5DRN B; 3EH1 A; 4W68 A; 4AGQ A; 5FW6 A; 3TNM B; 3C60 A; 4RDB A; 3KWT A; 1OGA D; 1L3N A; 5AW8 B; 4OZG F; 5F6H J; 2G5B B; 3MNM A; 1O5D T; 1JFH A; 3U4B H; 2FX8 H; 5BW7 A; 3HPJ B; 4RIS L; 4L8S A; 1UT9 A; 3OJV C; 3ZDY B; 4MGX A; 5KBL A; 4LLY B; 2QHR H; 3ZMR A; 4B95 C; 4WI9 A; 3WAJ A; 4OFP A; 1NGW A; 4F92 B; 2Z31 D; 1ADQ H; 5IP4 A; 3V6O A; 4OFY A; 2BIN A; 4CMH C; 1BM3 H; 3AB0 C; 1SM3 H; 2W65 A; 3TT1 H; 1AR2 A; 3TZV B; 4F1S A; 3DVG A; 1V27 A; 2C9Q A; 3ABM B; 4PS4 L; 3OZ9 H; 4BBW A; 1DAN U; 2QPU A; 4BK5 C; 3H7B A; 3VMP A; 3FFO A; 3LPP A; 5ITB L; 4I3R L; 4AYF A; 4CBZ A; 1KFA L; 4NXS A; 5AUM B; 4WHT B; 4LRN H; 2RAL A; 1MI5 D; 1UWG H; 3QO1 B; 4GIA A; 5CJO L; 1MCF A; 5F0L B; 4QTI L; 3O3A B; 2PLT A; 4P59 L; 1FNE A; 3DPD A; 4AKO A; 1T7X A; 2XVG A; 3BAI A; 3TEY A; 4NM4 H; 2IWG A; 2VWE C; 2KQM A; 2WQS A; 3H9S A; 3B37 A; 2GQH A; 4PBW D; 4PVB A; 1U8Q B; 3ZX1 A; 4BCY A; 2J3S A; 3U0W H; 4JBS A; 3VI4 F; 2LU7 A; 2XPZ A; 2KLT A; 5C7Z A; 3TJH C; 5E4A A; 3IDY B; 4R4N D; 4MDJ A; 4Z0X B; 5D2L B; 1WG3 A; 4XAK E; 2FWL B; 4AEN A; 1OAY H; 5D2L E; 1R3L A; 2BCK B; 2Y22 A; 4JPW L; 1FNS H; 4FP8 H; 4PW9 A; 2IVF C; 3DDF A; 5DLM H; 2XVB A; 3MRP B; 5DVK A; 2WO2 B; 3ZU8 A; 3ZE0 F; 3NIF E; 4MA7 L; 3M45 A; 4OJE H; 2ED6 A; 1UB5 B; 5D1Z I; 1DQM H; 4JE0 A; 5EU6 D; 4F1Z A; 2CHD A; 1C12 B; 3DMM A; 2J1X A; 4TUJ A; 3BJ9 1; 3SDA D; 2AV7 A; 2MCA A; 1LO2 L; 2ZCH L; 4OI8 A; 4PJ7 A; 2MRY A; 3U46 A; 3ARG D; 2DQI L; 1KN2 H; 5JYM A; 4PRN A; 5HGB B; 1FZJ B; 1QUP A; 4P5K B; 1A6U L; 2BP8 B; 3M46 A; 1WA1 X; 2BJM H; 1QLF B; 2DWT A; 3KBP A; 4IW4 C; 1BHG A; 1GHF H; 3T08 A; 2ZYH A; 4GW4 B; 2CDF A; 1J0J A; 3OAY H; 2OZ4 A; 4H8H A; 1OT2 A; 2NSX A; 4M9O A; 3IFP A; 3RGV B; 3UP6 A; 1T0O A; 2XWT A; 2C9X A; 3ABZ A; 5LXG L; 3MRM A; 5F7U A; 3FCS A; 1ET7 A; 2JJS C; 2V6H A; 3G5W A; 1GC1 C; 3H8N A; 3GMQ B; 1AGB A; 4BE5 A; 4HK3 J; 4DPA X; 4P2A A; 5CIN L; 3TNN B; 4OF7 A; 1MCR A; 4FQJ H; 4MAY C; 4F2M A; 3ZDZ F; 4IGB A; 1UFG A; 3MJ7 B; 5IWL C; 2XN1 A; 3ZIY A; 2HA1 A; 3GMP B; 1J18 A; 1ETA 1; 1KT2 A; 4WSK A; 5FUZ H; 5C0C D; 4R7L A; 1KIU A; 2XY2 A; 1JGI A; 4GP4 B; 4J3W A; 3B9K C; 3J07 A; 4WJG 3; 4QKX B; 2BY1 A; 5ETU B; 2IAM B; 3PP3 K; 4NKO B; 5HGG S; 1EUU A; 2Y36 H; 4E9Q A; 4GI9 A; 3LB6 D; 1HIN H; 2HTK D; 3I2C H; 1I53 A; 3FSV A; 5AFA A; 4UV7 H; 1ZTX E; 2H6P A; 2KL6 A; 3PPS A; 3FCG A; 1E43 A; 2WZ6 A; 1MCS A; 4QXW A; 3CLE H; 4K9E L; 4OZF B; 1RVF H; 3FVP A; 3GPE A; 3U7Y H; 2DJ4 A; 4C56 B; 1T20 A; 5HVH B; 3L3O C; 4JMN A; 4KJW C; 4X6C B; 4C56 E; 3ES6 B; 4HC1 H; 4UBD D; 1E7V A; 2G4G A; 4GMS H; 4JFE A; 4JKM A; 3OB0 K; 3F05 A; 1T4K A; 4NCD A; 2KS0 A; 4LVO B; 4XP1 L; 1Q0Y L; 2GK0 B; 4WFF E; 5J8O A; 1FNG A; 3KLA A; 4YHI B; 3VW3 H; 2CJS A; 2VDQ B; 4XP4 H; 1TXK A; 4N2M A; 1AKJ D; 5I1C L; 1JZ5 A; 5ID1 B; 3PDO B; 4K23 H; 3WDI A; 4KKN A; 4N5U A; 2OI9 B; 1MCP H; 4F7C A; 3OD3 A; 2NY0 D; 2W0K A; 1RZQ A; 3PAZ A; 1R3K B; 2R69 A; 2AJX H; 1MKF A; 3C5J B; 3L4Y A; 4N0F B; 3RNK B; 4S0S D; 3S0P A; 1UTD A; 4DGI H; 3GMR A; 4PJ5 A; 2NNF A; 1MFC H; 4AQE A; 4U7K A; 3CNK A; 2D8K A; 5EDX A; 3SYQ A; 4P2R D; 4RAV B; 4XRC A; 4AT6 A; 2YBR B; 4LOV A; 1D5R A; 3IPB A; 3M17 A; 4ABW A; 4I8D A; 2CMR L; 3IMV A; 2G0L A; 1JN6 B; 1AGF A; 4OT1 H; 4UOQ A; 5DMI L; 4KQ3 H; 3RDT A; 5FB8 A; 1E4X L; 3DHJ A; 4HKJ B; 1Z9M A; 5L8J B; 4DWH A; 5LGJ A; 1CDI A; 3RQ3 A; 4MB1 A; 2F19 H; 2PAB A; 5DO2 C; 2DYP A; 3QW9 A; 4KTY A; 4X83 A; 1QD0 A; 3AG2 B; 5K9Q I; 4WLC A; 2NY3 C; 1B4J L; 5DLL A; 2PO6 C; 5H35 B; 3Q48 A; 3UAF A; 3HZY B; 2VB5 A; 3FZU C; 5EU6 E; 3US9 A; 2ARJ Q; 5DUM L; 2Y24 A; 1OSE A; 4I48 A; 2J5W A; 4N0C D; 3IU4 L; 5E5M B; 1E6J L; 4PJC A; 2VIR A; 1OKE A; 2Y7O A; 5HHO D; 4HT1 H; 2KT6 A; 4LBE B; 5J3H D; 3L17 A; 2OMT A; 3LY6 A; 3CZU B; 1CT8 A; 3WFB H; 4J1U B; 4E5X A; 2I9T A; 3QFG A; 1U3Z A; 4LHJ B; 4LK4 A; 2VDR L; 4Y1S A; 2V7N A; 4LL4 A; 1DVZ A; 5CD3 B; 3HE7 B; 4PJD F; 3BRD A; 3EGD B; 3RKD D; 4MXQ D; 1KCV L; 1NGP L; 1F1G A; 4UT7 H; 1KPU B; 4CDG C; 2YGQ A; 1CFQ A; 1BAG A; 1WCB B; 2EE2 A; 4DW2 H; 1BX2 A; 4P05 A; 1PZ5 A; 1YJD C; 4F8N A; 1GS8 A; 5SWZ B; 3BKY L; 1XGY L; 2PP8 A; 5DYF A; 5F3F A; 5HGD A; 3D87 B; 3QRL A; 1URI A; 2CAK A; 4KDT A; 2DQC L; 3NAC H; 2LTQ C; 3K4Z A; 1PG7 W; 4IRS D; 1A7P L; 4O51 A; 1F3J A; 2DQJ L; 2PWD A; 1FJ1 A; 1O5P A; 3QVA A; 3ZQ9 A; 5D2N C; 1R3J A; 3ECQ A; 4HKB B; 3ML9 A; 2WQA A; 4I88 A; 4LCU B; 1RSY A; 3WXW H; 3E2H C; 3HJ0 A; 1L3W A; 2EXY D; 5DVO A; 1IBB A; 4MD4 B; 2APS A; 3BXN B; 1BJ1 H; 1NUD A; 5HI4 D; 1LMK A; 4NWT L; 2HWZ L; 1FSK C; 4WI0 A; 4QPE A; 1PLC A; 4XMU A; 2UUD K; 5ICZ B; 2O5X H; 2Y7S A; 2ZPK L; 1D9K D; 5J0B A; 5IQL A; 2G60 L; 2C9P A; 1DBM L; 1KPV A; 1P6F A; 5ECJ E; 5JIP A; 4N0Y L; 3AVE A; 1FG9 C; 1WZ1 H; 3D5O F; 5GSV B; 1R3I L; 4UOW 1; 3SPJ A; 1UM4 L; 1HWH B; 2J71 A; 3RE0 A; 5ISZ B; 1KYD A; 1F0L A; 3GHE L; 2YMT A; 1CF1 A; 1TEY A; 3HBQ A; 3KYO A; 4X0N A; 1PIF A; 3QJT B; 4XE0 A; 2N7A A; 4KK9 C; 4KNQ A; 3O3E A; 1A7N H; 3VXR D; 1DQQ A; 1KZQ A; 3MBW A; 1OQX A; 1J7V R; 2WB7 A; 5DGQ A; 1I82 A; 2X2U A; 3VSV A; 5K9K A; 2H6O A; 3PWU B; 3ZE1 F; 1OQO A; 4NDM B; 8FAB B; 2EXS A; 4ZUV B; 2BSB A; 4K3G A; 2GIM A; 1WIT A; 4G43 A; 5FM5 O; 2XZQ L; 4P2R E; 2H26 A; 4MQS B; 5B3N A; 3IDX L; 4JFQ A; 1W2K T; 4LNR B; 2P24 B; 1X5X A; 4AQ8 A; 1NL0 H; 2CM6 A; 3T1F A; 1WT5 A; 2MJW A; 3T2H E; 4QRT B; 1NWP A; 1P0S E; 4PRH A; 3U0P B; 2R0K L; 3QQN A; 4YSE A; 5DSC A; 2TMN E; 1NDM A; 4PBX A; 2BOC B; 1MJJ A; 3EJR A; 2V9Q A; 1KXQ A; 3K3Q A; 4FFZ L; 5I6O A; 3RZC D; 1OV8 A; 1JGV H; 3DC0 A; 4UQI M; 3WKN A; 3TES A; 1S7U B; 4F9L A; 1DVY A; 1YUK B; 2QQN H; 3O2V L; 5KG9 A; 3AXL A; 3KPM B; 3NZ8 B; 3KW7 A; 4NNX B; 3ZKS D; 2N4I A; 1LND E; 2AQ2 A; 5LGK A; 3VGF A; 1I1F A; 3HAE G; 4XMK H; 4FQ1 H; 5EUK B; 3U2J A; 1F3D H; 1RV6 X; 5HHO B; 1WAA A; 2P1Y A; 4FTD A; 2C4F U; 1Q0X H; 2VXQ L; 5FGB C; 5HHO E; 1BFO A; 3GFU A; 3LCY A; 1NC4 A; 5DZL A; 3GIN A; 1JXD A; 2H0E A; 3UTT A; 2YPV A; 4E9V A; 3OC8 A; 2JQZ A; 4BZ1 H; 4B6Z A; 2HG5 B; 4OV5 B; 3N0P B; 5DYO B; 3VSU A; 3HZK A; 4K3E L; 3E6F B; 1XF3 A; 5JW3 L; 5B3J F; 4F8T A; 2XV0 A; 5JPN A; 5AW9 B; 3RA7 H; 4I9X A; 5HD8 C; 4Y2D D; 4F24 A; 1AE6 H; 1AOZ A; 3DX6 B; 1DCE A; 1HZL A; 4OAW A; 2VXS L; 2WGN B; 5CWS B; 1E28 B; 3HZM A; 4Y2L A; 4DGW C; 3MCY A; 3MCG 1; 2MSP A; 2VN7 A; 5DK0 A; 2V2T B; 1KGJ A; 5FXN A; 2GYS A; 3MLY L; 4BQ8 A; 4LLU A; 4IRS B; 1W8N A; 1T1X B; 2RCS L; 1TTR A; 4HII B; 4PJ8 D; 4WSG A; 4LHU D; 4OM0 L; 5LNE A; 1TZA A; 1A47 A; 2PCX A; 2H9G B; 3POH A; 4POZ C; 5DK2 A; 3GXU B; 2VDR B; 4BKS C; 5D1Z A; 3SGD H; 4DPC X; 3T04 D; 3L5W B; 1PJ9 A; 5J5E A; 5DHE A; 2FJS A; 4P9H H; 5DJX B; 2E0P A; 1YUH B; 3HEI B; 3UAJ C; 1Q90 A; 3LN5 A; 2FR4 A; 3BN9 D; 1TJH H; 3P4O A; 2Q8A H; 5IVX B; 3OJD B; 3T0X A; 5L2K D; 4QRQ B; 1KLF B; 1KY7 A; 2HRL A; 4BKL B; 4L1B A; 3SDD B; 4GW1 B; 3LHA A; 3PFU A; 1ZA6 B; 3ZRF C; 5HYS A; 3VFR B; 4I85 A; 5ESQ B; 1GCF A; 3PE9 A; 2W60 B; 1TPX C; 4TF4 A; 5TE6 L; 4ZAK A; 3HBG A; 4JGM A; 5LN4 A; 4LEB A; 2UYL A; 5ICX A; 1BBJ A; 2ZON A; 4PG9 B; 5E6I A; 3FTY A; 2DIC A; 4JQI A; 2VQU A; 1KFG A; 3AW5 A; 2VOT A; 4WEB L; 1JXJ A; 1AO7 B; 1FLR H; 3EIM A; 2AXW A; 4FFY H; 4I2X B; 1CCZ A; 5HY9 A; 4QHM A; 1AO7 E; 1CZ8 L; 1SJV A; 4FXK A; 1CBV H; 3VJ6 B; 5JTW A; 4NN6 B; 4QNP F; 5JZ7 D; 3TBT A; 3VUA A; 1KJO A; 4FOM A; 2BY3 A; 4YSC A; 3L1F A; 3H85 A; 2DMB A; 1KB9 J; 4JFO B; 4KKZ A; 4A7Q A; 1BF8 A; 5KZC B; 6PCY A; 3B9V A; 1TCR A; 3BGM A; 1ODA A; 1ZLW K; 3G5V A; 3RAJ H; 1ZDQ A; 2YD6 A; 4HBC H; 5KOM C; 5FDW A; 5KS9 B; 3QI9 D; 3SYO A; 2OTW A; 1AS6 A; 5KS9 E; 3OJM B; 4LDO B; 3KJ4 B; 1SHS A; 4HG4 K; 2E9I A; 3TFB A; 1ROW A; 2XG4 A; 3WG7 B; 1F4X H; 1Z9S A; 1HKF A; 3MLP A; 3F58 H; 3N1Q C; 1I9I L; 5HDQ L; 1HL8 A; 3KYN A; 4Z3G A; 5EC1 B; 4CAD A; 2A4C A; 3DPB B; 4LEO B; 5IEK A; 2JKW A; 4AR8 A; 4UAH A; 5I15 L; 1IGY A; 2P42 B; 1G7H A; 3ZZS A; 4EBQ H; 2WNG A; 2WLI A; 1OE3 A; 2D03 L; 3CPL B; 5KVE L; 1RZK L; 5E8O B; 3VX1 A; 4HIE A; 3UPC A; 1OT1 A; 1WFT A; 2ERJ B; 3VFT A; 4L2Y A; 3ARE D; 4GP5 B; 1NAM L; 2K9F B; 3FN0 H; 5FSP A; 2UY7 A; 2NNA A; 1JNJ A; 3RJQ B; 3WKQ A; 1OTT D; 1JTT A; 3D07 A; 4BFN A; 3VU1 A; 3CXD L; 5DUB B; 3SEP A; 4M3H A; 1CN4 A; 3RWI A; 1SQM A; 2B2X L; 2Y5T B; 4L9L B; 1RIV L; 4H0I A; 1QYG H; 2VZU A; 4Q6I B; 5BV7 C; 5I1A B; 5IT2 H; 3AQZ A; 3H6A A; 4KZD H; 1IGJ B; 5FA3 A; 3D69 B; 4PJ7 F; 1GGB L; 3HS0 C; 1QBM H; 3L3J A; 2I5Y L; 4O9H H; 3HNS H; 1SYV A; 4QO1 B; 5LLI C; 4XVS L; 2XVL A; 1E6O H; 4DF9 A; 4EZL A; 3IMB A; 4KH2 A; 3BCF A; 5GSR A; 4GBX A; 4R5X A; 2R1W A; 2FLB T; 2ZJS L; 2BXZ A; 5IRO A; 4PJ8 B; 4FQL L; 4DAG H; 4RGO L; 3UTZ B; 5L88 H; 1HK6 A; 2MTP A; 1NFI A; 2VH5 H; 2IBZ X; 1B9Z A; 1JCK A; 2JK5 A; 1H3T A; 4M43 H; 2BIQ A; 3BKC H; 3LS5 H; 4ODW B; 1MRD L; 2FBR A; 3O6F D; 4FL4 B; 5D71 H; 5K1J A; 1ZEA L; 5LGH H; 3DJR A; 2YAH A; 3FOM A; 4S0T D; 1WIP A; 1OHZ A; 2OX5 B; 3NFN A; 4A7U A; 3A6C H; 3TN0 D; 4XMT A; 2XV3 A; 1FHN A; 5I30 L; 3P7R E; 3ZOA A; 2ZUQ B; 4WNQ B; 4KVP A; 1NPC A; 4XT1 C; 5L2K B; 4RBP K; 4HF5 L; 5L2K E; 4ODV L; 5FOJ A; 1DBJ L; 4X5W B; 2HT2 C; 4LCV A; 5I6N A; 4DKF L; 3H3P H; 4ODX B; 2ZT9 C; 3HAE H; 4MMX B; 1AD0 B; 2WHV A; 5EOC J; 1NJ9 B; 2NY6 D; 3ML8 A; 3T8F A; 5E8E B; 5CIP H; 4QD2 E; 4PR5 B; 1FDL H; 1XVS A; 2CGR L; 4CUC A; 5I0X A; 4HOZ A; 4R0L B; 2CJU H; 5AAP A; 2JF1 A; 3CLW A; 2RR3 A; 3BUP A; 4NUG H; 2G9K A; 2Q8B L; 3C9N A; 3JV4 B; 2GEE A; 1QG3 A; 5EA0 L; 