The genus trace: a function that shows values of genus (vertical axis) for subchains spanned between the first residue, and all other residues (shown on horizontal axis). The number of the latter residue and the genus of a given subchain are shown interactively.
Total Genus |
129
|
sequence length |
349
|
structure length |
349
|
Chain Sequence |
MLDNSFYTAEVQGPYETASIGRLELEEGGVIEDCWLAYATAGTLNEDKSNAILIPTWYSGTHQTWFQQYIGTDHALDPSKYFIISINQIGNGLSVSPANTADDSISMSKFPNVRIGDDVVAQDRLLRQEFGITELFAVVGGSMGAQQTYEWIVRFPDQVHRAAPIAGTAKNTPHDFIFTQTLNETVEADPGFNGGEYSSHEEVADGLRRQSHLWAAMGFSTEFWKQEAWRRLGLESKESVLADFLDPLFMSMDPNTLLNNAWKWQHGDVSRHTGGDLAAALGRVKAKTFVMPISEDMFFPVRDCAAEQALIPGSELRVIEDIAGHLGLFNVSENYIPQIDKNLKELFES
|
The genus matrix. At position (x,y) a genus value for a subchain spanned between x’th and y’th residue is shown. Values of the genus are represented by color, according to the scale given on the right.
After clicking on a point (x,y) in the genus matrix above, a subchain from x to y is shown in color.
| publication title |
A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
pubmed doi rcsb |
| molecule keywords |
Homoserine O-acetyltransferase
|
| molecule tags |
Transferase
|
| source organism |
Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730
|
| total genus |
129
|
| structure length |
349
|
| sequence length |
349
|
| chains with identical sequence |
B, C, D
|
| ec nomenclature |
ec
2.3.1.-: |
| pdb deposition date | 2015-08-12 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| A | PF00561 | Abhydrolase_1 | alpha/beta hydrolase fold |
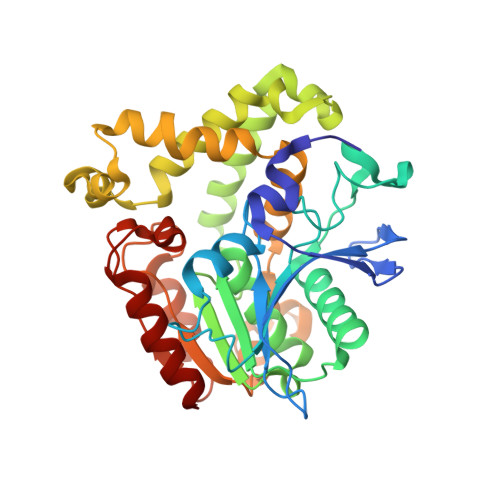
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)
cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Alpha Beta | 3-Layer(aba) Sandwich | Rossmann fold | Rossmann fold |