2VUY A; 1YCS A; 4HCR A; 2JEL H; 1H6E A; 5IFA L; 1SF3 A; 3HBP A; 1I3V A; 5IUC A; 4K5P A; 4GW5 B; 3VWK C; 3QYM A; 5MEQ B; 1FZK B; 1WFU A; 4CW1 A; 3EYS L; 2KZW A; 4P4K D; 3QEU A; 3INO A; 2I69 A; 4YHP A; 1H5B B; 1JYN A; 3HI5 H; 1I9J L; 4NYL A; 3A47 A; 4M6N H; 2AEW A; 3BVW A; 1J88 A; 5LO7 A; 3W5N A; 1L7I L; 5E9A A; 4GLR I; 4JUS A; 5C1M B; 3R1G L; 2CGY A; 2HT4 D; 5I18 H; 4XXW A; 3L3K B; 3OAZ H; 5ACL A; 2R2H B; 3QA3 A; 2WC7 A; 4UTA H; 5FYL L; 4TQE H; 1M56 B; 5FO2 A; 5TE4 L; 1YN6 A; 1G1O A; 1TZG H; 3G5Z B; 3UX9 B; 3QC5 X; 4OZH B; 4HIH A; 2FGW L; 1XF4 A; 1OK8 A; 2N7B A; 2Z6F A; 3IB4 A; 3QOR B; 4OZH E; 1TMN E; 3TYG L; 2P44 B; 5E8P A; 2R1Y A; 5F2F A; 1S7Q A; 1G6R B; 1OGT B; 3GSN H; 4U05 A; 1MFB L; 1Z32 X; 2K0R A; 4IHO B; 3ATF A; 1A4B A; 2YMX H; 4RFN C; 4YHZ L; 4L2L A; 2FYY B; 3E1F 1; 4BCZ A; 1XGT B; 3EDF A; 1BCI A; 4M5Z L; 2QVT A; 5IKC B; 4J4P D; 2DD8 L; 3G5Y B; 3WFE L; 3ECB B; 1CQK A; 5D70 L; 5TF0 A; 4G6K L; 3BPN C; 3VG0 L; 1BGX H; 3Q5O A; 5FYL D; 5T3Z E; 3HR6 A; 1J1P L; 4KK6 C; 5JXE C; 4XXC A; 2WVU A; 3LKQ A; 2XPG A; 2C7U B; 4NUJ B; 2CXG A; 4IOF C; 3JXA A; 5D9S A; 1RP9 A; 1L3F E; 3RIF K; 2NQ3 A; 4HJE A; 3BUD A; 4PJC F; 1P1Z A; 1LMI A; 3I85 A; 2R8S H; 2P3Y A; 3P30 L; 1S78 C; 1PW3 A; 4XOA A; 1RJC A; 3EFO B; 3S8G B; 3L16 A; 1A3Z A; 3DYO A; 1A9V A; 3FON A; 1K2D B; 2VAB B; 3HLA B; 4LF3 A; 3MKK A; 2Y1A A; 3MJG X; 5ES4 B; 1KCU L; 3ZVV A; 4WTW A; 4G8E A; 3LDB C; 5EC2 B; 1QBB A; 3ZZ1 A; 4Z77 A; 5LDU A; 4CBP A; 2JKI S; 4N8V G; 5E0Q A; 3CXF A; 2UXV A; 3B8S A; 5CCK L; 2DLH A; 3T65 A; 1C5C L; 1GZP B; 1MOE A; 2WXJ A; 3LX9 A; 4NQS A; 4X5P A; 1YY8 B; 4WTZ G; 4G6F D; 1N7M L; 4PRE A; 5IFH H; 1G9E A; 2K4A A; 1TJG L; 4HC1 A; 2A6I A; 3QNF A; 1GW0 A; 2WY3 B; 4LOU C; 4U1S A; 4JDZ A; 3C5Z B; 3SGK A; 1HH6 A; 4Z8F H; 4FNP A; 4OLY L; 1EJ8 A; 1XBR A; 3O8X C; 3MRO B; 3TBY B; 5GRY A; 2EE9 A; 1ZGL B; 4EQ2 A; 1Z5Y D; 2IWF A; 3IBO A; 2FVN A; 3BT2 H; 3ULU F; 3GYR A; 1F6A B; 3L4Z A; 2I24 N; 5D7L A; 3VRJ A; 3NME A; 3F7Y B; 4WUU A; 3IFO A; 5FW8 A; 2F1B A; 4L4J A; 3D33 A; 1YO4 A; 5IHW A; 1QSF D; 4K7P L; 3OSK A; 1JF1 B; 4XCE A; 3WEB A; 2A73 A; 3ZTD C; 1H3V A; 4EIU A; 4KVN H; 2X3C A; 4KNU A; 1U8C A; 3WTY A; 2QJD A; 1P2C B; 1YEH L; 2V2W B; 3B5H A; 4IEF B; 4WTQ A; 3WN5 C; 1N3N A; 2X40 A; 3INU L; 3OR7 A; 3IRP X; 1IAI H; 3KNB A; 2PYE D; 3IGU A; 4CC4 A; 2UXT A; 4MSW B; 1BVK B; 4YQX H; 2R7E A; 5I17 A; 2F53 A; 4P3D A; 3P4L A; 1WFM A; 2E9G A; 1RUA H; 5FS5 A; 4LSS H; 4ZZF A; 1OE1 A; 2QL1 A; 3FB8 A; 3RI5 K; 4YE0 A; 1AXT L; 4TVP H; 3U1Q A; 4MQT B; 4NPJ A; 4X4X A; 1DJS A; 1WWC A; 2ICC A; 4MDI A; 2BVQ B; 3NTC L; 3MJ8 B; 4WNJ A; 2CJ3 A; 3EG9 A; 2XQY G; 3UCR A; 3ZY7 A; 4UIL L; 3MLV H; 2VKX A; 5K39 B; 4GKZ A; 5KVD E; 2H0P A; 3MS3 A; 1F3R B; 4FZE L; 4GPV A; 3OKO B; 1IT9 H; 2XU9 A; 1N6Q L; 2WO3 B; 3MHZ A; 2HT3 C; 4WHC A; 5C09 D; 3B8Y A; 3VMN A; 1A8J H; 1FYT D; 1U3H A; 4XX5 A; 3HC4 L; 4YC2 C; 4FQX B; 5D8J L; 4L8S C; 3GUF A; 3K74 B; 1DBB L; 3CFD A; 3LRM A; 3VGH A; 5LWD E; 1OP4 A; 2WKG A; 3VQ1 C; 1TET H; 2YD3 A; 3F7Q A; 2DI8 A; 4X6E A; 2FSD A; 5AB1 A; 3BC9 A; 5HHM D; 2XKN A; 4XN1 A; 3VXP B; 3VDC A; 4G9F B; 4ZOB A; 1LD9 B; 4G9F E; 5IRE A; 3IN2 A; 1HHH A; 1NDT A; 3K81 A; 1WAA F; 4FEM A; 1YC8 A; 5FYJ H; 2F7I A; 3TBW A; 4BFG A; 1BF5 A; 4F37 K; 4AIT A; 4RH4 A; 3NJN A; 2FH6 A; 4L5F H; 4V1D D; 2NY2 D; 2YRL A; 5GGQ H; 5F96 L; 3BP6 A; 5BJT B; 1CVS C; 2Q6W A; 3MVS A; 2ZCL H; 4PLJ D; 5DD1 L; 3QQ9 C; 4PRB B; 4JDE B; 1R6V A; 3O45 A; 5J9P B; 4C59 B; 2VR9 A; 3HZV B; 4I0P D; 3DV4 B; 4G6F B; 3V9E A; 1GL4 B; 3MOA H; 4OSU L; 5J6G B; 3U1S L; 4X2U A; 4FNR A; 5GIR B; 3WIH L; 2AF2 A; 2NY1 B; 2LHA A; 3ART A; 3PL6 C; 5J6S A; 1IBD A; 3VFV B; 5CMA A; 4TNF A; 3KYM B; 4PG2 C; 4EUP A; 1NBQ A; 2P8P A; 2B4C C; 5DEG A; 4OLZ H; 3FTZ A; 2QPE B; 4UF7 C; 4OD1 L; 5CD5 D; 1VER A; 1HCY A; 1CID A; 1KN4 L; 4O58 H; 5E4I A; 3KED A; 5SX5 H; 1JNN L; 2J7L A; 3W1Z A; 5KSA B; 4JZN A; 2J2Z A; 2QQK H; 3U1P A; 4MAY D; 2V9R A; 1BM7 A; 5DVP A; 3MYJ B; 2Z92 B; 2YR3 A; 1HXY B; 3GGW A; 3D3V A; 1R47 A; 1V3L A; 4BJL A; 4PWJ A; 1A7O L; 3WCY A; 4W9H C; 2R51 A; 1I3G H; 2PYE B; 4Y19 D; 1DJZ A; 2CJT A; 2NLJ A; 3JVF C; 3MRK A; 4PJX A; 2PYE E; 3V5H A; 4QLW A; 3SO5 A; 5FV2 A; 3NN8 A; 5ACZ B; 4FMK A; 1SHW A; 3ARF C; 2L0D A; 5ERW A; 1MEX H; 2MCP L; 4FXT A; 5IR1 A; 4GXU N; 4HWB L; 2FQF A; 3JTS A; 4IGB C; 3ROO A; 3K91 A; 1M6O B; 4HJJ H; 4GRL B; 4OJD H; 3SGJ C; 1A7Q H; 2YHG A; 1KTL A; 4M65 A; 3V0G A; 2F2H A; 1NM9 A; 3OGC A; 3UBH A; 3FUK A; 4F3F A; 5FUO L; 3UJI L; 1EHA A; 2X2J A; 2E4F A; 3FV4 A; 3ATE A; 4AGM A; 4AYI D; 1FPT L; 2FL5 A; 3SE8 H; 1YMM D; 3HG2 A; 2LAB A; 1VFA B; 5T1K A; 5E4E C; 2D7T L; 1D2P A; 4XCY A; 3BH9 A; 3K8L A; 4H7V A; 1HN1 A; 2KH2 B; 1MJ7 H; 2ZSV A; 1CFV L; 3QIW A; 2D7N A; 2XV2 A; 1CLZ L; 2QV4 A; 2BVP A; 2WVT A; 3FJT A; 1S7R B; 1ONQ B; 4QJ5 B; 3DX7 A; 4L8D B; 1XF5 A; 4AVJ A; 1JGL H; 3MJ6 A; 5F7E H; 4E9W A; 1DQK A; 5HDB E; 4YXH L; 1E8X A; 4TNV F; 2JI1 A; 3C5S A; 2IAN B; 4YDZ A; 1KY6 A; 3W9D A; 1MW1 A; 2X10 A; 4TKW A; 5AOK A; 3UTP D; 1C9S A; 3MRH B; 3MLZ H; 4OJC A; 1UHV A; 4KI5 D; 5EWI L; 3APM A; 3D0V A; 3QO0 B; 4JVU A; 1KGI A; 3CX2 A; 2A4E A; 2VRA A; 1JRH H; 4WO0 A; 3EJZ D; 4AJV A; 4DEP B; 2B8O T; 3HNT H; 3V4V L; 5AYU L; 4P9M H; 4AMX A; 5E5L A; 4OZF G; 3C5X A; 1JGE A; 5F7D A; 4CVW A; 1AGE A; 1T1Z A; 1KB5 L; 5CS7 A; 1NEP A; 1R17 A; 2ADG A; 4V1D E; 3UAE A; 1DBK H; 4R8W L; 4TL5 A; 4FTV A; 2R2E B; 5DD3 H; 3RUG B; 4OD2 A; 5JSS E; 3RUG E; 2NDG A; 1SXB A; 1CLV A; 2EYR B; 3V7M A; 3RKP A; 2E8Z A; 2QPF A; 3OB8 A; 4YGV A; 3QJF B; 3RDT C; 3ARQ A; 4MMZ A; 1F3D J; 1B0W A; 1P5U C; 1QLE H; 1QOK A; 3RUR A; 3QUM L; 3X1G A; 3T0E B; 4ZMW A; 4U6T A; 4CVX A; 1T7V A; 2AC0 A; 1CWV A; 3T0E E; 3E9T A; 4PP1 D; 3B5G A; 2H32 H; 4UTC A; 1HOC B; 2ESV D; 4MNG A; 1T1W B; 4PJ9 B; 3WLB B; 2A1W H; 1YDP A; 4AE0 A; 2FX7 H; 2DWE A; 4KXD A; 3D6G A; 4I9W E; 3EOT L; 1CFM A; 5IB3 A; 4K73 A; 2WBJ C; 4M1D H; 1HYT A; 1JDA A; 2HUH A; 3K4A A; 4N68 A; 3LQA C; 1KLG B; 3BIK B; 4ZQK B; 4IMK A; 2K4W A; 4JG1 L; 1N64 L; 2CDE B; 3VQL A; 2AI0 H; 4WW1 B; 2WB1 A; 4E41 A; 4G83 A; 1MCK A; 5CBA B; 1CR9 L; 4RA3 A; 3S6C A; 1EO7 A; 2IGT A; 3ZQW A; 2WIN A; 1YY9 C; 4EI5 D; 3EO1 B; 4HKX B; 3AQX A; 1C5D A; 2VTF A; 2W0F B; 4L3C B; 4P2Q D; 1YEK L; 2LAA A; 2HKH L; 5H94 B; 1Q9K A; 4Y19 B; 3BZF A; 4YSD A; 4TNW F; 2NY5 C; 4HIJ A; 4Y19 E; 3GXN A; 4HD5 A; 5FO7 A; 1T21 A; 3D85 D; 3BMW A; 4QLR A; 5K59 F; 1X5A A; 2DN7 A; 1TVH A; 2RFX B; 4HJ1 A; 5KYW A; 1IVT A; 1WRF A; 3WO4 C; 3RDJ A; 4FKH A; 3G04 B; 4NP9 A; 4B97 A; 1QRK A; 2OXH B; 1AZB A; 3K0Y A; 1B90 A; 1QNZ L; 3EYV A; 4P5T C; 3TM6 A; 1JZG A; 1MY5 A; 3WTW A; 1VEO A; 5CP3 H; 3TGX A; 3GSQ B; 3GUT B; 2X6M A; 3O2W H; 2NUT A; 5ANM A; 4FHK A; 2VQX A; 3WHE 1; 1YMM B; 3V4P H; 1EQW A; 1MDT A; 3ULU C; 3QIU D; 4YDI H; 1YMM E; 2ITC B; 4AL8 C; 4PJI B; 1NBV L; 4V2B A; 1EX0 A; 1VFO A; 4YUE L; 4PJI E; 4HUL B; 1Q1J L; 4KO9 A; 4URT B; 5IMO A; 1BLN B; 3ATB A; 3QUZ A; 4R7N A; 5DRX B; 3IKC A; 3V0J A; 1A1M A; 2XET A; 1P4U A; 5HGA B; 4JFD B; 1LP9 F; 4JFD E; 1OCW H; 1B4R A; 5C2B L; 4K3J H; 5FGC B; 5BXF B; 2ADJ B; 5TTR A; 1RZF L; 1PD6 A; 3S8F B; 4L4T B; 5FGC E; 4L29 B; 3UTP E; 4L4T E; 2WXO A; 1LE9 B; 3MGT B; 1NC2 D; 3SBP A; 3VM5 A; 3VXU D; 2WXK A; 3O9W D; 3GI8 L; 1QI3 A; 4FDW A; 4CA4 A; 4IBW A; 4KI5 E; 5T5N F; 3V0W L; 1F4W L; 1J8S A; 3PQY D; 4YEE A; 3ZYO A; 3FPY A; 1J05 A; 5IVO A; 4Y4F C; 1D2O A; 1IE5 A; 1U8P A; 2MKL C; 1ZSH A; 3EO9 H; 4XBE H; 4WIQ A; 1AD9 B; 2ZUT A; 1IU1 A; 1PEZ A; 1VEM A; 5HHV H; 2B0S L; 4FNU A; 1P9M A; 1M7I A; 1SBS L; 1SDY A; 4LCY A; 3HI1 B; 3R4D A; 2R1X B; 4UM9 D; 4P46 D; 4AEF A; 4OCR H; 4YL9 A; 1WT5 C; 2WMP A; 3ZTC C; 1R24 B; 3CFN A; 2FJU B; 2BS9 A; 4KTE H; 4HUX B; 4YHK H; 4HWN A; 3PWP A; 3WMS A; 2AJV L; 4TSA H; 4HSA C; 1OS0 A; 4MFH A; 3CFC H; 3LZP A; 3CHP A; 4AVI A; 5CHN A; 3HAE A; 3VFP A; 1DLF H; 4MCY B; 4QCI B; 3IT8 D; 1QF2 A; 5H37 G; 3SKM A; 3TFK C; 3TO4 B; 1LNF E; 2F0C A; 1FFO A; 5MEN B; 1N26 A; 1SOK A; 5FA4 A; 3PGF H; 3TA3 A; 4ECN A; 4D8O A; 3UAC A; 2BNR D; 3J7H A; 3EUD A; 3MRN A; 3Q5Y A; 3C2A H; 3A5V A; 4JFU A; 4PP2 B; 4UOW 0; 1D5I L; 1T2X A; 3BH4 A; 3EH2 A; 3MRL B; 4ZS6 C; 5C0J B; 1KYO J; 1QSE B; 4JKN A; 1WPC A; 4M7L H; 4NKM A; 1UA7 A; 4PJE A; 5E08 L; 1QSE E; 1LJM A; 2DI9 A; 3GIV A; 1E27 A; 4FHB D; 2VDM L; 1MFA H; 4EI5 B; 3GK8 L; 4LLM B; 1QBA A; 1KJ2 H; 5DT1 L; 5DHM C; 1JNH B; 4YDL C; 5E2V H; 5GGR B; 1F90 H; 3FCT B; 1DJY A; 1VPO H; 3NCX A; 2K6Z A; 3L3I A; 1ZTX H; 3OC5 A; 3X14 A; 4B5E A; 2PTT B; 1XGP A; 4LO9 A; 2WLM A; 3D85 B; 3UXF A; 3ISW A; 1JK8 A; 3VXO B; 4BSW B; 4B9Y A; 1QQD B; 4HHG A; 4P99 A; 5SV3 A; 2EDF A; 1ZWI A; 1V7M H; 2DQU L; 2W9D H; 4PH8 A; 1JL8 A; 3OKE A; 3B56 A; 5J1S C; 3RFZ C; 32C2 A; 3N55 A; 3FB6 B; 3ZGI A; 3X12 A; 1UJ3 C; 1IM9 B; 2Y0S A; 4G71 B; 1R5I A; 3D9A H; 4KZE L; 2ORB H; 1EUT A; 4R7D A; 5A40 E; 3FTG B; 3SPG A; 2PMZ A; 2AQ3 A; 5KSV B; 4PTT B; 4OCM C; 1KTC A; 3S7G A; 3GBN L; 5F3B B; 1TU2 B; 3DJT A; 5AW2 B; 5C0B A; 2Z8R A; 3GSN A; 2CK2 A; 3ARB B; 1NIB A; 1G0X A; 3FFD A; 5JA9 A; 5LIF E; 1I8L A; 4QAW A; 5D1Z C; 1BDY A; 1L9P A; 3H9H B; 4PGD A; 2GJ6 D; 4PGC B; 1SVC P; 1M2V B; 3DVN B; 4X4K A; 5C0A D; 1MLB B; 1GA2 A; 2VLJ D; 4XNZ B; 4JNX A; 1SFH A; 3D18 B; 3KL5 A; 5HSF A; 4RIS H; 4UNI A; 3OVU B; 4EIB A; 1UUS A; 3ESP A; 2DQF A; 2HJL A; 3IWV A; 1R5V B; 2FTA A; 4DKT A; 1DN2 A; 4S1Q L; 2XQB L; 4HK3 N; 4NHU A; 4Q7D A; 4ZMA T; 3S5L A; 1LNU B; 2H4E A; 3FU3 A; 4MNQ A; 2V8M A; 1U8N A; 2RQ8 A; 2X70 B; 3O9W B; 3D2U B; 3EKC A; 5C08 D; 4Y16 C; 3TBV B; 3WMQ A; 1P5V B; 2YUZ A; 4HQQ H; 4PJB A; 4WF7 A; 2NNC A; 2A9M H; 3PQY B; 4PS4 H; 3PQY E; 3NCF B; 4ZUU B; 5GRV K; 2HRH A; 3DBX B; 1JWU A; 2E4T A; 4KOC A; 4KRP B; 5B1J A; 2EAE A; 5TQ2 L; 5ITB H; 1KFA H; 1UAS A; 2DKM A; 1X5L A; 5CJO H; 5EN2 B; 1SMD A; 1HWG B; 2FIK B; 3IJH B; 2Q2L A; 4I18 B; 4QTI H; 3S37 L; 5EUO F; 4P59 H; 4UT6 L; 5EU7 C; 2LU5 A; 2WQ8 A; 5J1T C; 1MHC A; 4FXK C; 3IWU A; 3AXH A; 1NIC A; 3OTT A; 1N8Z B; 3HNV L; 1KDJ A; 3GDC A; 3RL2 A; 2PW1 A; 4M3K B; 2WKL A; 3F5W A; 4X7T A; 4BFR A; 2BOB A; 4Y5Y A; 1H3P L; 3HLK A; 5TH2 B; 3UTO A; 3LKO B; 4ACP A; 1NBC A; 4JFX A; 1G5G A; 4DOH B; 2FBJ L; 4JZJ A; 4YZF E; 4DOH E; 2XN2 A; 5DWP A; 5TQ0 L; 4AZ8 A; 3KPL A; 3FQ4 A; 1UOL A; 5HVF B; 5AZE L; 2R0Z L; 4Q89 A; 3ZO0 A; 4H8P A; 3OG4 B; 4XAW L; 3ZT6 A; 4HOW A; 4U6H A; 1NFD B; 4PCK A; 1NFD E; 1PBY A; 3N5A A; 3WKP A; 1XL4 A; 4F9P A; 1JZF A; 1OHQ A; 5HYF A; 2EDM A; 3QUG A; 3VGD A; 3GPS A; 2ICE A; 1XIW A; 3I3D A; 4YDK L; 5LBS L; 3BLB A; 1LO2 H; 3PXJ A; 2ZCH H; 5D7J D; 2AS5 M; 4APQ D; 2DQI H; 1FL5 B; 1M71 A; 2H0F A; 4NQD A; 4OCL C; 2FCB A; 1A6U H; 3HE6 B; 5FYJ U; 4LOF A; 3L3D A; 4AR9 A; 3AS3 A; 4AV4 A; 3U82 B; 5DMK B; 3ECV A; 4WO4 B; 1B9K A; 1HCF X; 2NZ9 D; 2IJ0 C; 3ISY A; 4MB5 A; 4QOK B; 3ZE2 F; 3FB5 A; 4QOK E; 3ZKX B; 2ZUX A; 4QRR B; 3EHB C; 4QRR E; 3MUU A; 4H1L B; 2HKA A; 2WUB L; 1ACX A; 3HG1 A; 3X1E A; 5LXG H; 1RZG A; 4WV1 A; 5D6D A; 3U30 B; 3FAX A; 3LRH A; 4XCF L; 1RZJ L; 2EH7 L; 4X7S L; 5JIH A; 4OGA C; 3I3B A; 5T8I A; 5CIN H; 4TUO A; 3GRB A; 2O1A A; 4Y1A A; 4U6X B; 5EUL V; 3ASN B; 4GBX C; 4P2O C; 2HT2 D; 4PYG A; 3EFD L; 4LLW A; 4LGP B; 2ROY A; 1MCD A; 3EXJ A; 2VDM B; 4DER A; 5C97 A; 3JVG C; 4BGL A; 4ZX4 A; 4JY4 A; 4RFE A; 1J5I A; 5SWS B; 1NCD L; 5F3H B; 3LPO A; 4CJ0 A; 4JY6 A; 1CFS A; 1S8D B; 1INQ A; 2VLJ B; 3D8M A; 4GT0 A; 3M1B B; 3FO9 A; 1PD1 A; 5C0A E; 1KEG L; 2VLJ E; 1KEI A; 4K9E H; 1HNF A; 3FRU A; 4KGL A; 1G0Y R; 4KMQ A; 2FWU A; 5H37 H; 2G2R A; 5DZV A; 5EG6 C; 5FEC A; 5E95 B; 2QAD D; 4QYO A; 2BFG A; 4B3E A; 3W57 A; 2VSM B; 4HCG A; 2Y23 A; 2AXG B; 3QDM D; 1G1C A; 5C08 B; 2QFR A; 5HDO A; 2YPL D; 5I1E L; 5C08 E; 4XP1 H; 1Q0Y H; 4KFM A; 3EGS B; 3BD4 A; 4PJG A; 5IFJ A; 1CXH A; 4S0H A; 3EJU A; 2HLE B; 3JTT B; 3S9D B; 5I1C H; 1VKX B; 4Y4K A; 4II7 A; 4MHH G; 4K4K A; 5C6T L; 1IIK A; 1U3J A; 1HHI A; 2Q5B A; 4BQ6 A; 3O4O B; 1ZS8 B; 3BN3 B; 3NID E; 3U7W H; 2EO7 A; 5KUM A; 1TEG A; 1FP5 A; 2X4O A; 5HDW A; 4NFC A; 1CFN A; 3JV0 A; 1BZE A; 5JHL A; 2V2X B; 1D5M B; 2CPC A; 3U4A A; 4RZC A; 4AVK A; 3LH2 L; 3BZE A; 4A7G A; 2CMR H; 1MUJ B; 5GZN C; 2ZXQ A; 5DMI H; 4E42 A; 5JO5 B; 5FKA B; 4QGY A; 4Y5X B; 3GEF A; 3O0R L; 1BVN T; 4DUW A; 4C83 B; 3H4H A; 3QEH B; 5DMG C; 4XXW C; 4ZJ9 A; 3PAV A; 4YPG B; 1B4J H; 5F6I C; 1X4Z A; 3T2L A; 1FVE A; 2YD2 A; 4F27 A; 5B8C A; 3OHX B; 4KK6 D; 3WNI A; 2QSQ A; 2A0L D; 5E1I A; 2DJM A; 5DUM H; 4JRE C; 3ASW A; 3FQR B; 4X7D C; 1KYF A; 3QNX A; 4G7Y L; 2M1K A; 1HSB A; 3KPR A; 3MV9 D; 1U4E A; 2QMJ A; 3SZK C; 3IU4 H; 1RU9 L; 3ARD A; 3IDM B; 5IHU L; 4O2E A; 1YNT A; 4NRY B; 2E75 C; 5CJQ L; 1PQZ A; 2J6W A; 5ABA A; 3HPL A; 4Y88 A; 3EJC A; 3GXF A; 4GEJ A; 5T33 L; 2X4S B; 3LXA A; 2XXC B; 4U1L A; 3F62 A; 2VDR H; 4PD4 K; 2EIZ B; 4LMQ E; 1JUF A; 2EDQ A; 3K6D A; 4PRI D; 3DO3 A; 3QXT A; 4PW4 A; 1KCV H; 1NGP H; 2XG5 B; 2FEE L; 4UQ3 A; 4L3E D; 2H8P B; 5AWN L; 1DVS A; 1DVF C; 3DXA B; 3DXA E; 1KGX A; 1I7Z A; 3SKU D; 3O6K H; 2HVJ A; 1Q9L B; 2H41 A; 4GG6 F; 1R5W A; 3T8G A; 4H8W L; 3BKY H; 2WBK A; 1XGY H; 3L48 A; 4L1T A; 4N8V A; 3Q1S L; 4W9C C; 4JJ5 B; 1ZLD A; 3F12 B; 1NMC C; 2VN5 A; 2DQC H; 2YOC A; 4YSS A; 1A7P H; 1HHJ B; 1QA9 A; 2AAB H; 3SDC D; 1G7Q B; 4I0C C; 3WBD A; 2D7Q A; 2WTO A; 3I02 B; 3SE3 C; 4LAJ H; 2F1A A; 5D5M E; 3QIB C; 3B2M A; 1A9B B; 3LND A; 1SY6 A; 4Z5R A; 2EIE A; 4GSD L; 3GRW A; 2HWZ H; 4NWT H; 3U0T B; 3HC0 B; 5IM7 B; 1I85 A; 1LEG B; 2QAD B; 3CMG A; 4BHU I; 1AAN A; 4M6M L; 4LRI C; 5EBL B; 2Y07 L; 4RWY H; 1T7W A; 4Z3J A; 2G60 H; 3QDM B; 2XFG B; 1DBM H; 3FPX A; 3FCQ A; 3QDM E; 4H5S B; 4RDQ G; 3ZKN D; 3ABK B; 2N01 B; 3RTQ A; 2QKI C; 4NP4 L; 4Q9R L; 2GJP A; 1R3I H; 1RZ8 B; 2XZA L; 5A2K H; 2QZQ A; 2NR6 A; 4FG6 C; 4O2F B; 1F11 B; 5HVG B; 4DO5 A; 1E2Z A; 3CVD A; 4XN8 A; 1ND0 A; 1JK9 B; 3PLY A; 4B52 A; 1AQD A; 4RGN C; 3VXS B; 4ZNE A; 3RX5 A; 2JKR M; 2X0V A; 3IDI B; 4PGB B; 3CQF A; 1THL A; 4OV6 F; 3I6K A; 1FJQ A; 1FJT A; 3GS4 A; 1BZ8 A; 1K5N A; 3Q8B A; 4XNQ B; 2Q87 A; 4APX A; 2ZWL T; 3QGO A; 5I16 A; 3MN8 A; 2H0J A; 4JM2 D; 2XZQ H; 5CJX C; 1AHW A; 2EXW C; 3C60 D; 3DGD A; 3MV8 B; 5JMR A; 3MV8 E; 3CZG A; 5DEF B; 3CFT A; 1D5Z B; 4BFF A; 4M8Q A; 4Q74 A; 2WIM A; 1E5Z A; 3ZDW A; 1BD2 A; 2OKM A; 4FE9 A; 1S9X A; 5C0F A; 2AQR A; 2DMC A; 3R06 A; 2J8U B; 5CN3 A; 2J8U E; 2R0K H; 2ICW B; 1JS4 A; 5J13 B; 3MRD A; 3APF A; 3BQU C; 3WUW G; 3W39 B; 4OBR A; 5JPM C; 2CE6 A; 3IBE A; 1FZM B; 3PXL A; 4JHB A; 3F01 A; 4FFZ H; 5A1A A; 4NEH B; 1OF2 B; 1A6W L; 1IV8 A; 5C0N D; 2CHN A; 4J3X A; 5CZX C; 5CUS H; 4AUU A; 3VNY A; 3O2V H; 3MX0 A; 4JRY A; 5C3I A; 4MQX D; 2WPG A; 2Y5E A; 2AKR B; 4NAQ A; 4U3S A; 1CFB A; 3P3U A; 2NUP A; 3OPZ L; 1FBI H; 1QLR A; 4FFW D; 4G72 B; 4ABU A; 4MWP E; 3VXT B; 3DSN B; 3CDC A; 2VXQ H; 4OCW L; 5DRN A; 1XME B; 2L7E A; 3F85 A; 3CFK B; 4QTH B; 4GNK B; 1ZLW H; 3TNM A; 4Q0X L; 3CIQ A; 4L23 A; 3H9S D; 5CN2 A; 4YSP A; 3IRC A; 5D4I A; 2G5B A; 4K3E H; 4PRI B; 5JW3 H; 3UBF A; 1ZDS A; 3Q5T A; 4PRI E; 3HPJ A; 5I6X B; 5T5F H; 2CXK A; 4LLY A; 1DFB L; 5BQ7 B; 2V8L A; 3QFJ B; 2P28 B; 3KLH C; 2VXS H; 5C07 A; 3SKJ L; 3WJM B; 5DI8 B; 4LAQ H; 3WI9 A; 4QTF A; 2GSG B; 1D5B B; 1LO3 L; 1J86 A; 4C4K T; 3TZV A; 2HN7 B; 3MLY H; 3WIF B; 3MSN A; 3L4C A; 4IDL A; 5KBN C; 1TZO A; 2RCS H; 1PYW B; 4HIP A; 2YOA A; 3DMM D; 1KL6 A; 4F91 B; 1ZGL P; 1ZGL M; 2C9A A; 2F5B L; 4ODH L; 3DOS B; 5AUM A; 2P5E B; 4WHT A; 2WUC L; 4NNY B; 5D1Q B; 4WSJ A; 3QO1 A; 5D1Q E; 4ATT A; 2F58 H; 3BVN B; 3O3A A; 5GGS Y; 3WMP A; 3LOW A; 4KK5 D; 1QI5 A; 1P4B L; 4FLR A; 3UO8 B; 1T7Z A; 3EBA A; 4AQA B; 1NGY B; 2E3V A; 2I60 H; 1RK0 B; 5AW7 B; 1IAK B; 1UG9 A; 3CX5 J; 4KKA D; 4KTD L; 4AIO A; 2Q3Z A; 2NXY B; 2UVE A; 4QI3 A; 5D2L A; 5E7B A; 2FIR T; 1EVT C; 3QH1 A; 4K3D L; 2M8X A; 2EUV A; 1PE8 A; 1AZV A; 2BCK A; 1K8D A; 2Z1K A; 3P9W B; 5HHN B; 1N0X L; 2YGO A; 2X2Y A; 5TE6 H; 3MRP A; 3L3H B; 5CEY B; 4ZW6 A; 3GJE B; 2KUT A; 1UB5 A; 4MKM A; 4DP6 X; 1ET5 A; 4NIK B; 5D96 C; 1B2W H; 4RRP A; 3WD5 L; 1C12 A; 5HHQ B; 1XSI A; 1DAN T; 3QUK A; 1AY1 L; 4W6X B; 4WEB H; 5DUP L; 4Q2Z B; 3FHQ A; 4JM2 B; 1FNL A; 1CZ8 H; 2VJX A; 3VVW A; 5I82 A; 5HGB A; 5AOZ A; 2R69 L; 2J4W L; 1FZJ A; 1J0H A; 4P5K A; 2A77 H; 3JQO B; 2BP8 A; 4K15 A; 4LVN C; 2XWB B; 3RIL A; 2SEB B; 1QLF A; 4PJB F; 3REW A; 3D4Y A; 2EEB A; 3OXR B; 4JN1 H; 2ZX5 A; 2MQC A; 4Z3I A; 1RQ5 A; 3NSX A; 3QUX B; 4GW4 A; 5HGH B; 5I0W A; 1JPS T; 1HAU A; 3CK0 L; 1DSF L; 3OX8 B; 2OMV B; 4JHW L; 3S2A A; 2DTM L; 1KXV C; 3P0Y L; 4JFE D; 5CGY D; 3NCY S; 2WSK A; 4JFV A; 1X7T A; 2CM5 A; 2VL5 B; 3CFE B; 3GMQ A; 1KTD B; 3ULV L; 1L9M A; 1I9I H; 2IFF L; 2WQU A; 3BDY L; 3TNN A; 4QJ3 B; 3H7B B; 3VB8 A; 3MUJ A; 3MJ7 A; 4TPR L; 4Q4I A; 5I15 H; 4JQV C; 3GMP A; 1A14 L; 2UZP A; 3LKP A; 2DL2 A; 2EIN B; 5I6P A; 5GS0 L; 5KVE H; 1RZK H; 5ANH A; 2XXG A; 4PJ5 D; 4BSV B; 5EII B; 2IAM A; 2E56 A; 4NKO A; 4NI6 A; 2NNX A; 3VFO A; 3W11 C; 3WDJ A; 3PWN B; 2YUX A; 4BK8 A; 2NX5 B; 3WJL C; 2NX5 E; 3CXD H; 4YSU A; 2B2X H; 4OZF A; 1RIV H; 1EK3 A; 2VAA B; 4C56 A; 1I8U A; 4X6C A; 2A9N L; 1K5W A; 3SB3 A; 3ES6 A; 3ULV D; 2GBV A; 1GGB H; 3UBG A; 5JHD B; 4TOY H; 2DF3 A; 2I5Y H; 5F1I B; 2VW7 A; 2ZOU A; 3PGC B; 4XVS H; 4PJH B; 3HHS B; 3S4S B; 2GK0 A; 4PJH E; 2DI7 A; 3E0G A; 2YD7 A; 1E65 A; 4HCF A; 5BS0 D; 5C0D B; 4YHI A; 5GS0 D; 3I50 L; 2YD5 A; 2HG0 A; 1CI3 M; 1WTL A; 1S7W A; 2ZJS H; 4L0P B; 4Y4H D; 4RGO H; 3W82 A; 5J11 B; 3TCT A; 2AV7 B; 1A2Y B; 1A6P A; 3NJF A; 3PDO A; 2OR9 L; 2BO0 A; 4HYT B; 5ID1 A; 3G6J B; 3L1O L; 3P69 A; 4MKT A; 3ME0 B; 3G6J E; 4XBM A; 1R3K A; 3WEX A; 4P9H C; 3ZKM D; 4RMU A; 1MRD H; 4DKE L; 3C5J A; 1CGV A; 4N0F A; 4X99 B; 1ZEA H; 5HHX L; 3RNK A; 5I0Z A; 2Y4S A; 2YGP A; 2ZSW B; 4BSJ A; 2XHN A; 3SO3 C; 1L9R A; 4EVN A; 3ULU L; 2E5E A; 5IHH A; 5I30 H; 2YBR A; 4H20 L; 5AOL A; 2B14 A; 4LNV A; 1KTJ A; 3R7Q A; 2YZ8 A; 1Z1W A; 1JN6 A; 4HMD A; 4HF5 H; 3NJK A; 4ODV H; 1DBJ H; 4HKJ A; 4DKA A; 4AR1 A; 3MRB A; 4FG0 A; 3FCS B; 3STB A; 3PY9 A; 4DKF H; 3JWO L; 3AJ7 A; 4MCN A; 4R3S B; 1JGD A; 2Y1Y A; 3CVI L; 4G11 A; 4A6Y A; 4ZML A; 3SOB L; 2ZCK L; 4L3A A; 4NN7 B; 4PJ6 A; 3MAC L; 1WC7 B; 5TE7 L; 4G7V L; 2CGR H; 4LP4 A; 5T3X E; 4LLA A; 3HZY A; 3QPQ C; 5ENW B; 2Q8B H; 4F15 C; 4PGJ A; 3OKK B; 3WTU A; 5EA0 H; 3KGT A; 4GAY A; 3A67 L; 5L9D L; 1AH1 A; 2YAF A; 5KVD H; 2V37 A; 5IFA H; 1F13 A; 1AJ7 L; 2ITD B; 4OCS H; 2E7C A; 5E5M A; 3ULU D; 5CBE A; 4AWJ C; 4HZA A; 3EYS H; 4LBE A; 1T83 C; 3I6L D; 2DJU A; 1Z3G L; 1EJF A; 1OT4 A; 5I76 B; 4J1U A; 1NEU A; 1I9J H; 3ETS A; 3UG3 A; 4UU9 B; 1L7I H; 3R8B B; 5CD3 A; 3HE7 A; 2W9E L; 3V95 A; 3EGD A; 2IPT L; 3ET9 F; 2EEF A; 2FE0 A; 3LN4 B; 3R1G H; 2YST A; 5JXN E; 1KPU A; 4KBP A; 5TE4 H; 3ZSS A; 1FNG B; 1IFR A; 4B9C A; 1G7J B; 2H3N B; 1F58 