#chains in the Genus database with same CATH superfamily 4B7Z A; 3S2Z A; 5D6O A; 4NZZ A; 2WJ6 A; 3FYU A; 2HA7 A; 5JAR A; 1MT3 A; 4UFP A; 3DL4 A; 5JAP A; 2PPL A; 2ES4 A; 1ETH A; 2JGK A; 2XMR A; 1FFA A; 2XMC A; 1XRR A; 4XX1 A; 4Q3O A; 2H7C A; 4BAG A; 4K6H A; 3O0D A; 4UHF A; 4G5X A; 2XMZ A; 1SCQ A; 1LLF A; 4D9J A; 4ZXF A; 1ZI8 A; 5JZB A; 4J0K A; 4XJV A; 2G63 A; 5CML A; 4QDU A; 1VKH A; 1E89 A; 1JJF A; 2WTM A; 3CN7 A; 4CIA A; 5C8V A; 2CBG A; 3FCX A; 4KEA A; 1JMK C; 2PSJ A; 3OPM A; 4X94 A; 4KJX A; 2YH2 A; 2C58 A; 3D2A A; 4BYM A; 1Q83 A; 5LKA A; 4ZI5 A; 1H2Z A; 2HU7 A; 5HFN A; 3OM8 A; 1QID A; 3AZO A; 2J3D A; 4X95 A; 1BN7 A; 3LCR A; 1QIK A; 3FSG D; 3QMV A; 4U2B A; 1CIJ A; 5HF9 A; 4OYL A; 3MUN A; 4B85 A; 2BGR A; 2QS9 A; 3DD5 A; 2YXP X; 2O7R A; 1MO2 A; 4BDT A; 5EFZ A; 3EF3 A; 4X92 A; 2XUA A; 1PV1 A; 1XZF A; 3S8Y A; 1ODS A; 3HAC A; 2WIF A; 4KRX A; 5DJ5 A; 5FOQ A; 1S8O A; 2JBW A; 4BRZ A; 2Y2U A; 3WL5 A; 3ILS A; 1XZM A; 3WZL A; 4UBN A; 1QOZ A; 3BDI A; 2Q0X A; 1XLW A; 1W75 A; 1TAH A; 1MAH A; 1BS9 A; 4BTV A; 4QZV A; 1CPY A; 4FBL A; 2HDW A; 2HA5 A; 3FYU C; 3CCC A; 1VE6 A; 3I6Y A; 3G9Z A; 2AJL I; 4BAU A; 1ACL A; 5ALT A; 3F8S A; 4AZ3 A; 2VSQ A; 1CFJ A; 1AX9 A; 1DX4 A; 5HKA A; 3C8D A; 2WLS A; 1USW A; 3PFB A; 1XZJ A; 3BL8 A; 1IMJ A; 4U2E A; 3O4J A; 3GFF A; 3HZO A; 1KEZ A; 4OUK X; 4B9A A; 4W1R A; 1LPB B; 2WFM A; 1QIF A; 3BJR A; 3LLC A; 5HF5 A; 4MEA A; 3WMR A; 1XZI A; 2GBC A; 4KE6 A; 1K6E A; 2G5P A; 1SC9 A; 2ROQ A; 1FFB A; 3T52 A; 3SC2 A; 1JI3 A; 2JGE A; 5HF8 A; 2D5L A; 4J3J A; 3W2T A; 3KDA A; 3P2M A; 3HAB A; 5EIE A; 1E8M A; 3K9B A; 4Y7D A; 3FAK A; 5ALO A; 2E3J A; 4EB0 A; 3A2L A; 3WA6 A; 2HU8 A; 2JEZ A; 4TGL A; 4CG3 A; 1W76 A; 2V98 A; 4YX9 A; 3DOI A; 2I03 A; 3KD2 A; 4HRX A; 3BIW A; 5CXU A; 3HLK A; 5JAL A; 1ORV A; 3H17 A; 4HTA A; 3STV A; 1EI9 A; 4X3C A; 5ALG A; 2WG0 A; 2H1I A; 3IA2 A; 1W4L A; 2PUH A; 1TCB A; 4MJ3 A; 3GRO A; 2WTN A; 4H7H A; 1VXR A; 1Q0Z A; 1TKR A; 3FBW A; 1ZGB A; 1RYY A; 4AMY A; 4AX4 A; 3R41 A; 3V48 A; 1XRL A; 1CR6 A; 1VLQ A; 4W1S A; 3V1K A; 3FSG A; 1XZB A; 3UUF A; 4H7K A; 1YA4 A; 2WUG A; 2QMQ A; 1BRT A; 2ZVD A; 4DTC A; 1BCS A; 4HXE B; 1LBS A; 5K3D A; 3IDA A; 3G9X A; 3WKD A; 3VKF A; 5AKJ A; 5EHZ A; 2C7B A; 4EUS A; 2I3D A; 3C70 A; 1ZD3 A; 4N8E A; 5HFA A; 4CCY B; 1EXW A; 3D3N A; 5AKZ A; 1YA8 A; 5ALI A; 4UHE