L; 2FGW H; 5GRW A; 4BHU A; 2OV0 A; 4MD5 B; 1IGI L; 3TYG H; 4GIP D; 4A7G F; 5SWZ A; 1WTG T; 4K7P Y; 4ACT A; 4AXW B; 1FG2 B; 3SYP A; 2R9H C; 1MFB H; 4F7C B; 2CR3 A; 1H6U A; 2BVE A; 4N2D A; 5EEW A; 1BWM A; 3EDJ A; 5IBL D; 1MPA L; 4YHZ H; 2FO1 A; 3P4M A; 3MOD L; 2OMU B; 4S1S L; 4U1K B; 1V5J A; 5DJA A; 4HKB A; 4LCU A; 4M5Z H; 1C7T A; 1YN7 B; 3LEY L; 4MD4 A; 1JVO A; 2DD8 H; 4PBV C; 2Q7Y B; 3KJ6 L; 3BXN A; 3CC5 B; 3WFE H; 1BW8 A; 3NJJ A; 4G6K H; 4DES A; 3VG0 H; 4UQC A; 5JHA A; 2PXY C; 3WNO A; 3T74 A; 3BOV A; 1J1P H; 2YPV L; 3QDA A; 2A7G E; 4PRH D; 4XBG B; 5ICZ A; 2XRA H; 4YWG L; 3ULV E; 1U91 B; 2W38 A; 3G3L A; 3PWJ B; 4NC2 B; 2X5P A; 2G3Z A; 5AAW A; 4NB7 A; 1YNT F; 2DBJ A; 4PRA B; 3P30 H; 5GS1 B; 5GSV A; 5BS0 B; 1JZD C; 5AKV A; 2HFG L; 5FV1 L; 4Y4H B; 5BS0 E; 2IAD A; 5ISZ A; 4IRJ B; 2B1O A; 1KBP A; 5EIV A; 4NQV B; 1P9M C; 2CSP A; 1BWW A; 1KCU H; 5KVF L; 1N8O E; 4ARF A; 4LPU A; 3KM1 A; 2OMZ B; 3I9P A; 1W8O A; 4IU2 B; 3MR9 B; 1NA8 A; 4K8R L; 3WHE 0; 2NY4 D; 3H2W A; 3PWU A; 5CCK H; 3ZRC C; 3FUL A; 1C5C H; 1CGX A; 2DRU A; 3FMG L; 3MFG B; 4IBS A; 4NDM A; 4YK4 C; 4ZUV A; 1N7M H; 1NC4 D; 4JG0 L; 3RGV C; 3T87 A; 3UTT D; 3IRZ A; 3LXB A; 1TJG H; 4JS7 A; 4YSA A; 2XC8 A; 2V7N B; 3EYQ C; 4LNR A; 4K71 C; 2P24 A; 1QFU L; 1NIN A; 4N2K A; 3KYK L; 4IRZ A; 4R2G B; 4OGX L; 1CIC B; 6TLI A; 3QJZ A; 1XV8 A; 4QRT A; 1KYA A; 5I0Y A; 2O6C A; 4OLY H; 3WE6 B; 1YYL L; 4HLZ H; 3TA3 C; 5D7K D; 3CHR A; 2VDO B; 3E8U L; 4WWK B; 4FNQ A; 3BLK A; 2JTD A; 3LV3 B; 1OTU C; 4NOF A; 1NBZ A; 5F9O L; 2JFC A; 3PHQ A; 5HGD B; 2GK2 A; 4TNL A; 1A4J B; 3ULS B; 1JF5 A; 1S7U A; 1VRS A; 4A63 A; 4O51 B; 4K8R D; 2BCM A; 3KPM A; 1FJ1 B; 1L9S A; 3NZ8 A; 4NNX A; 4KTG A; 3TEX A; 1BQH B; 2PF6 A; 1YEG L; 2JWY A; 3T3M B; 4CN4 A; 5EUK A; 1YEH H; 1HSA A; 3EW3 B; 3T3M E; 5HHO A; 1PZA A; 1AR1 D; 4JQI L; 3INU H; 5I1I L; 1D7B A; 2BER A; 2GUO B; 2VJ8 A; 3I6L E; 3STL B; 4UT9 H; 3V7A E; 1X4Y A; 3ARV A; 1SEQ L; 3L5J A; 5F7M C; 2RHE A; 2BDN L; 2HJF B; 4FA7 B; 2HG5 A; 4OV5 A; 1SDA B; 5DYO A; 4F8L A; 3E6F A; 1NOL A; 2X3A A; 1LK2 B; 3EO0 B; 4XN4 A; 2WZP A; 1AXT H; 4PWG A; 3NIG B; 4YE4 H; 1XCQ B; 3DV6 B; 3NIG E; 2VE6 B; 3KYO B; 4XTR D; 4UIL H; 5CWS A; 1E28 A; 3NSJ A; 4ZAK D; 5FAA A; 4H2A A; 4FYR A; 3O3E B; 3P9M B; 4FZE H; 4LEE A; 2V2T A; 5E6I D; 1OP3 H; 1NUN B; 2E9W A; 1N6Q H; 4TQH A; 4HII A; 5CU5 A; 4FYT A; 2IH1 B; 2OA9 A; 3HC4 H; 2OLD A; 3P73 A; 2WWM D; 5D8J H; 1X5J A; 1JI2 A; 1BYN A; 1DBB H; 1TQS A; 2B08 A; 1KCS L; 2JCW A; 4FXG B; 5TD4 A; 1ZJA A; 1ROC A; 2FLM A; 4ZPV A; 5DJX A; 1YUH A; 1V54 B; 4NJA L; 4X5R A; 2X4P A; 1CDH A; 3WAK A; 4ADJ A; 3C12 A; 1EGJ H; 1HXM B; 1BMG A; 3ESM A; 5HQD A; 1H8L A; 1NDM B; 2DIJ A; 3D51 A; 4NM8 L; 5IVX A; 3BFO A; 1KLF A; 4HS6 B; 3VO0 A; 1KB9 K; 1T0P B; 1GSM A; 2IDF A; 4BKL A; 3SDD A; 4GW1 A; 3C6L D; 3GSV B; 1KXQ E; 4OF5 A; 3WS6 C; 1ZA6 A; 4F7K A; 2ATK B; 2PN5 A; 3WGV B; 4ALA L; 3PAY A; 3VFR A; 3BP4 A; 1W0V A; 2CA4 A; 3TWC H; 5KG9 B; 2W60 A; 1CE1 L; 2FLR T; 3BGF C; 5F96 H; 3QT2 A; 2QGD A; 4PG9 A; 1DSW A; 4ELM B; 1ECY A; 5DD1 H; 4ELM E; 4P04 A; 1TTA A; 5IK3 B; 2NY4 B; 1B0G B; 5DU7 A; 1UZ8 B; 2D4F A; 1BFO B; 1AO7 A; 3FLF A; 4I2X A; 1XCT B; 4AEH L; 2B5I B; 2DBL H; 1CB4 A; 3UTT E; 3U1S H; 5DFW H; 5MER B; 4J7R A; 3WIH H; 2J8O A; 4N8P A; 4AV0 A; 3P3Y A; 3HZK B; 4OJF L; 1HU8 A; 1INE L; 4JFO A; 4N90 D; 5JPN B; 5BCA A; 4LST H; 2YIH A; 1OM3 L; 4Z7N B; 2N56 A; 4W9D C; 5D7K E; 4P23 B; 4ILD A; 5F7F A; 4OD1 H; 3CN2 A; 5KS9 A; 4R6U A; 1KN4 H; 2CHX A; 1JNN H; 3VVV A; 2QSC L; 2HVK B; 1EDQ A; 2VDK L; 5KVF E; 3HZ3 A; 2R0W H; 1MEL A; 3OZ9 L; 2JB6 B; 5IRO D; 5EC1 A; 3DPB A; 1PNC A; 4LEO A; 2XZZ A; 4NWU L; 1Z66 A; 5MEO B; 3B1U A; 1A7O H; 5D2L F; 5FXC H; 2CK0 L; 3CPL A; 4UIM B; 2IWE A; 1IGT B; 5E8O A; 2Q2E B; 1QFP A; 4HPY L; 4HCR L; 4B53 A; 4ZTO L; 2J55 A; 3NP4 A; 4NQC B; 2Y7L A; 5ILT L; 3NIF F; 1N5A B; 5AL8 A; 1WZA A; 3DEC A; 3P4O B; 5IHL B; 2E7L A; 5G55 A; 4W9Z A; 4QXT B; 2MCP H; 3DTX A; 3F2P A; 1YEE L; 2TSB A; 3FT3 B; 4HWB H; 4KQ0 A; 4DVR L; 5DUB A; 1TIT A; 2EE3 A; 1UA6 L; 4L9L A; 2Y5T A; 4L18 A; 4D9W A; 1WE5 A; 4BKT C; 1GS6 X; 5I1A A; 5HDL A; 1IGJ A; 2CUA A; 2W91 A; 1ORQ A; 5FUO H; 3UJI H; 2ZID A; 4ERS H; 5I1K L; 4K5N A; 3HTL X; 5F9W B; 1FPT H; 1MCC A; 5B22 A; 4ZAK B; 1DLH B; 2AGJ L; 2O5Y L; 4IIQ C; 4QO1 A; 1BBJ B; 1YPZ B; 2DQM A; 1J9R A; 1F4H A; 1PDK B; 1DQM L; 1QP1 A; 2D7T H; 1PG7 X; 2A6K B; 2FX9 L; 3UPR B; 2RQE A; 4J8R A; 1U8L B; 3TUR A; 4PJ8 A; 5IZ7 A; 3UTZ A; 1EEY B; 3X2Q B; 2ZOL A; 2F1E A; 1CLZ H; 1HL9 A; 5HY9 B; 4QHM B; 3DUU B; 3BJL A; 1KN2 L; 2QGC A; 1G9H A; 4AQ1 C; 4YXH H; 4ES9 A; 5DC0 A; 3F7R A; 4AKP A; 3TVM C; 2ZNX A; 3W81 A; 5T5F A; 1THC A; 2OX5 A; 5DS8 B; 1J5H A; 4N0R A; 4Q8B A; 2V4L A; 1TCR B; 1X6N A; 1XGQ A; 4WNQ A; 5C6W H; 4ZT1 A; 3Q44 A; 3MRJ B; 5EWI H; 5L2K A; 4I89 A; 5E51 A; 5G4O A; 2ZG3 A; 4EJ1 C; 4X5W A; 4JPK L; 5CNH A; 1D7C A; 4Y4K C; 5JQ6 L; 3C6L B; 1ZIA A; 3V4V H; 5KYX B; 5AYU H; 6TAA A; 1BQB A; 3BD5 A; 4N6Q A; 4ODX A; 1M1X A; 4MMX A; 2ADI B; 1NJ9 A; 3FB7 B; 3V9F A; 5KW9 L; 3BVU A; 4G59 C; 4XX1 E; 5GZO C; 5E8E A; 4R8W H; 3B7Y A; 1FUN A; 1OAL A; 4PR5 A; 1LO5 B; 4R0L A; 1GCY A; 3IFN L; 2QTE A; 1G7H B; 5DJ0 B; 3HC2 A; 3K71 B; 3JV4 A; 5DHV A; 5I5M A; 1XH1 A; 4PIR F; 1KXH A; 2J5L B; 1AMX A; 4OLW L; 1RY7 B; 5AKS A; 2QEJ A; 2EDD A; 3M86 A; 5FCS H; 3QUM H; 4FJZ A; 1TZH B; 4GW5 A; 3I6G B; 3I2C L; 4NPK A; 4TNG A; 3F7V B; 2F7O A; 1S9W B; 2UY7 B; 2BSS A; 2DEW X; 4JPI B; 5MEQ A; 4CMH B; 3GMN B; 3AB0 B; 4XB7 A; 1KTB A; 1THA A; 2XTJ D; 1EFX B; 3T77 B; 2CRM A; 4N90 E; 5C2U B; 1H5B A; 2ZXW B; 5B8C C; 1F1D A; 2ENP A; 4N22 A; 1JMX A; 1S5I L; 3GS7 A; 3MRR B; 4QGT A; 4HC1 L; 1N5Y L; 1NFK A; 2CO6 A; 4JG1 H; 2BM3 A; 3WKY A; 1N64 H; 3L3K A; 3T6X A; 3R0N A; 1P7Q B; 4DN4 L; 3L4T A; 4ZMP A; 1QX1 A; 3G5Z A; 1CR9 H; 2ZXA A; 4OZH A; 3QOR A; 3VW3 L; 1A25 A; 4XP4 L; 2X6V A; 1ID5 I; 5I24 A; 1I1Y B; 3SPC A; 1G6R A; 1OGT A; 3T4A C; 4ZMZ A; 7PCY A; 2XJK A; 1MCP L; 1HT6 A; 4JW3 A; 4IHO A; 1QVO B; 4DH2 A; 1QMU A; 1ZVN A; 2HKH H; 3T6Z A; 2Y69 B; 4GIO A; 2ZKM X; 2IDQ A; 2FYY A; 1H3G A; 4A37 A; 1XGT A; 3GLE A; 2MPA L; 2X1Q A; 4OFI A; 2LLG A; 3R8Q A; 1T7Y A; 5IKC A; 1GMI A; 1PKQ B; 4IJT A; 1F41 A; 3G5Y A; 1PKQ E; 1JGU L; 2Q9O A; 3ECB A; 4OT1 L; 4KQ3 L; 3RIA K; 3SKO B; 4EN3 C; 5CAY B; 1QNZ H; 3QUL B; 1BVO A; 4NO2 B; 3TT3 L; 4OD3 L; 2C7U A; 4NUJ A; 4PHZ A; 4LPX A; 4HEP A; 3VYP A; 1VA9 A; 4N85 A; 3OLD A; 4BK4 A; 5FX3 A; 4P06 A; 5CLZ A; 5D7L D; 3KE0 A; 1CXF A; 1IJN A; 2X6U A; 4KV5 E; 3VAC A; 4MB4 A; 4M00 A; 4WUU D; 4N4E E; 2WN2 A; 4RCA A; 1NBV H; 3U2I A; 5CP7 B; 4YUE H; 3VA2 C; 2E7M A; 2BP3 A; 3VFN B; 3EFO A; 4KUZ L; 1Q1J H; 1A7R L; 2ZF8 A; 5H2B B; 2DIA A; 3RBS A; 4CL7 A; 2VAB A; 3HLA A; 3CVC A; 1QHQ A; 1JHK H; 1LNA E; 5EC2 A; 3OV6 A; 3QG7 L; 2KMW A; 4ZXB C; 3B2U D; 3T8X B; 5C2B H; 1H0D A; 5DD5 L; 3HHR B; 1GZP A; 1H9D A; 1RZF H; 1YY8 A; 2MQ0 A; 4HK0 B; 3GXI A; 1OCO B; 4XO9 A; 3IS1 X; 1O7D C; 2EYT B; 3GI8 H; 2F53 D; 3FAW A; 3V0W H; 1F4W H; 3C5Z A; 4OLX L; 1T5K A; 4A7V A; 1UC6 A; 3B9K B; 1QF1 A; 3MRO A; 3GF8 A; 3F7P C; 5BZX A; 3D4Z A; 3TBY A; 4AK2 A; 1ZGL A; 4HIH B; 3AU0 A; 4TLN A; 1F6A A; 3K80 A; 3BDX A; 3ZL2 A; 3ILD A; 5PAZ A; 3F7Y A; 2B0S H; 1KBB A; 2R4R L; 5TK2 A; 1L7T L; 2X1P A; 1CUO A; 1SBS H; 4HGK C; 4GIN A; 3KH3 A; 3L3O B; 4GY4 A; 5EF2 A; 1KDI A; 2K70 A; 1S7X B; 1U3H D; 1DVX A; 4GFX A; 4ELK B; 2DKU A; 3ZL4 H; 1P2C A; 1FJ3 A; 3P7T E; 2V2W A; 4Z3H A; 4XZU B; 4IEF A; 5CGB A; 2EEC A; 3EDK A; 2AJV H; 4MA8 L; 2XTJ B; 3DRT B; 2HUE A; 2KC0 A; 2XTJ E; 3CXH K; 2X4Q B; 3MYZ A; 4FGM A; 4MSW A; 1BVK A; 2VDP L; 4HAF A; 4XN9 A; 4ZUS B; 4GOS A; 1QWN A; 3DET D; 3QEQ B; 4M5S A; 1HVX A; 3MV7 B; 1J0K A; 4U6V L; 3MV7 E; 4WHY G; 3QXA B; 1ESO A; 1S3R A; 3CD4 A; 1AHK A; 3WSQ L; 3VRI A; 1PE7 A; 25C8 L; 2BVQ A; 4JKK A; 3MJ8 A; 5E08 H; 4X6B B; 3KBH E; 2VDM H; 2B20 A; 3GK8 H; 2G4Z A; 2KBG A; 3AM8 A; 2NTS P; 4HFW B; 4LF3 B; 5DT1 H; 1DYZ A; 1NBY A; 3OKO A; 4FG4 A; 4ZFO A; 3PIQ B; 2EJ1 A; 3S35 X; 2EED A; 2OJE B; 3ORJ A; 3T65 B; 1C5B L; 3MGO B; 4FQX A; 1T2J A; 4LLD A; 4L8C A; 1NIF A; 3N1G C; 2NY3 B; 3HB3 C; 2DQU H; 1Q9W B; 1WEJ L; 2A6I B; 2JJT C; 1YHP A; 3LKS B; 3DUS B; 3LH9 A; 2CLR B; 4JAM A; 4OLU L; 4D94 A; 1FFR A; 3G08 A; 3TZV C; 1FH5 L; 2WOB A; 1GKO A; 4G9F A; 4B7L A; 3QZO A; 1DO5 A; 1LD9 A; 3VDA A; 2DV6 A; 2EEX A; 4PVN A; 3GBN H; 4OF0 A; 2A2Q T; 3VXM B; 4ELM F; 2F15 A; 1ITB B; 3FFC B; 5D7L E; 1JZJ A; 3MBE C; 4XNA A; 3FFC E; 4KDW A; 4WUU E; 4ZO8 A; 5AOM A; 4LPV A; 2J73 A; 4KX8 A; 1C7S A; 2ZYS A; 4FNT A; 5ADP L; 5BJT A; 4MD0 B; 1G7I B; 3NAB L; 1O7V A; 3D3V D; 4B9Z A; 3LJ3 A; 5E6R B; 5BZD B; 1KEM L; 1Z2K A; 1E3Z A; 1K8I B; 4JDE A; 4LB8 A; 4PRB A; 4YDJ B; 4C55 A; 3NJG A; 3NCJ L; 3IGL A; 1Q0X L; 5J9P A; 1U2C A; 4OFK A; 2IPU K; 4MBO A; 4S1Q H; 4PB0 L; 3VST A; 2XQB H; 3DV4 A; 4BFE A; 2YQ2 A; 1MY7 A; 1J8K A; 1K9T A; 3RY6 C; 5J6G A; 2ZWY A; 2IDC A; 5GIR A; 1DP0 A; 3C8K B; 1I7U B; 4IOS A; 1AYR A; 4WUK L; 3DHM A; 3VFV A; 4JHZ A; 2EC9 U; 1RP8 A; 2PET A; 4OLV L; 3M88 A; 2EAC A; 1H3W M; 3I9G L; 3KYM A; 4D8P B; 5TQ2 H; 3VTA A; 2WQX A; 5AW0 B; 4HJU A; 1CDU A; 1NGZ B; 1LK3 L; 3PWL B; 3S37 H; 1CU4 L; 3E2H B; 4KKL C; 1GZH C; 4UT6 H; 1GY1 A; 5KSA A; 2O5Z L; 1KF9 B; 3RRQ A; 5EEV A; 3HNV H; 1FSK B; 2B77 A; 3MYJ A; 2Z92 A; 3DX4 A; 2EE6 A; 4OZI B; 1HXY A; 3QIW D; 4X9I A; 2W92 A; 7TLN A; 1OE2 A; 1J1X L; 3CCH B; 3UDW C; 4OZI E; 3N41 B; 3CFD B; 3K5R A; 4FQ2 L; 1U93 B; 1PAM A; 2FBJ H; 2GI7 A; 3DID A; 3MA9 L; 5LDS A; 3QR2 A; 4G70 B; 4HXA L; 5F7L B; 5TQ0 H; 7TLI A; 5AZE H; 4XB8 A; 2R0Z H; 5TF1 L; 1ITC A; 2A74 C; 4A7T A; 4XAW H; 4GYB A; 5DA0 B; 1TT6 A; 1SJM A; 4N5E D; 4AGP A; 5ACZ A; 3K6I A; 1HQ4 B; 2DTS A; 5I0D A; 1XB8 A; 4XO3 A; 3KPQ B; 4WRL A; 3KPS B; 1UH4 A; 3KPS E; 1M6O A; 4GRL A; 1BOG B; 4XNM B; 1XSK A; 1CXE A; 5IHZ B; 4YDK H; 1F45 A; 5LBS H; 2OSW A; 1Q72 L; 2TSA A; 3P9L B; 3WTS A; 1GR7 A; 3P7S E; 3IJS A; 2KDY A; 1IGF L; 1FNH A; 1LE5 B; 2IXQ B; 3MLT B; 3NK4 A; 3BE1 L; 4FTV D; 3VGA C; 2C1O B; 1VFA A; 1VZH A; 2JB5 L; 3VRL E; 1HCZ A; 3UIY A; 3BIA X; 3DI2 B; 1R4X A; 43C9 B; 3T8C A; 3DKH A; 3KW8 A; 4UT9 A; 2R0L L; 1ONQ A; 4ODU A; 3ZHD A; 4L8D A; 5HVH C; 5KS9 F; 5DPE E; 3HS8 A; 4I7Z C; 4Q9Q H; 2VNC A; 4WW2 B; 4XCF H; 1RZJ H; 4X7S H; 2Z31 C; 4J2Q A; 2A6D B; 2IAN A; 1QFH A; 4JFP B; 1IEB B; 4CSP A; 4BEL D; 2J72 A; 3MRH A; 5EII G; 1JZ2 A; 3EFD H; 2OMN A; 3QO0 A; 4TM9 A; 3B7S A; 2E6Q A; 5EBG A; 3F7L A; 1D5X B; 1DTU A; 1FI8 C; 4OUU B; 3NA4 A; 4ZW8 A; 2W0P A; 4ODT L; 3G9V A; 3S34 L; 4XNB A; 1Y07 A; 2BIO A; 2X9W A; 2ZWN A; 3AUW B; 1D6V L; 2JH2 A; 3GGW B; 2R2E A; 1YAZ A; 2YZ1 A; 1OWW A; 3L9R B; 3RUG A; 2E74 C; 3UMT A; 3WKM L; 4GKO A; 5KAN I; 1KEG H; 1BBD L; 2AQT A; 2XUW A; 1SJX A; 2EYR A; 1ZPU A; 3KPT A; 1JV2 B; 5IMM B; 3S4S G; 4G6M L; 2OYM A; 4R4N C; 1YZZ A; 1TEF A; 4PJX B; 1MXD A; 3V5H B; 3MRK B; 4LL9 A; 4WHY H; 3T0E A; 4PJX E; 1GGT A; 4YBQ D; 1EGJ A; 1FGN L; 5I1E H; 3URO R; 5C9J B; 1HOC A; 3VXQ B; 1T1W A; 1I9E A; 4PJ9 A; 2WCG A; 3WLB A; 2FT6 A; 1JHT B; 2Q7N A; 2LLL A; 4CN1 A; 3K6S B; 2VL4 A; 2ZAI A; 1RCY A; 3GJF K; 1KTL B; 1CLC A; 1MCJ A; 1QYG L; 5CN1 A; 1UCB L; 1OTS D; 7FAB L; 1ZLV K; 5C6T H; 2OL3 B; 2AAA A; 1KLG A; 3BIK A; 3SDA C; 3KXF B; 4ZQK A; 5A2F A; 3BRF A; 1QBM L; 3HG0 A; 2WTP A; 3GLD A; 3KXF E; 1TQW A; 1SEB B; 2CDE A; 3MXW L; 3BRG C; 3ARG C; 4WW1 A; 4YHN B; 3ROL B; 15C8 H; 3IMW A; 8CGT A; 3KYA A; 4XN7 A; 3KL3 A; 4HZ1 A; 2VM9 A; 4ATW A; 5CBA A; 1FCG A; 5FWR A; 1QTS A; 1U4F A; 1UUN A; 2P8M B; 5J6H B; 3IJ8 A; 1FCC A; 2XWR A; 5IQ7 L; 1PFO A; 3EO1 A; 3ZHK A; 4AEE A; 2VH5 L; 1SVZ A; 3DX7 B; 2VLR D; 3QUZ D; 1URZ A; 2LU4 A; 2W0F A; 1DUY B; 3PHO B; 1J87 A; 4L3C A; 4QHL B; 4UAN A; 2CO1 A; 5AKR A; 5F1K C; 1I3R B; 5H94 A; 4H26 B; 3C5S B; 5IND A; 2CUI A; 4ENE D; 2WWN A; 3W9D B; 1QKZ L; 4O02 B; 2Z8V C; 2RFX A; 1TPX B; 3FUF A; 1NGX A; 3EFF B; 1MWA B; 3G04 A; 2FNB A; 2EVF A; 4D0B B; 2OXH A; 3MI6 A; 2XWE A; 5A2C A; 1BZ9 B; 4ZMO A; 4GXV L; 3ETB F; 3NK3 A; 4G7Y H; 4MXW L; 3H3P L; 2Y5Q A; 3US0 A; 2BNQ B; 3GUT A; 4ODS H; 1RU9 H; 1B4L A; 2P5W B; 4NGH L; 5IHU H; 5L6Y L; 2ADG B; 2P5W E; 3NSC A; 1VAC B; 4OIB A; 1LEK A; 3LB6 C; 2ITC A; 2HTK C; 2CJU L; 4QY8 B; 3BVD B; 4O5L H; 1HZK A; 5FKP B; 5JDS B; 3UZV B; 3GJF B; 2UV3 A; 4YDV B; 4WU7 B; 3VYO A; 1BLN A; 4GUP B; 4AGL A; 5B1C A; 2Y1V A; 5DRX A; 5GRJ L; 3U4K A; 5EEX A; 3A4F A; 1FHG A; 5HGA A; 3II1 A; 4UYQ A; 3Q6F B; 3HC3 L; 4JFD A; 1DE4 B; 4UBD C; 5AWN H; 3PWP D; 3PYC A; 1XDT T; 1OL0 A; 5BXF A; 3Q9Q A; 1ODU A; 2ADJ A; 4OZH F; 4L4T A; 2V5R A; 4H8W H; 4L29 A; 4LSU L; 1LE9 A; 3TON A; 5AZ2 L; 3MGT A; 4LCW A; 4P5S A; 2VR4 A; 4U49 A; 3Q1S H; 5TDO B; 1TVD A; 4YR6 B; 1X44 A; 4Z7W A; 4DVB B; 5IJK A; 3TA3 D; 4JHU A; 5GGT A; 3UJJ L; 2EUX A; 4KNT A; 4XPH L; 2NY0 C; 4Z7U B; 1Y18 B; 1TQV A; 3GSX B; 2BRQ A; 4FMV A; 2DS4 A; 4U0R C; 4Z7U E; 1B0I A; 2LT9 A; 3B2D C; 3VWJ A; 5I9Q C; 1JXK A; 4K9E C; 4MTW E; 3PXH A; 4Y89 A; 1AD9 A; 2V3F A; 1BAF L; 3GSN L; 2F6A A; 4GSD H; 3HCV B; 1KYO K; 1HWW A; 5CEZ E; 1BAW A; 2J44 A; 1TYJ A; 4JY5 L; 3FT0 A; 3HI1 A; 2WZ0 A; 2R1X A; 5BV7 B; 5BRZ B; 4Q58 C; 2Y07 H; 5J4M A; 4HSC X; 2VQ1 A; 1R24 A; 1T21 B; 3HS0 B; 1IKO P; 3B6S B; 2WWO A; 1CIU A; 4HUX A; 3OF6 A; 4NP4 H; 4Q9R H; 4M3J A; 2O72 A; 2WBW B; 2XZA H; 5K1V A; 1GSK A; 5EHF A; 4YHO L; 2IQ9 H; 2YRZ A; 4UZG A; 4S1D D; 5CCJ A; 5IVZ B; 4MCY A; 4QCI A; 4UMS A; 3TO4 A; 3NFS H; 5KSU B; 5MEN A; 5JYL A; 1E5B A; 2Y6T E; 1HQR A; 5CR8 A; 2OW7 A; 4G8I B; 2RH0 A; 5EBM B; 2VR8 A; 3LZQ A; 4N0C C; 1MJ8 L; 5C0B D; 4ZOE A; 1DL7 L; 4IS6 B; 1YRK A; 4PP2 A; 3MQI A; 4B41 A; 3QWR A; 3MRL A; 5C0J A; 1QSE A; 2QST A; 3E1Z A; 4FI7 A; 5J3H C; 1DUZ B; 1B88 A; 2ZZU T; 2VAJ A; 1RLB A; 1PLB A; 1CG9 A; 1LNE E; 1IBY A; 4LLM A; 1ZVS B; 2D3V A; 2VLR B; 5KN5 B; 3IKC B; 3L6F B; 2VLR E; 1JNH A; 3QS2 A; 4N1H B; 5IIE B; 3FCT A; 1PMY A; 3K8M A; 3LNI A; 4HBQ A; 3RKD C; 5JII A; 4MXQ C; 4YSO A; 3M1C A; 3AT8 A; 3TMN E; 1EZV Y; 1I4F B; 3D85 A; 2APF A; 4BSW A; 1I7R B; 3VXO A; 2W95 A; 1KTK E; 1QQD A; 1BYP A; 5IFH L; 4MNQ D; 1BQH G; 1A6W H; 5LUM A; 3FB6 A; 3FCU B; 3O11 A; 2Z35 B; 5DDH B; 2KDG A; 2XN9 B; 1IM9 A; 2XN9 E; 3JZY A; 3A6B L; 3PHA A; 3V7A G; 4XWG L; 1ZY9 A; 2JIX B; 2XDH A; 3HS9 A; 4IRS C; 4W93 A; 1M05 B; 3FTG A; 3OPZ H; 4G1E A; 5KSV A; 4PTT A; 4QJ4 B; 2AXF B; 3VWK B; 2WWK T; 1CL7 I; 4LP8 A; 4TUK L; 3RIF F; 3CFU A; 3C87 A; 4KDV A; 5E03 A; 3SE9 L; 1VHP A; 3ARB A; 1JD9 A; 1E9P B; 2EXY C; 4NEN B; 4Z7I A; 5EBW B; 2Q1E A; 1IEH A; 3H9H A; 4UB0 L; 2W9L A; 2ICW I; 4PGC A; 4TLT A; 4NZ8 A; 1M2V A; 3DVN A; 1MLB A; 5GGV Y; 5DR5 L; 3GEW B; 5I26 A; 2GIY A; 4KY1 L; 3D18 A; 4DQO L; 1D9K C; 3PWP B; 2V5Y A; 4TL4 A; 1L9Q A; 3LMJ L; 1N70 A; 2DWX A; 3PWP E; 1R5V A; 3TEW A; 4LE8 A; 1DFB H; 3U36 B; 5AVQ B; 1LNU A; 3SKJ H; 1WQV T; 5CHN B; 1CE6 B; 3O3U N; 4KPH L; 1LO3 H; 4PU2 A; 2X70 A; 2XT1 B; 3D2U A; 3O9W A; 3GM8 A; 4MJ5 B; 1P5V A; 1FNA A; 5LB1 A; 1RIH L; 1QAC A; 3ZXA C; 1SGK A; 3PQY A; 1JDD A; 3LD8 C; 3D30 A; 5GQB A; 1WD8 A; 1CFT B; 3NSD A; 3MRN B; 2H2S D; 3DBX A; 2AK4 B; 3KWX A; 1UXM A; 4RFN B; 1XIW D; 2AK4 E; 2F5B H; 4ODH H; 3KLS A; 4JQX A; 2WUC H; 3KWW B; 4PJE B; 1TLP E; 1AGD B; 4I18 A; 5EN2 A; 1E27 B; 3NAA L; 4NT6 B; 4PJE E; 4AYS A; 1FOR L; 4MMY B; 1GVI A; 4NQD D; 1N0L A; 5EOR L; 1P4B H; 1WZL A; 2WTR B; 1HIM L; 3G5X B; 1L9O A; 4EIG B; 3BPN B; 4JFF B; 1N8Z A; 1KAC B; 4RGM C; 4JFF E; 1WS7 A; 2KKQ A; 4JMX A; 1DED A; 1W72 B; 3X14 B; 4R5V A; 4KTD H; 4OW6 A; 2JXM B; 3B2P A; 5JOP E; 1O0V A; 3K0W A; 2UXG A; 3BH8 B; 4PJF B; 5DZP A; 3LKO A; 3HG1 D; 3OMZ A; 4PJF E; 4E9L A; 3H53 A; 1WFO A; 4PMF A; 1PX4 A; 3JWD L; 3QCT L; 4K3D H; 1U8I B; 3KJ4 C; 3CZF B; 5EMY A; 1N0X H; 3RZC C; 4XML L; 2OR8 A; 3P54 A; 5FWQ A; 1R5I B; 4Y1A D; 3Q7J A; 1NFD A; 4XH2 B; 4R7D B; 4JCM A; 3TOP A; 2I26 N; 3DO0 A; 1F18 A; 3WD5 H; 1H6F A; 3RI5 F; 2FJF A; 3GXD A; 4NJ9 L; 1AY1 H; 1WFJ A; 5C0B E; 4XN5 A; 2FEC L; 1E50 A; 2YBG A; 3ARX A; 2QZ5 A; 4J3B A; 3WFD L; 5IOK A; 4EXK A; 1FL5 A; 4L5F L; 4MXJ E; 1HAW A; 2ERJ C; 2ZUY A; 3HE6 A; 1DBA H; 1XR8 B; 3TTY A; 1F5W A; 5DMK A; 4QME A; 3AML A; 4WO4 A; 5JHL L; 4OQ1 A; 3CE1 A; 4QOK A; 2EVG A; 5DUJ A; 5H8D A; 4QT5 B; 3RG1 C; 4HWU A; 4QRR A; 1B6U A; 4XPB L; 1BJ8 A; 3QWN A; 4JAN B; 4H1L A; 5HJD A; 1FJW A; 1DSF H; 4Y2D C; 5JID A; 4Q6I C; 4JHW H; 4R26 L; 4GUQ A; 2DTM H; 3P0Y H; 4YBH A; 4X8X A; 5E6S B; 5GGU B; 1M7U A; 2FT8 A; 3ULV H; 5MHS D; 3ILE A; 2IFF H; 1YNL H; 3BDY H; 3BX7 C; 1ILU A; 1Y3G E; 2EVI A; 1MJU L; 4U6X A; 2BC4 A; 1S7T B; 4MAU L; 4HUK B; 4TPR H; 4BIK A; 4ZO9 A; 1TMC B; 3IVK B; 1DQI A; 2AJ3 A; 1Q1P A; 1RK1 B; 3EYO B; 3DBK A; 1A14 H; 1U8J A; 1U21 A; 3D39 B; 3E3Q C; 1Q38 A; 3QFD B; 2IGL A; 4PJ8 C; 3D39 E; 5FLU A; 4O58 L; 4WWO A; 3FRP B; 5GS0 H; 4H0H B; 5HYQ B; 3SBR A; 1JYW A; 3IMS A; 3WN5 B; 2EKS B; 4P2Y A; 4F33 A; 4OQT A; 5DVM A; 2QQK L; 5KBP A; 1ACC A; 1ZT7 B; 4F37 F; 3BQV A; 4FL4 C; 4M02 A; 3RL2 B; 5SWS A; 1DZB A; 4MAZ A; 5F3H A; 2A9C A; 4DZB B; 4ORZ C; 1S8D A; 2VLJ A; 3V6Z A; 4LDE B; 3M1B A; 4ZFQ A; 3TID B; 2NY7 H; 3BN9 C; 3VQM A; 1LM8 V; 2IC2 A; 1TSH A; 1N2R B; 2VGL M; 2Z0B A; 2A9N H; 3DNK A; 1I3G L; 2DQG L; 2AP2 A; 2AXG A; 5D93 C; 2QVM A; 3WTX A; 2YUV A; 1AXI B; 1PCX A; 3N6Y A; 5E5R B; 1L0P A; 3RFR A; 5AYT A; 3ARZ A; 4MTM A; 1QNI A; 2GAZ A; 5EO1 A; 4W4O A; 3F6I A; 4AYD C; 1G9N L; 4YIQ B; 3R19 A; 3SBW A; 1VKX A; 2OR9 H; 4KJP D; 1Z5H A; 3L1O H; 5KAQ G; 4BH7 B; 4YC2 B; 3KGR A; 1OW0 A; 4PB9 L; 1ZS8 A; 5A7E A; 3ARD D; 5HDB F; 1AXS B; 4DKE H; 5HHX H; 4HW6 A; 2B9B A; 1QO3 B; 2ROX A; 3WOV A; 4NKI L; 5AEN A; 2V2X A; 3MRC A; 1D5M A; 5JZ7 C; 1LDP L; 4ZFF A; 3D50 A; 5DHY B; 1HS6 A; 4H20 H; 3VGE A; 4MA3 B; 2P80 A; 3DHP A; 1KIP B; 1MUJ A; 4XX1 G; 4BRJ A; 3U83 A; 1G84 A; 5JO5 A; 5FKA A; 1SY6 L; 1KUL A; 1GPO L; 4JFT A; 3MJW A; 4Y5X A; 3HG1 E; 3GRW L; 1JGL L; 4ZRG A; 3V0A C; 5I23 A; 4AW4 A; 3JWO H; 3QEH A; 3QI9 C; 3VFU B; 2FGZ A; 3CVI H; 5MI0 C; 2X0U A; 1U95 B; 1OAN A; 4NQE B; 4XR8 C; 3SOB H; 2ZCK H; 1FRG L; 4NQE E; 4PLN C; 2VJ0 A; 4YPG A; 2YS4 A; 3MAC H; 5TE7 H; 2ZOW A; 3S3D B; 1NFA A; 2W88 A; 2X87 A; 4G7V H; 4KFZ C; 3SE4 C; 4E68 A; 3IMT A; 3RUG F; 1AG6 A; 3OHX A; 2ZWZ A; 4Y9C A; 3GS0 A; 3NDZ E; 2F61 A; 3FQR A; 4JGJ A; 4AIX A; 3A67 H; 1N4X L; 2EDH A; 5I8C B; 1AJ7 H; 1FFQ A; 3IDM A; 2BXY A; 4I77 L; 1CFS B; 4NRY A; 5DF0 A; 3AFL A; 3ARE C; 3PL6 B; 1RUL L; 8TLN E; 1Z3G H; 2W94 A; 3FRU B; 4NFB A; 5FEC B; 1TTU A; 1MCQ A; 1UXL A; 1OTT C; 2X4S A; 2F5A H; 1TQB C; 3B9E A; 3V8O A; 1GS7 A; 2EIZ A; 3QJV B; 2HLF C; 2W9E H; 4L4V D; 2IPT H; 3LS7 A; 4OM1 L; 1HXK A; 4QWW D; 2XG5 A; 4U7M A; 1UM5 L; 3BAW A; 3EOB A; 2EDY A; 3RTQ D; 3QEG L; 3P0G B; 1E4K A; 3DXA A; 4KH9 A; 3HI6 L; 4PJG B; 1F58 H; 4M1C D; 4PJG E; 