A; 1EHY A; 5ALN A; 3IUN A; 1IUP A; 4Y9S A; 4DMF A; 5EGN A; 5LZ5 A; 1NX9 A; 1AZW A; 2RIP A; 2X5X A; 1QFS A; 1H2Y A; 4UHB A; 4ZV7 A; 3MGA A; 5A71 A; 4BCD A; 4WY5 A; 4C6H A; 4C01 A; 3JWE A; 3V1L A; 1Y37 A; 4X91 A; 2WIG A; 1T4M A; 1J2E A; 3NOX A; 2ACK A; 4EY6 A; 4L9A A; 2O7V A; 4H7J A; 3H1A A; 4I3F A; 1HQD A; 4DNQ A; 2DCM A; 4BP8 A; 4Q34 A; 1H22 A; 1RP1 A; 1HKH A; 5AM5 A; 2O2I A; 2IIV A; 4UHH A; 5IVH A; 2XUP A; 4N8S A; 5CT8 A; 4WCV A; 4KR0 A; 5CXX A; 1JMY A; 4Y2J A; 3ESC A; 5DYW A; 4PV7 A; 1ORW A; 1H2X A; 4B84 A; 1EH5 A; 2C0Q A; 3III A; 3HYS A; 4X00 A; 3EBL A; 4ZWN A; 5AO9 A; 1FFD A; 5LKR A; 4YAS A; 4ETW A; 2XLB A; 3WK9 A; 3FYU B; 2CUT A; 4HAI A; 4A23 A; 1CUW A; 5JAO A; 3DLT A; 4EBB A; 4AO7 A; 3M3D A; 1JU4 A; 3QZU A; 2WHP A; 2QXU A; 5AM4 A; 5JZS A; 2XUD A; 3PF8 A; 3HXK A; 3W06 A; 4KE9 A; 5JYC A; 4UBJ A; 1LGY A; 2G4L A; 1Q0R A; 4FBM A; 4OB6 A; 1CQZ A; 5LLS A; 4J7A A; 5JD6 A; 5HF6 A; 1H23 A; 4GXN A; 4LXH A; 1VA4 A; 2WJ3 A; 1AQL A; 1XRN A; 2VAX A; 3I6M A; 1A8U A; 1ZIX A; 2VT6 A; 1XLV A; 4PNZ A; 3AIN A; 1CUX A; 4H7E A; 5AI0 A; 1QGE D; 3H04 A; 1QO9 A; 1N5M A; 2ZYH A; 2C5F A; 1XQW A; 4Q3K A; 1CUG A; 1LPP A; 1MPX A; 2XUH A; 4OSE A; 1Z68 A; 3IUR A; 5EIH A; 2ZJ7 A; 4Y2X A; 2BGN A; 4KE7 A; 4BVL A; 2RHW A; 3AIM A; 1P0P A; 4DM7 A; 3I2K A; 5CT9 A; 2VJD A; 3AFI A; 2DQY A; 2VEO A; 3GUU A; 5T4H A; 3G0I A; 3E3A A; 1XKL A; 3RKS A; 2QOE A; 3UUE A; 3YAS A; 3WWP A; 1JJI A; 1SFR A; 4GW3 A; 4EY8 A; 4KAA A; 5AOA A; 3W04 A; 3G8Y A; 3F67 A; 2QJR A; 4N5H X; 4DMH A; 4F21 A; 2RON A; 5SYN A; 5AI5 A; 1F6W A; 3ZI7 A; 2VTV A; 1WB4 A; 4UBM A; 1B41 A; 4JYP A; 5FXM A; 4AN1 A; 1ZIY A; 4FNM A; 5LZ8 A; 2VJB A; 4DIU A; 1JU3 A; 1CQW A; 3BXP A; 2OGT A; 5BOV A; 2J4F A; 2XQG A; 2AJC A; 4BRS A; 2XMG A; 3H2K A; 5A7G A; 1DWO A; 3ZV7 A; 4ARA A; 4HVT A; 2CEK A; 5JAU A; 4WFJ A; 4KAF A; 2X76 A; 2OGS A; 3G02 A; 3H2H A; 1TQH A; 2QR5 A; 1F8U A; 4LXI A; 1CUU A; 4IQ4 A; 3IUQ A; 5JAH A; 3DJY A; 3WZM A; 2DSN A; 3A6Z A; 5CT6 A; 1UXO A; 1GPL A; 3I1Y A; 3H19 A; 4G4J A; 4V2I A; 4C4Y A; 3QPD A; 1KU6 A; 2FUK A; 2O2G A; 3WKA A; 4AQD A; 5ALR A; 