4NHC L; 5MHS E; 2FX7 L; 1KCL A; 1NKR A; 5IB4 B; 1EZL A; 1Q9L A; 2NR6 D; 1IGI H; 3UYR L; 1XL6 A; 1KCW A; 2BIM A; 4JJ5 A; 3FH5 A; 1RZK C; 3F12 A; 3L7E B; 3N44 F; 2PR4 L; 3TH4 T; 2JTY A; 1GOF A; 1HHJ A; 4L0Z A; 1MPA H; 1CFN B; 3WUZ A; 3MOD H; 1G7Q A; 2TRY A; 4JO5 A; 4U1M A; 4S1S H; 1MCB A; 3O6F C; 3BWU F; 3LEY H; 1RER A; 1R9F A; 3NZU A; 1A9B A; 3KJ6 H; 3ARF B; 4G6A D; 2J88 L; 3TN0 C; 4DJM A; 5KJ7 E; 4Y9E A; 2VLK B; 4AV5 A; 1R0A L; 3U0T A; 1BD2 D; 1G0D A; 1LEG A; 1KIQ B; 2C3V B; 3FW6 A; 1UD3 A; 3HC0 A; 2MCG 1; 5IM7 A; 1WLH A; 2XWJ A; 2NYY D; 4ZO7 A; 2IAL B; 4NRZ B; 4YWG H; 3AAB A; 5EBL A; 4F58 L; 4ABQ A; 4JDT L; 4CGT A; 1LQS R; 3A23 A; 3GZQ A; 3QDM A; 3H56 A; 4PV1 C; 2BVT A; 1MG3 C; 4H5S A; 4LAR H; 3QUH A; 2NY6 C; 4F57 L; 3TBS B; 3UC0 L; 4KO5 A; 2HFG H; 3C2S A; 4PJI F; 4O2F A; 1F11 A; 1AG0 A; 4JRY D; 2AJZ A; 1PY4 A; 1JPF B; 3T71 A; 3BP5 B; 4NAM A; 5EB9 A; 2IQA B; 1FJU A; 1JK9 A; 2ISD A; 1EPF A; 2GSI B; 1PGR B; 3LN9 A; 5KVF H; 3VXS A; 3ARD B; 3IDI A; 4PGB A; 5FYK L; 1XD0 A; 4K8R H; 2XZC L; 3CU7 A; 5B3S B; 2DM4 A; 3FMG H; 3S5Z A; 3DIF B; 4LGR B; 4PLK D; 4ZFZ A; 4JG0 H; 1E4W L; 4P4K C; 3KCP B; 3WXV L; 2XKU A; 3MA7 B; 3MV8 A; 3VI3 B; 3DJZ A; 2V5M A; 1WUN T; 4KI5 F; 3VI3 E; 5DEF A; 2DB8 A; 1JUF B; 4UUJ B; 1QFU H; 3KYK H; 4FKE A; 2OP4 H; 4OGX H; 4UQ3 B; 2J8U A; 5FK9 B; 1YYL H; 4Z7O E; 2ZXE B; 1FO0 L; 2HT4 C; 8PAZ A; 3CFM A; 3E8U H; 4K3J L; 2NCM A; 3X3F L; 4FI8 A; 2GBT A; 2ICW A; 3FN9 A; 2HLA B; 2EDT A; 1R5W B; 5C07 D; 3T3P B; 2H3X C; 1Z5L B; 1FZM A; 3MLW L; 5AOI A; 1ZLE A; 3HVN A; 5F9O H; 3T3P E; 1IO4 C; 3MNV B; 1LNB E; 1JG9 A; 1TLX A; 1ANU A; 1N18 A; 1OF2 A; 1S2E A; 1IFG A; 5FYK D; 5BOJ A; 3RWJ B; 3ZT7 A; 1MSP A; 1UD5 A; 2AKR A; 1IFH H; 4PGE B; 1P5U B; 1YEG H; 4RQQ B; 2CLZ B; 4LSQ L; 3S36 X; 5B1A B; 3VXT A; 3DSN A; 3A0O A; 4JQI H; 3WL9 B; 4PD4 J; 1DFX A; 2BH0 A; 4QRF A; 5I1I H; 1WWW X; 2MIZ A; 3CFK A; 4J4P C; 2A83 B; 4QTH A; 4GP8 B; 4YH6 A; 1SEQ H; 1GME A; 1OSZ A; 2WII B; 3T6Q C; 5TLI A; 2BDN H; 3FO1 B; 3V6B R; 3HRP A; 5KWY C; 4L4V B; 3UG5 A; 4J2X A; 1ILS A; 4L4V E; 1M57 B; 3HRZ C; 4XAK D; 3V9C A; 3MV0 1; 3RTQ B; 1NPU A; 1S7V B; 3CX5 K; 4NKQ B; 1CS6 A; 1PLG L; 5BQ7 A; 3QFJ A; 4FEI A; 4NQX B; 3KWU A; 3WJM A; 4CIL A; 5DI8 A; 3ENE A; 4FYS A; 2GSG A; 2ICW J; 4QRP B; 5D72 L; 2HN7 A; 4QRP E; 5DRZ B; 3VFM B; 3WIF A; 1E8W A; 5D1X D; 4Y5V B; 2WCP A; 2N7S A; 5LND A; 3I6K B; 4APX B; 1PYW A; 5KYU B; 2BS8 A; 2X6A A; 2E46 A; 3RU8 L; 2P5E A; 4NNY A; 5D1Q A; 3N1F C; 1KCS H; 4X6D B; 3ZJA A; 4YJZ L; 1XB6 A; 3BVN A; 3TS8 A; 3KVQ A; 1X5Y A; 4LCC C; 4X6D E; 3WO4 B; 1NMA L; 3IAG C; 4NLA A; 1T04 B; 4AGN A; 1PND A; 1EVU A; 2I25 N; 4AQA A; 5TGB B; 1F90 L; 4P5T B; 1RK0 A; 3AAC A; 1IAK A; 1NCH A; 3I9A A; 1ZTX L; 4NM8 H; 1EO5 A; 3K2U L; 4FQH B; 3UTQ B; 3MRD B; 1ZLS L; 3WTT A; 4HUV B; 4UM8 B; 4ALA H; 5AVU B; 4WI4 A; 2VO5 A; 1KTR L; 5HHN A; 4N86 A; 2HH0 L; 1CE1 H; 1HEZ B; 2VDN B; 1YEJ L; 3LNE A; 1QWU A; 3L3H A; 5CEY A; 2ORB L; 3IPF A; 5CU9 A; 4P8Q A; 2WQV A; 2P8L B; 3N9G L; 1QLR B; 2WLJ A; 3D25 B; 5HHQ A; 4J12 A; 2VDQ L; 4HJG B; 5I1D A; 3SY6 A; 4QXG L; 1ZVH A; 4Q2Z A; 4JM2 A; 1EZS A; 2IAM D; 4AEH H; 2WM0 A; 3LPW A; 3OKL A; 1O9Z A; 3MUY 1; 2EZ0 D; 2XWB A; 4OJF H; 2BSC A; 2SEB A; 3L4U A; 1AZU A; 4Q6Y A; 5D7E A; 2LVE A; 5FOB B; 1INE H; 3OXR A; 3KLK A; 3QUX A; 1OM3 H; 2GTQ A; 5HGH A; 4C56 D; 2X6B A; 4P57 A; 1BQ5 A; 3ZDY E; 4Y4F B; 2ZAG A; 4LKX B; 5C07 B; 3MV1 1; 3OX8 A; 3WFH B; 3VFG L; 1NOF A; 2D0F A; 2OMV A; 4JGU A; 5C07 E; 4AM0 A; 3SJV B; 2QSC H; 1I7T B; 4QF1 B; 2GG1 A; 4U33 A; 4WFF D; 3SJV E; 2YLH A; 3WE1 A; 2VL5 A; 2VDK H; 5ICU A; 4HQQ L; 3CFE A; 4A6Y L; 2A9M L; 1A4A A; 1KTD A; 5FYK E; 2A5U A; 1KG0 B; 1J9C T; 2DQT L; 1T03 H; 4LPY A; 2CIK B; 3MU3 A; 3ZTN L; 2AAN A; 1NCT A; 4NWU H; 4BH8 B; 4ZSO B; 1CDJ A; 1USZ A; 2J43 A; 2OSL A; 4ZSO E; 4YZF F; 2CK0 H; 4F2F A; 2WN3 A; 1RZI B; 3D0L B; 1DR9 A; 4BSV A; 3IET B; 1PF3 A; 4HCR H; 4HPY H; 2G3X A; 5DD0 L; 4ZTO H; 5EII A; 4M5Y L; 5ILT H; 4WAF A; 2YJL A; 4IBL T; 4NX0 A; 1FO0 B; 2A9D A; 5DJ8 A; 1EEQ A; 4TXV B; 2IPU G; 4TLI A; 1NFD F; 4ZNC D; 3PWN A; 1OK0 A; 2UX7 A; 2NX5 A; 1YEE H; 2A38 A; 2VZ3 A; 4DVR H; 2AXH A; 1GC1 H; 1UA6 H; 4NN5 B; 5HJ3 A; 3T0B A; 3VU2 A; 4WFE D; 2VAA A; 2ZG1 A; 2G3N A; 9PCY A; 1NMB L; 5DJ6 A; 3QCV L; 1H9V A; 1K8D B; 4ZX3 A; 5JHD A; 5I1K H; 3BDB A; 2M2D A; 2BFV L; 5F1I A; 3RB7 A; 3RNQ B; 1GH7 A; 1XR9 B; 2AGJ H; 2O5Y H; 2YQB A; 3PGC A; 4XOC A; 4PJH A; 3HHS A; 5HI3 D; 3S4S A; 4TTG A; 4AMK L; 2X4T B; 2FX9 H; 2NY2 C; 5D1X B; 2HX7 A; 5C0D A; 5IOP B; 3NT0 A; 5D1X E; 5IB5 B; 3APN A; 1KJM A; 5DVL A; 3QUK B; 2J6E L; 1UOV A; 4PLJ C; 1A2Y A; 4EI0 A; 3W2D L; 3EBI A; 3G6J A; 2N1K A; 4EUP G; 3ME0 A; 1CD9 B; 3DGG B; 2PPF A; 4I0P C; 4GKN B; 1YEI L; 2DYR B; 4I0K A; 4X99 A; 3MRG B; 3UC2 A; 4BUG A; 5DCC A; 4QHP A; 5C3T A; 2ZSW A; 3CQQ A; 4H88 L; 2ZX7 A; 3REW B; 3SGE I; 1UJ3 B; 3WEM A; 4PIB A; 4MZ3 A; 5E8D L; 2COV D; 1RI8 A; 2ITF A; 5IBU G; 1KOA A; 4DSY A; 1RMF L; 1TTF A; 3HE7 D; 4M62 L; 4JPK H; 5JQ6 H; 4NER A; 5I8E B; 1T4W A; 3CMO L; 3JT2 A; 4R3S A; 3RYM A; 5CD5 C; 3FGD A; 1IBG L; 2UWE A; 3MOB L; 5CLX A; 4LEX L; 4N2H A; 5KW9 H; 1LL1 A; 4LOE A; 5KSB B; 3FTX A; 1JPE A; 1YYM L; 3P2T A; 4IJ3 C; 1WC7 A; 4FLO A; 5KSB E; 3IJ7 A; 3BES R; 1JXG A; 5SWZ D; 4U1G C; 3IFN H; 3LKP B; 4GI6 A; 4WKX A; 5ENW A; 1BVL B; 2JE8 A; 3TJE L; 3OKK A; 4JPQ A; 3IPE A; 1K72 A; 4OLW H; 1M7X A; 3K6N A; 1JZ4 A; 1A3L L; 4TV3 A; 2ITD A; 3GMO B; 3VFO B; 2FQG A; 3CHX A; 1GGY A; 4PPT A; 4Y16 B; 2BSR B; 1ZD6 A; 3I3E A; 2GJZ A; 3MNW B; 1H19 A; 4KO6 A; 3FYI B; 3VG9 C; 3SD5 A; 5I76 A; 3AY4 C; 2DEY X; 4S1R L; 3O2D A; 4W9E C; 1S5I H; 3BWA B; 3DJ9 A; 3GZK A; 4UU9 A; 4X6C E; 2NXZ B; 5IG7 A; 2XIC A; 5FFG A; 3DIV A; 5IW6 B; 1N5Y H; 3NID F; 1VEN A; 3FQT B; 3LN4 A; 2GIT B; 4B5Z A; 3MJ9 A; 4NZF A; 4U0Q B; 1WHO A; 4IRZ L; 4DN4 H; 1BEY L; 3CDG B; 4IKN A; 1G7J A; 2H3N A; 4Z0B L; 1A1N B; 4MD5 A; 1S7W B; 3S3C B; 5ISZ D; 1DVT A; 1KNO B; 4XNX H; 4AXW A; 1FG2 A; 4PWI A; 2Q86 B; 2EDK A; 2ZOO A; 3POC A; 2H2P C; 3WEX B; 4GUC A; 4ISV B; 3HJE A; 4GQQ A; 1ORS B; 3BVX A; 4HIS A; 3SDX B; 3U7W L; 5DHD A; 3UER A; 4GS7 C; 3TF7 C; 3SDX E; 2WGX A; 4RAH A; 3CH1 B; 2OMU A; 2NCE A; 5DVN A; 1EJ6 A; 1W16 A; 2F8O A; 4U1K A; 4XP5 L; 4AUY A; 4EVN B; 2MPA H; 2QGB A; 1UEY A; 5LWY L; 2AQN A; 2W80 C; 3FS9 A; 1YN7 A; 3LXC A; 4RAU B; 4WHD A; 2Q7Y A; 2G2N A; 3ALX A; 4JX0 A; 4HVB A; 3CC5 A; 5F88 B; 4G1M B; 4LES A; 1JGU H; 3MRF B; 4B9I A; 4YO0 B; 5JNY B; 4H8Q A; 3MRB B; 4GEI A; 4KI5 C; 3EYF B; 2IIJ A; 4WFE E; 4A6Y B; 2NQD A; 1JGD B; 3AT0 A; 4OD3 H; 4HGW B; 4XBG A; 2O5N A; 3TT3 H; 5BVJ B; 1EMT L; 5FO9 B; 3EJZ C; 1A0Q L; 5IJK C; 1OD7 A; 1U91 A; 3PWJ A; 4N9G A; 2ED7 A; 1SF5 A; 4FC3 E; 2CKB B; 4FNS A; 5F71 A; 3JBG 7; 4LDL B; 1EWH A; 3VQ2 C; 4YVU A; 3N50 A; 4PRA A; 4GQ9 L; 5F9D C; 3UO1 L; 3ZTE A; 5GS1 A; 5BS0 A; 4Y4H A; 1OLL A; 2CKN A; 2MOV A; 3DVI A; 3T8H A; 3N04 A; 4KUZ H; 4MRC A; 1A7R H; 4GAY B; 4IRJ A; 5ABD E; 4XPT L; 4JO1 L; 1QR4 A; 4NQV A; 5EZI L; 2Z8S A; 2K8M A; 4JLR A; 3EHB B; 2EEA A; 5CBE B; 4IU2 A; 2OMZ A; 2CCV A; 3L7F B; 3QG7 H; 3V0H A; 1CXL A; 3MR9 A; 4B7I A; 4F7E A; 2BWI A; 2A3T A; 3LS4 L; 4OH2 A; 1S46 A; 4MCZ B; 4NIY E; 5DD5 H; 4PP1 C; 2O4E A; 5AVX B; 1V55 B; 1FAI L; 5IW1 B; 3EH4 B; 4BZ2 H; 1JZI A; 3S96 B; 1G7P A; 1VJS A; 4P2O B; 4KC3 B; 2VN4 A; 1BMW A; 4ANU A; 4OLX H; 2X41 A; 1AZZ C; 1J12 A; 1CIC A; 2XXF A; 2WZP D; 5DJY B; 3Q6G L; 3WE6 A; 2VRQ A; 1OEZ W; 2YAQ A; 4WWK A; 3QJR B; 1UD8 A; 2BPB A; 3O6K L; 3T2P A; 1WYJ A; 5D5M F; 3PWV B; 3LV3 A; 4F80 A; 1XH3 B; 4O2C B; 3FQU B; 5SWZ E; 4DFI A; 1A4J A; 3GMM A; 2R4R H; 3QPX H; 1L7T H; 3ULS A; 4ETQ B; 1F2Q A; 4EI5 C; 4QKT A; 5JQI B; 3MLX L; 1HH9 B; 1OZT G; 2AAB L; 4ZK8 A; 3P4M B; 3SYC A; 4K3H A; 6PAZ A; 3VD5 A; 1AI1 L; 5DJA B; 1TUP A; 1BQH A; 2ODI A; 4N2F A; 4GO8 A; 3WNL A; 3CST A; 4TN9 A; 4IIZ A; 3PJS B; 4MA8 H; 1BPL B; 2UXF A; 3OAY K; 2GUO A; 3STL A; 1HDM B; 2VDP H; 3APC A; 2HJF A; 1YEF L; 2ZWS A; 2PTU A; 4PTU B; 4RWY L; 4MBR A; 1M1S A; 4GSX A; 3EO0 A; 1LK2 A; 4U6V H; 3FXS A; 4X42 A; 1O6V A; 1WVZ A; 3WSQ H; 4EUP H; 3SDD D; 3TBC A; 2GCY B; 1MFN A; 4XWO D; 4ZPT B; 3QJH B; 25C8 H; 1C8P A; 2IAD B; 1XCQ A; 3DV6 A; 3QIU C; 2VE6 A; 1GZQ B; 3KAA A; 5ISZ E; 5L4I A; 3FUE A; 3RWH A; 1G5A A; 3P9M A; 1OAK H; 5M94 B; 1DN0 B; 3ESU F; 2J8U F; 3Q3G B; 6FAB H; 3ZUN C; 4PYH A; 4ZMV A; 1DQJ B; 4X0K B; 2IH1 A; 4YLC A; 3B43 A; 4GXF A; 1C5B H; 1AO7 D; 1LP9 B; 2UY6 B; 1LP9 E; 4FXG A; 1MCO L; 5F6I B; 3F2L A; 1CTN A; 1WEJ H; 5EU5 B; 1OZU A; 3N40 P; 1WBX A; 1PY9 A; 4OLU H; 3O9W C; 1EER B; 4JJO A; 2BSE A; 2L4O A; 3IFL L; 4JRE B; 2NZ1 A; 1FH5 H; 3KMD A; 5KZC D; 2O6P A; 1HXM A; 1G7L B; 2J1Z A; 3AAZ B; 1YED B; 3Q8A A; 4LVH B; 5G4N A; 5D7I B; 2FO4 B; 4HS6 A; 1DEE A; 4OGQ C; 3BVV A; 4HJI A; 3QZW B; 5D7I E; 1J9S A; 4EIZ C; 1CQY A; 1NBZ B; 3PHQ B; 3GSV A; 3FUM A; 5HYJ D; 4A2M A; 4UQB A; 3FCB A; 1RPK A; 2ATK A; 4EDW L; 3FT1 A; 5CUS L; 3NA9 L; 5C0E B; 4HV8 B; 3P0V L; 3QZL A; 5ADP H; 3NAB H; 3VB9 A; 1YMH B; 4V2C B; 4ELM A; 2V17 L; 2B15 A; 1FBI L; 1X7Q B; 2Y2T A; 4P46 C; 1KEM H; 1PG7 L; 1HSA B; 1XMW A; 1KGW A; 5IK3 A; 4EEE A; 1B0G A; 3NCJ H; 1SJH B; 4G9D B; 1UZ8 A; 4OII L; 3V0V B; 1FN4 B; 2ZG2 A; 5JDJ A; 3P11 L; 1DVF B; 3NGB B; 4PB0 H; 4RA0 C; 5JXA L; 2CHZ A; 1XCT A; 2YRP A; 4FG2 A; 1EA9 C; 4LCI L; 1NCG A; 4G2A A; 4A2H A; 4Z3F A; 7CGT A; 5MER A; 4M5T A; 4WUK H; 1WWA X; 3ESV F; 2YXM A; 4OLV H; 3I9G H; 1EAP B; 4GO9 A; 5T5F L; 5ALB L; 1FC2 D; 3GC3 A; 4R96 B; 3MTX A; 4P23 A; 4YDM A; 1NQB A; 1LK3 H; 4PFE A; 3KZ8 A; 3LPF A; 1CU4 H; 4LAQ L; 5B38 B; 4ONH B; 3QIB B; 3T8M A; 2O5Z H; 2HVK A; 3NRQ A; 5I1L B; 2VXV L; 1XCX A; 5KYY B; 3QSK B; 2VXT L; 3CZK A; 5JMO C; 4HME A; 2YGV A; 2JB6 A; 1Z0N A; 4WO4 C; 4HWE L; 5K9J L; 2C9V A; 1GKE A; 1J1X H; 2FUQ A; 3P73 B; 4GLR H; 3BSG A; 1ZHN A; 5MEO A; 5TPW L; 4DP1 X; 4FQ2 H; 1KEN L; 1T89 A; 2F58 L; 3MA9 H; 1FL6 B; 3G6A B; 4HXA H; 4UIM A; 1IGT A; 2QKI B; 3UFI A; 3GJF H; 4ZPV B; 3QZB A; 4BFO A; 5F70 A; 5TF1 H; 1ZLV H; 4NQC A; 1S5H A; 2I60 L; 1IKN A; 3MDJ A; 2X4P B; 1N5A A; 3VH8 B; 2JKS A; 4OFQ A; 4QXT A; 2XYB A; 5DUR B; 5AB9 A; 3PNW B; 1NLD L; 3FT3 A; 3DI3 B; 4RGN B; 5IVX E; 2WZA A; 4RGN E; 2JQA A; 4DN3 L; 4PLO B; 4RES B; 3UZE C; 3L9Y A; 4XVJ H; 4ZMN A; 1BVN P; 1Q72 H; 4UWQ B; 1WIS A; 2D9Q B; 1BII B; 3CGT A; 4M8U A; 4DP7 X; 3BP4 B; 3MO1 B; 4JM2 F; 1IGF H; 1W0V B; 4R97 B; 3ZDX B; 4HS8 L; 3H42 L; 1DLH A; 2AER T; 1MHP L; 3BE1 H; 3QB7 C; 3ZDX E; 2EIK B; 3S5L G; 1YPZ A; 2JB5 H; 1PDK A; 2A6K A; 3VNZ A; 1B2W L; 3GBL A; 4P3C L; 3UPR A; 1UOW A; 2WXN A; 1U8L A; 3MUZ 1; 4U1J A; 1JWM B; 4R90 L; 1EEY A; 5TLN A; 5LGY A; 3MSP A; 4WU5 A; 4BFJ A; 1VFB A; 2R0L H; 3NPS B; 1KMG A; 1G6R L; 3E7J A; 1BXU A; 3DUU A; 4I2X E; 5I73 C; 4R4B B; 4J8Q A; 2EAB A; 3PRZ A; 3BQU B; 5JPM B; 4N0U B; 2A77 L; 4Z78 B; 4N0U E; 3FRP G; 3WJJ C; 3GXM A; 4JN1 L; 1KCK A; 5DS8 A; 3R7R A; 2AFN A; 2AHI A; 3NQI A; 2JKJ A; 2VZS A; 3GNM L; 2XVK A; 4LSR L; 2OMY B; 3MRJ A; 1VCB C; 4KX9 A; 3BUT A; 2J12 B; 2OXG B; 3TP4 A; 3IJY B; 5CFH A; 2Q7A A; 4N26 A; 1NAN I; 1YAI A; 2ZYI A; 3C6L A; 5T3Z L; 5HYJ B; 5KYX A; 5IQ9 B; 5HYJ E; 2ADI A; 1DSY A; 3FB7 A; 4ODT H; 4ONG L; 2CRY A; 3S34 H; 3QOR D; 2F54 B; 2FZ3 B; 1D6V H; 2F54 E; 2QGE A; 3WKM H; 1LO5 A; 1BBD H; 2DQD L; 1HI6 B; 1AOH A; 5AOJ A; 5DJ0 A; 2ENS A; 3F6J A; 4G6M H; 2BZC A; 1KLU B; 3UGX A; 1U6A L; 4D2N A; 3K71 A; 5I1D B; 2BNU B; 4ZOD A; 4XC3 H; 1WP6 A; 5ELI A; 3DK2 A; 2YXW A; 1TZH A; 3I6G A; 4GKS B; 3W5M A; 4G5A A; 1FGN H; 3F7V A; 4PRP B; 1S9W A; 1AQH A; 4JPI A; 2PP9 A; 3GMN A; 4K7P X; 3DTX B; 4PJH F; 5L8K B; 2A9B A; 4ETY A; 1SPD A; 5IJV B; 4AC2 A; 1EFX A; 3T77 A; 2HAZ A; 4BRV A; 1JP5 A; 5T3Z D; 2X7L B; 2HLF D; 4BK5 A; 1UCB H; 7FAB H; 3MRR A; 5E1A B; 4FKK A; 2MHA B; 1ORQ B; 5KEZ A; 3G6J F; 1MVY A; 4TOY L; 4XXD B; 4HUC A; 3WIA A; 3SCM B; 5D7J C; 3MXW H; 4A04 A; 4APQ C; 1P7Q A; 1HOE A; 2R2C A; 1DQD H; 2MTA A; 4A7S A; 2J57 A; 3KPN B; 4J8R B; 1FYH B; 1I1Y A; 1BP3 B; 2ZOL B; 2NZ9 C; 3QC6 X; 3QAZ C; 5IQ7 H; 4FN0 A; 2C8N A; 2X2I A; 3CZN A; 3ULA B; 3KHQ A; 1ZHL B; 2P9R A; 1H15 B; 5CKA A; 1QVO A; 4IQH A; 1TSR A; 4XOD A; 2G3M A; 4B9G A; 3CLF L; 3AQD A; 1QKZ H; 2E47 A; 2WI0 A; 1CK0 L; 6CGT A; 1XGQ B; 1PKQ A; 2CGZ A; 4TVS a; 5HHP A; 5D96 B; 3SKO A; 4UV4 H; 2C7F A; 3S3B B; 1FV1 A; 3C5Z D; 2H3K A; 1OAQ L; 4GXV H; 4U7S A; 3QUL A; 3RX8 A; 4MXW H; 4JK0 D; 4NO2 A; 1M1X B; 1GAF L; 2YKK A; 3L5X L; 5JW4 Q; 2UX6 A; 4NUP A; 3SN6 N; 2DQE H; 1WK0 A; 2G4E A; 1RD6 A; 1P6A B; 4L05 A; 4NGH H; 4OF6 A; 5L6Y H; 3E9U A; 2NSQ A; 4LVN B; 5JOF B; 1LWR A; 5CP7 A; 1F8T L; 4Y9F A; 4PY7 A; 4U4B A; 5DHV B; 2CNZ A; 2G9H B; 3VFN A; 5H2B A; 5L41 E; 5KVD L; 3FF8 A; 2ZCZ A; 2QAD C; 3IAP A; 4OCS L; 5GRJ H; 1RZJ C; 4L1H A; 1FH2 A; 3AAD B; 3HC3 H; 2BSS B; 2CO2 A; 3S35 L; 4ZFO L; 1B6D A; 3ILP A; 3T8X A; 3OR6 B; 4AH2 B; 1IKF L; 3TV3 L; 1R46 A; 1RLW A; 1XFP A; 3EJY D; 4HK0 A; 4FZ8 L; 4LSU H; 4NSK B; 2GUY A; 2QQL H; 5AZ2 H; 5IWL A; 2EYT A; 4N20 A; 3IMR A; 3KEH A; 2CO6 B; 1K6Q L; 2QEL A; 3DX3 A; 3T0V A; 3RVT D; 3UJJ H; 2H7W A; 5K33 A; 5JIM A; 4EPS A; 4XPH H; 4YMP A; 3DYM A; 2AJS L; 1IEA A; 3B9K A; 3IGF A; 1CI5 A; 1MRE L; 2AZU A; 5F89 B; 1BAF H; 5E1H B; 5JIQ A; 2WXF A; 3GO1 H; 5E94 A; 2JKX A; 1VFU A; 2QQQ A; 1LGV A; 4JY5 H; 5HP5 A; 2CLV B; 3GB7 B; 2F9B T; 4B0M B; 3OMI B; 3L5I A; 1M3J A; 2DEX X; 3L3O A; 3ULV F; 1S7X A; 4ZQT A; 4ELK A; 4FQX D; 2Q76 B; 3BW9 A; 1RL1 A; 4XZU A; 3QHF L; 4HEM E; 2WNX A; 3DRT A; 4YHO H; 1BPV A; 3TH2 T; 1A6Z B; 4OKD A; 1QEW B; 2X4Q A; 1AYO A; 5BOP A; 1HL5 A; 2WXP A; 2A0L C; 3K72 B; 1JHL H; 3P2B A; 2Y6S D; 4G9F D; 4MZI A; 4WGK A; 5FC9 A; 4YS4 A; 2XRA L; 4ZUS A; 2X12 A; 3QEQ A; 3MV7 A; 3QXV A; 2XIB A; 4WI6 A; 1MJ8 H; 2YC1 B; 3QXA A; 5A16 B; 1QEX A; 1DL7 H; 4PCY A; 3T2J E; #chains in the Genus database with same CATH homology 3O0L A; 3I6S A; 3TEY A; 5CU5 A; 4FYT A; 2OA9 A; 2WQS A; 3B37 A; 4PVB A; 4AQ1 A; 4JBS A; 1TQS A; 2XPZ A; 2KLT A; 5C7Z A; 4F8O A; 2Z1P A; 3UGU A; 1YVL A; 4JO8 A; 3WAK A; 4ADJ A; 3C12 A; 3ESM A; 2Y22 A; 4PW9 A; 2IVF C; 3D51 A; 3DDF A; 4MDR A; 4OJE H; 2ED6 A; 5D1Z I; 4JE0 A; 5AAW C; 4EAE A; 4F1Z A; 2PN5 A; 3PAY A; 5GZO A; 2J1X A; 2CA4 A; 2YA1 A; 4BCX A; 2MCA A; 1ECY A; 2CO4 A; 4P04 A; 5DU7 A; 3M46 A; 4IW4 C; 4N2N A; 2ZYH A; 4MS6 A; 1F00 I; 3UP6 A; 2F18 A; 2C9X A; 3EF7 A; 1HU8 A; 4Y2O A; 2MEJ B; 2ZOT A; 3DA0 A; 4BE5 A; 4HOM A; 4P2A A; 4ILD A; 3QZP A; 4N25 A; 4IGB A; 2BOL A; 3VVV A; 2ZAH A; 4R7L A; 5KVF E; 2DW5 A; 2NDF A; 3J07 A; 4WJG 3; 3T4A A; 5IRO D; 4KKP A; 4BV1 A; 1Z66 A; 3B1U A; 2CO3 A; 1UZG A; 2F08 A; 1ZTX E; 2KHA A; 3FCG A; 3ELQ A; 4YN3 A; 2Q2E B; 2ATA A; 2O6E A; 5IJD C; 2Y7L A; 4JMN A; 4BZ2 A; 5KVG E; 5FO8 A; 4ZW5 A; 2Q10 A; 4KQ0 A; 1CO1 A; 4NCD A; 2KS0 A; 4L18 A; 2YD0 A; 1JSY A; 1M42 A; 5LCV A; 4AK1 A; 1Y1U A; 4P0D A; 4N2M A; 4K5N A; 3WDI A; 3HTL X; 4NLH A; 3LZN A; 2R69 A; 2DQM A; 1MKF A; 2YQ3 A; 3NRP A; 3Q06 A; 2O6F A; 2RQE A; 3Q8E A; 3TUR A; 5IZ7 A; 2NNF A; 2O6D A; 2IXQ A; 2ZXG A; 3M6D A; 1D5R A; 4AQ1 C; 3CZS A; 2RA1 A; 2MPV A; 4ES9 A; 4Z3T A; 3FXI C; 3FUH A; 5T5F A; 2O30 A; 4N0R A; 2YEV B; 5IJB C; 2CG9 X; 2NNR A; 3Q44 A; 3QW9 A; 3EJP A; 5E51 A; 3RB5 A; 4FX5 A; 1SLX A; 5DLL A; 5G4O A; 4ES8 A; 1D7C A; 3IS0 X; 3UAF A; 4XMV A; 1F02 I; 4N6Q A; 4CGU B; 2OMW A; 3US9 A; 3Q8C A; 3BVU A; 4G59 C; 4PW3 A; 4JG5 A; 3KCW A; 1OKE A; 2R29 A; 2Y7O A; 1XHH A; 3HC2 A; 2KT6 A; 5K69 A; 3Q43 A; 2OMT A; 3BUB A; 3FSO A; 2FWS A; 1AMX A; 3D4G A; 3M86 A; 2XQY A; 3QZM A; 3QFG A; 2F7O A; 2DEW X; 4LL4 A; 2XWC A; 3ETT A; 2CO7 A; 2YGQ A; 3FUJ A; 4N22 A; 4P05 A; 3GHP A; 4F8N A; 2CO6 A; 2BM3 A; 2MH4 A; 5DYF A; 3QRL A; 1QX1 A; 3EJQ A; 3R4R A; 4H4N A; 3QOR A; 3B7R L; 1ID5 I; 2MQG A; 3CBJ B; 3K2K A; 2L04 A; 4I88 A; 4Q56 A; 2KZB A; 3SZ6 A; 4DH2 A; 3UAJ A; 4GIO A; 4WI0 A; 4A37 A; 4QPE A; 4XMU A; 3FTV A; 3GLE A; 1WDA A; 2LLG A; 2Y7S A; 4IJT A; 5IQL A; 1EXG A; 2C9P A; 4A38 A; 4RMB A; 3FUI A; 3HKZ A; 4KX7 A; 2X9Y A; 2J71 A; 3N3E A; 1KYD A; 4HEP A; 2YMT A; 1CF1 A; 3HBQ A; 3VYP A; 3PNR B; 3DOI A; 3FTS A; 4ADI A; 4KNQ A; 4P06 A; 2YO3 A; 3VAC A; 2WN2 A; 2WB7 A; 1I82 A; 2H6O A; 3FTW A; 2XBD A; 2ZF8 A; 2FAU A; 2Y7N A; 4RVB A; 2KMW A; 3UZV A; 1H9D A; 1P0S E; 3IS1 X; 2ZWK A; 3FAW A; 4IGB D; 2X9X A; 3EJR A; 5LXV A; 1TQU A; 3VAE A; 3LSO B; 3LZL A; 4UQI M; 4AK2 A; 5BZX A; 3GF8 A; 3D4Z A; 1YUK B; 4ZW3 A; 3AU0 A; 1GZH A; 3EJS A; 1IX2 A; 3ILD A; 4FTD A; 3L1G A; 3NRF A; 4N2L A; 3GIN A; 2YPV A; 2K70 A; 3OC8 A; 5DZ9 A; 4GFX A; 2JVU A; 5FPI A; 4B6Z A; 1PS3 A; 2FHF A; 3CV5 A; 4DGU A; 4IEF A; 2JI3 A; 4J39 A; 5JPN A; 2A74 A; 2KC0 A; 2KZK A; 4I9X A; 4FGM A; 4F24 A; 4XN9 A; 3HS0 A; 4AYB A; 2WGN B; 1QWN A; 1R19 A; 4AYF B; 4M5S A; 4Y2L A; 4DGW C; 4F5C E; 4Q52 A; 2XBZ A; 1AHK A; 2YMO A; 3BUQ A; 2BP5 M; 3KBH E; 4IBY A; 3QOR C; 2PCX A; 5J5E A; 5DHE A; 1SLW A; 1KY7 A; 3DOH A; 3LH9 A; 4D94 A; 3LHA A; 3QYN A; 1EAQ A; 3QZO A; 3HBG A; 5LN4 A; 4LEB A; 4XNA A; 2HPO A; 3FTY A; 4XMZ A; 4JQI A; 3D34 A; 5AOM A; 2J73 A; 4KX8 A; 1C7S A; 2ZYS A; 3CBK B; 2AXW A; 3Q13 A; 2FYV A; 4FXK A; 5JTW A; 1X5M A; 4NN6 B; 2K8Q A; 3VUA A; 4JDE A; 4RSY A; 4LB8 A; 3IGL A; 3L1F A; 4Y2O B; 4MBO A; 3H85 A; 4AQ8 C; 4XMW A; 2YQ2 A; 2JSF A; 4IOS A; 1AYR A; 4IGB B; 3D08 A; 5LAP A; 4F2M E; 5LG6 A; 1SHS A; 3M88 A; 3D05 A; 3VTA A; 2WQX A; 2Y7M A; 3DPB B; 1GZH C; 4N2A A; 4AR8 A; 3Q9P A; 1EXH A; 3B1T A; 3KVP A; 2W1N A; 3DX4 A; 5F0J B; 3QDH A; 3N41 B; 2WTR A; 2VQ4 A; 3D07 A; 5LDS A; 4BFN A; 3VU1 A; 1SQM A; 2BSD A; 3CHQ A; 3UOA B; 4E36 A; 2RMN A; 3AQZ A; 3H6A A; 2VME A; 4AGP A; 4JRF A; 3HVD A; 4XO3 A; 4QO1 B; 4DF9 A; 3IMB A; 3BCF A; 3FNK A; 2JI2 A; 3NDY E; 4FJ6 A; 4R5X A; 3WTS A; 3Q05 A; 2KDY A; 4YOK A; 4AQE B; 3NK4 A; 1VZH A; 2BIQ A; 4XVO A; 2WYI A; 4FL4 B; 3UIY A; 3DTD A; 3DI2 B; 1ECZ A; 1OHZ A; 4UT9 A; 2OX5 B; 2G30 A; 4N24 A; 4XMT A; 3HS8 A; 4KVP A; 3U9W A; 4J2Q A; 1XWV A; 2LOE A; 2J72 A; 2N3J A; 4G82 A; 3B7S A; 2WOY A; 5I0X A; 1FI8 C; 3BUP A; 4ZW8 A; 1N67 A; 4XNB A; 1Y07 A; 1YCS A; 2BIO A; 2X9W A; 5H37 A; 1H6E A; 2JH2 A; 3HBP A; 3FRP A; 5IUC A; 4K5P A; 2ODH A; 4KKR A; 3QYM A; 3KPT A; 2WJ7 A; 4HU2 A; 3INO A; 2I69 A; 4I1R A; 3BVW A; 5DC2 A; 5LO7 A; 3L8Q A; 2VMC A; 1O6T A; 4JUS A; 4EMZ A; 2CGY A; 4XXW A; 4FD0 A; 2ZAI A; 5CN1 A; 3QC5 X; 3QOR B; 1OK8 A; 2Z6F A; 1QTP A; 3HG0 A; 2WTP A; 3GLD A; 1TQW A; 1TQT A; 3FU0 A; 4MJH A; 4XN7 A; 2VM9 A; 2X53 A; 1QTS A; 1HJB C; 4L2L A; 2XWR A; 4GSQ A; 2QVT A; 1URZ A; 2F7Q A; 4A39 A; 2CO1 A; 4YLB A; 1O6S A; 4Y2N A; 3HR6 A; 3FUF A; 4ALA C; 3P1V A; 2CNY A; 3F3H A; 4F5C A; 4BEI A; 4HJE A; 3BUD A; 1LMI A; 2WAQ A; 3NK3 A; 2Y5Q A; 1E42 A; 4N6R A; 3US0 A; 2CR0 A; 4HET A; 1A9V A; 3MKK A; 1QBB A; 2JKI S; 2Z4T A; 3VYO A; 4AGL A; 5B1C A; 2Y1V A; 4FCH A; 3U4K A; 4UYQ A; 3OPU A; 5A7B A; 3QNF A; 2WY3 B; 4JDZ A; 3VD1 A; 3Q9Q A; 4IBV A; 2FVN A; 2AMU A; 3PG2 A; 4GSR A; 2DQ6 A; 1X9L A; 3GD1 C; 2F1B A; 3ZMN A; 3D33 A; 5IHW A; 1TQV A; 2LT9 A; 3B2D C; 3WEB A; 5HJB A; 4ELD A; 2A73 A; 2ADY A; 1WFI A; 4EIU A; 4GAA A; 3M7O A; 2X3C A; 3WTY A; 2CWR A; 2F6A A; 3COO A; 1HWW A; 4L3F A; 2J44 A; 1TYJ A; 3IRP X; 4Z1P A; 4OR1 A; 4CC4 A; 2L3B A; 3QZN A; 2J1Y A; 2E9G A; 3F83 A; 3RFZ B; 4ZZF A; 5FR3 A; 2V33 A; 4YE0 A; 5K1V A; 2A7B A; 3U1Q A; 2F7R A; 4UZG A; 4UTB A; 4UMS A; 3V0F A; 3QS3 A; 2DPK A; 5DS1 A; 3ZY7 A; 1E5B A; 2Y6T E; 2OW7 A; 4IYK A; 5KVD E; 2H0P A; 2RH0 A; 3LZQ A; 4GPV A; 3MHZ A; 3E1Z A; 3P2D A; 3LXU X; 5AB0 A; 3CIA A; 3GUF A; 3VQ1 C; 3QS2 A; 3PUU A; 3V0E A; 4MZR A; 4Z7A A; 2FSD A; 3BC9 A; 3KCP A; 3M1C A; 4FFZ A; 4Y7O C; 4XN1 A; 4GQZ A; 2W95 A; 4LZK A; 5IRE A; 5LUM A; 4FEM A; 2MNJ B; 2J21 A; 5BPP A; 5DZJ A; 1BF5 A; 3B7T A; 3PHA A; 1ZY9 A; 2XDH A; 3HS9 A; 3FHE A; 1XEU A; 3N40 F; 3CFU A; 5F0M B; 3ZHN A; 5ECG A; 4JDE B; 1EAO A; 1R6V A; 4Z7I A; 3UH8 A; 3KD4 A; 4NZ8 A; 4X2U A; 3WDH A; 2A9A A; 5J6S A; 2WII A; 2DWX A; 3TEW A; 4LE8 A; 2BIH A; 3US1 A; 1Z9S B; 2BIP A; 3L81 A; 3FTZ A; 4PU2 A; 5LB1 A; 3ZXA C; 3KED A; 2LFE A; 3W1Z A; 1WD8 A; 5AB2 A; 2J1W A; 3BUI A; 3U1P A; 4IBZ A; 3BVT A; 3KLS A; 2BII A; 2E59 A; 3FK3 A; 3N42 F; 5DVP A; 2H1L M; 2WTR B; 3LIU A; 2R51 A; 1YYC A; 4JMX A; 3JVF C; 4R5V A; 3B2P A; 1S6N A; 4CC4 C; 3N43 B; 5DZP A; 4FXT A; 4IGB C; 4OJD H; 3V0G A; 3OSV A; 4ZZK A; 3P54 A; 5FWQ A; 3FUK A; 3Q7J A; 3EXL A; 4AYI D; 4AGM A; 4MYP A; 4XN5 A; 1E50 A; 2YBG A; 4J3B A; 5IOK A; 1OGP A; 3ETB J; 2K78 A; 4QME A; 3VYN A; 1DQK A; 4OQ1 A; 2VUK A; 4KXA A; 5DUJ A; 2JI1 A; 1VZG A; 3RG1 C; 4YDZ A; 1KY6 A; 5HJD A; 3QWN A; 5AOK A; 4OJC A; 5FOB A; 3B2X A; 4GUQ A; 3APM A; 4LBA A; 3D09 A; 3ILE A; 4AJV A; 5E5L A; 3C5X A; 3QJX A; 1DQI A; 1NEP A; 1R17 A; 2NDG A; 5KBP A; 1H6T A; 1ACC A; 3RKP A; 2E8Z A; 4FL4 C; 2A9C A; 3N43 F; 1P5U C; 3RUR A; 2AC0 A; 3BZ5 A; 1CWV A; 3OWR A; 3E9T A; 4ZFQ A; 3VQM A; 4UTC A; 2IV8 A; 2VGL M; 2FO8 A; 4EE2 A; 4K5M A; 4KXD A; 2QVM A; 4K73 A; 3WTX A; 2OPC A; 3N6Y A; 3IGK A; 4N2G A; 5E1G A; 5LBV A; 4A1R A; 4LL1 A; 3AWG A; 4MTM A; 3VQL A; 4AYD C; 2WB1 A; 3R19 A; 4G83 A; 3C6E A; 3RTL A; 1Z5H A; 2WIN A; 3WOV A; 1XO8 A; 3AQX A; 2CCL A; 3BWS A; 5AEN A; 1AHM A; 2JV6 A; 5D55 A; 4R5T A; 3D50 A; 1HS6 A; 3LH8 A; 4HD5 A; 5FO7 A; 4GSU A; 4BRJ A; 4HJ1 A; 5KBD A; 3PF2 A; 1WRF A; 4FKH A; 4PQX A; 4ZRG A; 2OXH B; 4AW4 A; 3K0Y A; 3FUN A; 2X0U A; 1OAN A; 4XR8 C; 3UIZ A; 2C34 A; 3G7T A; 4AGO A; 3WTW A; 2FHB A; 2VJ0 A; 4N6R B; 4WKZ A; 4E68 A; 5K39 A; 4QIR A; 3OHX A; 3NDZ E; 4AL8 C; 4QUO A; 2ZYR A; 3LVT A; 4N2B A; 3B7U X; 3VD0 A; 5DZA A; 3V0J A; 2XET A; 1P4U A; 2W94 A; 1WH0 A; 2I07 A; 4JDZ B; 3QJO A; 3SE6 A; 2VN6 A; 4XN2 A; 3HRZ A; 4L0Y A; 1HXK A; 3RJO A; 3UZQ B; 4FDW A; 4IBW A; 2OCJ A; 4KH9 A; 4R7X A; 2BIM A; 1ZSH A; 1OPO A; 3FH5 A; 3N44 F; 1IU1 A; 4QDG A; 4L0Z A; 2Z64 C; 4IU3 A; 2LGN A; 4PHX A; 3PJL A; 3JYB A; 4YL9 A; 1RER A; 3KLQ A; 1R9F A; 4UYP A; 2BLF A; 2P5P A; 2MOG A; 2LR4 A; 2XWJ A; 4HSA C; 3VTT A; 1EAN A; 3AAB A; 2ZF9 A; 3LZP A; 3CHP A; 4J3O D; 3QUH A; 2Y2Y A; 2F0C A; 2OFQ A; 2JQM A; 1YG9 A; 4ECN A; 4D8O A; 5FO9 A; 3EUD A; 2ZQK A; 4WZ9 A; 1Z3R A; 3R18 A; 4JG9 A; 4NAM A; 4XMX A; 4ADG A; 1LJM A; 1QBA A; 3CU7 A; 5DHM C; 3V10 A; 3NCX A; 2K6Z A; 3KCP B; 3OC5 A; 4LO9 A; 3UXF A; 3H1Z A; 1SLU A; 5AAM C; 4FKE A; 3ZME A; 4PH8 A; 3ZGI A; 2DWY A; 2Y0S A; 3GT6 A; 3J2P A; 2XSK A; 1ZRU A; 1GYW A; 4X8C A; 5AOI A; 1ZLE A; 2PMZ A; 1IO4 C; 5J12 B; 1ANU A; 2OZN A; 4HU8 A; 1IFG A; 4J5V A; 4FYQ A; 2NYZ A; 1EZU A; 1P5U B; 1RPU A; 3UYP B; 3AQY A; 4V33 A; 1HTY A; 2ITE A; 1DFX A; 4JNX A; 1OSY A; 2MIZ A; 3OVU B; 4R83 A; 4EIB A; 1GME A; 3II2 A; 4DKT A; 3T6Q C; 3QE5 A; 4X8G A; 5KWY C; 3B3B A; 3FU3 A; 1I31 A; 2V8M A; 5I0V A; 3WMQ A; 1P5V B; 2NNC A; 3LZO A; 4B9J A; 2VR3 A; 3Q8F A; 4FEI A; 1G4R A; 3EAD A; 4CIL A; 4FYS A; 1OM9 A; 2P52 A; 3WU1 A; 4N2O A; 2N7S A; 5LND A; 1ZV9 A; 4APX B; 3FH7 A; 2OW6 A; 5GZN A; 5G4M A; 3ZJA A; 3TS8 A; 2CA3 A; 3ZN6 A; 3HLK A; 4NLA A; 4AGN A; 3PJN A; 4XND A; 2X9Z A; 3AAC A; 3FQ4 A; 1UOL A; 3ZHF A; 3RX9 A; 4H8P A; 4N2C A; 4KXB A; 4PCK A; 3WTT A; 3FU5 A; 2EDM A; 3QUG A; 4F8P A; 2ICE A; 3FH8 A; 3BLB A; 1QWU A; 2NYK A; 3IPF A; 4P8Q A; 2WQV A; 3V4L A; 4LOF A; 4AR9 A; 3CHO A; 1B9K A; 3SY6 A; 3EBH A; 3ISY A; 4AYE C; 1EZS A; 3S48 A; 1R34 A; 1HEH C; 3MUU A; 2HKA A; 2XWB A; 3HN5 A; 5D7E A; 3ZK0 A; 3KWV A; 2X0W A; 4ZX5 A; 2OMX A; 2B59 A; 2F68 X; 3FAX A; 2GTQ A; 3BCD A; 2K6Y A; 1E5C A; 2O1A A; 2ZAG A; 3B34 A; 2OMV A; 3FU6 A; 4EN2 A; 5KVE E; 2GG1 A; 3EXJ A; 1SLV A; 2YLH A; 3WE1 A; 2XWX A; 5ICU A; 1DO6 A; 2XCM C; 5C97 A; 5CFA A; 4BGL A; 2C9R A; 3MU3 A; 4ZX4 A; 2J43 A; 4JRL A; 3D8M A; 4GT0 A; 2WN3 A; 1QZN A; 3ISR A; 2IOO A; 3U6X S; 2FWU A; 2IV9 A; 5F0P B; 2YJL A; 1WGV A; 2A9D A; 4IBU A; 1KZY A; 2QKI A; 1PJW A; 4NN5 B; 2VO8 A; 5LHD A; 5BUA A; 4CC1 A; 3EJU A; 1XBD A; 2M83 A; 4ZX3 A; 2ALA A; 3RB7 A; 4II7 A; 4K4K A; 5D1X E; 3APN A; 3LYR A; 5JHL A; 4EI0 A; 3EBI A; 1I8A A; 3G6J A; 3F84 A; 3UC2 A; 5DCC A; 4QHP A; 1SUJ A; 4PIB A; 4JZJ C; 4ZW7 A; 2COV D; 2Z65 C; 2ITF A; 4ZJ9 A; 5JPM A; 1TG8 A; 4WKZ B; 3T2L A; 4E5X G; 4F27 A; 1R33 A; 3DX2 A; 5CF3 A; 1T4W A; 5E1I A; 2DJM A; 4DPR A; 1HJC A; 3U6X A; 4N2H A; 2OMY A; 3ASW A; 1KYF A; 4LOE A; 3FTX A; 3SZK C; 3ZGH A; 2XQ0 A; 1ML0 A; 3N42 B; 4XO5 A; 1CMO A; 3DDG A; 2WQW A; 4JPQ A; 2J6W A; 5ABA A; 3EJC A; 1G4M A; 4GEJ A; 4PN6 A; 4IBT A; 3TXA A; 4BFK A; 4GUO A; 4PW4 A; 1H19 A; 4HSQ A; 5EV7 A; 1WD9 A; 3LIZ A; 2DEY X; 3N0A A; 3SIK A; 3RLS A; 4C4B A; 3J27 A; 3D4C A; 1E5U I; 4F20 A; 4B5Z A; 1M9S A; 3L48 A; 1ZLD A; 2VN5 A; 4IKN A; 2YOC A; 4B60 A; 1VZI A; 3LZR A; 2D3J A; 2A99 A; 4IU3 B; 2WTO A; 3BWZ A; 2F1A A; 3POC A; 3B2M A; 4GUC A; 3BVX A; 5DHD A; 2WGX A; 2OMU A; 4BHU I; 4CC0 A; 5K5Y A; 2W80 C; 3AWE A; 1BG1 A; 3EBG A; 4R84 A; 4LES A; 3K2I A; 4B9I A; 2ALW A; 2N01 B; 4H8Q A; 4GEI A; 2NQD A; 2UZX A; 3AT0 A; 2O5N A; 3FTU A; 1O75 A; 2NR6 A; 4C4U A; 1SOX A; 2MNW A; 3D06 A; 3WEA A; 4D7P A; 4FC3 E; 4XN8 A; 1LYQ A; 3VD2 A; 3VQ2 C; 4JM0 A; 2JKR M; 2X0V A; 2YGN A; 5BUP A; 2HVB A; 3Q8B A; 4QRB A; 3N04 A; 4R7V A; 2MOQ A; 2WYH A; 2XTS A; 3FUD A; 4IU2 A; 2OMZ A; 2CCV A; 3V0H A; 4BFF A; 1D7D A; 4N2E A; 4NIY E; 2OKM A; 4FE9 A; 3K6O A; 2O4E A; 2GEQ A; 2WW8 A; 3V0D A; 1AZZ C; 2CE6 A; 2Y1Z A; 3DX0 A; 3Q01 A; 2BPB A; 4NAQ A; 4U3S A; 4B3V A; 3L1E A; 3DSN B; 4BZ1 A; 4DIX A; 2R59 A; 2L7E A; 3F85 A; 3D0A A; 1TUP A; 5D7H A; 5CN2 A; 2ZTB A; 2ODI A; 3UP1 A; 3IRC A; 4N2F A; 3RPK A; 4XO4 A; 2E9B A; 2VMD A; 2V8L A; 2P28 B; 2ZWS A; 4HEM A; 4MBR A; 3TEZ A; 4GSX A; 4QTF A; 2KLS A; 4X42 A; 5DS2 A; 1O6V A; 4FCA A; 4N28 A; 4Q6K A; 2N59 A; 2FHC A; 1TZO A; 2RTT A; 3DOS B; 1SVB A; 2E7D A; 3FUE A; 1XXD C; 5BUG A; 5D1Q E; 3T8V A; 2JKT M; 4OU3 A; 3SD2 A; 3WMP A; 4AYM C; 3NUK A; 4PBD A; 3UO8 B; 4YLC A; 4AQA B; 4AZ8 B; 1UG9 A; 2IOI A; 4FXG A; 3F2L A; 4QI3 A; 3N40 P; 2IOM A; 2QVK A; 2N0K A; 2BSE A; 3CHS A; 3I74 A; 2NZ1 A; 3KMD A; 2O6P A; 3E80 A; 2J1Z A; 2YGO A; 3Q8A A; 5G4N A; 3BVV A; 4HJI A; 4ZW6 A; 1ULV A; 4MKM A; 3FUM A; 4Q4E A; 4FFY A; 3EJT A; 3N44 B; 4ZX6 A; 3QZL A; 4N2I A; 3VVW A; 5AOZ A; 2Y2T A; 3JQO B; 4K15 A; 4I1P A; 3D4Y A; 3NSX A; 4G2A A; 1W80 A; 5I0W A; 4M5T A; 4M8R A; 4UT6 A; 3GC3 A; 3MTX A; 2WQU A; 3VB8 A; 3KZ8 A; 4Q4I A; 4QR7 A; 3NRQ A; 2W81 C; 3II3 A; 2E56 A; 3IE4 A; 4IBQ A; 4NI6 A; 2FUQ A; 3VTM A; 3WDJ A; 3KVD D; 2E8Y A; 4BK8 A; 3UFI A; 3QZB A; 4BFO A; 1I8U A; 3MDJ A; 3SB3 A; 4OFQ A; 5LBS A; 5AB9 A; 3DI3 B; 2ZOU A; 4K5L A; 1XXF C; 2WZA A; 2JQA A; 3AWF A; 3UZE C; 4L5F E; 2Y3N A; 4UWQ B; 5IJC C; 2XPY A; 4TMP A; 2HG0 A; 3HBZ A; 2ZXC A; 5J11 B; 2XTL A; 5LN8 A; 1IM3 D; 3P69 A; 4MKT A; 4XBM A; 2FUT A; 2YGP A; 4BE6 A; 4QRA A; 4IHQ A; 5LGY A; 2KLR A; 4BFJ A; 3E7J A; 5AOL A; 4J8Q A; 4LNV A; 1KTJ A; 1Z1W A; 4P07 A; 4AR1 A; 4FG0 A; 3PY9 A; 2Y1Y A; 2AHI A; 3NQI A; 4AQZ A; 4KX9 A; 4L3A A; 3CWG A; 2OXG B; 4PJ6 A; 4NN7 B; 2Q7A A; 4N26 A; 2ZYI A; 3WTU A; 3QOR D; 1GYV A; 2HPT A; 1AOH A; 5AOJ A; 3UGX A; 2KYI A; 5BZZ A; 1EJF A; 1OT4 A; 3L2N A; 2F7P A; 3ETS A; 4UTA A; 4G5A A; 4AM0 Q; 1HES A; 1GYU A; 2A9B A; 2EEF A; 1NM4 A; 1BXX A; 4BRV A; 4BHU A; 4FKK A; 4Z23 A; 2OAA A; 5G5D A; 2OUL B; 4JUT A; 4HUC A; 4AXW B; 1H6U A; 1GW6 A; 4N2D A; 4KKQ A; 4RZK A; 4G8A C; 3GLA A; 3QC6 X; 3CZN A; 3ULA B; 2P9R A; 1C7T A; 1BW8 A; 1TSR A; 4B9G A; 2HB0 A; 4K5O A; 1O7D E; 2WJ5 A; 2CGZ A; 3D52 A; 1PL3 A; 3TX4 A; 2H3K A; 2X3B A; 1G1K A; 4JK0 D; 2LW3 A; 1HEJ C; 3NCW A; 3UL4 A; 4RLD A; 3E9U A; 1XX9 C; 1N8O E; 4ARF A; 4IU2 B; 2J20 A; 2UZY A; 1NA8 A; 3V55 A; 2B39 A; 2CNZ A; 3FUL A; 4IBS A; 3N41 F; 3IRZ A; 4JGN A; 2CO2 A; 2XC8 A; 5LBG A; 4N2K A; 5I0Y A; 2O6C A; 4KXC A; 1T6B X; 2FEJ A; 3CHR A; 4H40 A; 4N20 A; 3V0I A; 3WTV A; 2K6W A; 3DX3 A; 3LSO A; 2H7W A; 4EPS A; 4YMP A; 2CZN A; 3IGF A; 5JHM A; 4A63 A; 4AGQ A; 3DX1 A; 4KTG A; 3TEX A; 4BIN A; 4RDB A; 3MNM A; 5HP5 A; 1D7B A; 4B0M B; 2VJ8 A; 2DEX X; 3L3O A; 3E38 A; 4ZQT A; 1RL1 A; 3WAJ A; 4F8L A; 2X3A A; 2BIN A; 2PR9 A; 4XN4 A; 2WZP A; 2C9Q A; 2L7Q A; 4BBW A; 4MZI A; 4WGK A; 4YS4 A; 2X12 A; 4CBZ A; 4H2A A; 2RAL A; 4FYR A; 5F0L B; 1KYU A; 4LEE A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...