3PDC A; 1BRO A; 4EY7 A; 4Z49 A; 4FMP A; 2HM7 A; 1P0M A; 2ZJF A; 4EZI A; 2AJB A; 1QIJ A; 1BCR A; 5LYY A; 1B6G A; 3BF7 A; 4FFV A; 2AJ8 A; 4BC0 A; 4TVK A; 1WPR A; 5AKK A; 5CW2 A; 4BC1 A; 2OQI A; 1DIN A; 3CXU A; 2QUB A; 1EDB A; 2ZYI A; 4RPC A; 3PI6 A; 2WUE A; 5CUR A; 5FV4 A; 2YYS A; 2WIJ A; 2PKY X; 3C6X A; 1GPN A; 4PSD A; 3O4I A; 2PL5 A; 2Z3W A; 1UT6 A; 5J3J A; 3WL7 A; 1SCI A; 3DNM A; 4P9N A; 2BCE A; 1A8S A; 3IUM A; 5AI9 A; 5T4E A; 4KAC A; 2WFL A; 5AKE A; 4WDQ A; 4MYS A; 5ALH A; 4AZ0 A; 1DQY A; 4Y2Y A; 3LP5 A; 3C5V A; 5AOB A; 4G8C A; 1R4Z A; 4J03 A; 1H2W A; 1HL7 A; 5LP1 A; 1XRP A; 2AJD A; 3E4D A; 5SYM A; 4AO6 A; 2PSF A; 4ITV A; 4U2G A; 1SOM A; 4Q3L A; 5KRE A; 2WHR A; 1CUA A; 4QDO A; 3HJU A; 4Y2P A; 4CG2 A; 2QJW A; 1LNS A; 2HRR A; 5ALS A; 5AM1 A; 3FOB A; 1LZL A; 4K6K A; 5AKX A; 1IUN A; 4DSA A; 3WKC A; 5HK9 A; 3F98 A; 4EA6 A; 5CTA A; 5CH5 A; 3WYD A; 3SK0 A; 1VOT A; 1CUI A; 1A7U A; 2K2Q B; 4RGY A; 5ALL A; 1XZD A; 3HC7 A; 3GA7 A; 4BAZ A; 4NVR A; 4B0O A; 3Q0T A; 1VA5 A; 1LPM A; 4N5I X; 2XLC A; 4KAJ A; 3N2Z B; 3G9T A; 1ZD2 P; 4QWW A; 1CUS A; 1D07 A; 4A5S A; 2WQZ A; 1CUZ A; 1LPS A; 1R50 A; 3AJA A; 2WUF A; 3R40 A; 4WFI A; 2JF0 A; 1UOO A; 3R3V A; 2I3Z A; 5ALD A; 1FSS A; 2OG1 A; 5C8X A; 1YCD A; 1VJ5 A; 3E0X A; 2WIL A; 4LYD A; 1XQY A; 4UPD A; 5CT4 A; 5FP0 A; 1YAJ A; 5ALE A; 2HA4 A; 1MJ5 A; 4Y2U A; 3WK5 A; 2RI6 A; 4X97 A; 4X85 A; 3WA7 A; 4F5Z A; 2JGF A; 4ARB A; 2WKW A; 3NWO A; 3BF8 A; 3D59 A; 2XMD A; 3QYJ A; 3ANT A; 2JGJ A; 5C81 A; 3AZP A; 5CRI A; 2DHD A; 1YAH A; 3GEL A; 2HA0 A; 3VJL A; 1G4H A; 4UBO A; 2R11 A; 1K4Y A; 3X3H A; 3ZWQ A; 2OAE A; 2R8B A; 2V97 A; 2DQZ A; 3WKE A; 5K3F A; 4W63 A; 4FTW A; 5FLK A; 3T4U A; 2WG1 A; 4U2F A; 4GWL A; 2XDW A; 4S0X A; 3D2B A; 2Y1K A; 3CN9 A; 2WHP B; 5IH7 A; 2XT0 A; 3C5W P; 5LZ2 A; 1DTE A; 2IX9 A; 3AZQ A; 5ALJ A; 2H9Y A; 3IUL A; 4J0C A; 5ESR A; 5T4F A; 5FDF A; 4J0G A; 3WL8 A; 4AMZ A; 5DNV A; 3C87 A; 4X6U A; 4B80 A; 5HQ3 A; 2QKY A; 5AM3 A; 5D7B A; 1WHS A; 4X93 A; 2JEY A; 1IZ8 A; 4G4G A; 1AUO A; 3WWD A; 1THG A; 4IH9 A; 2PM8 A; 5A7F A; 4E14 A; 2OCK A; 3WK4 A; 1N8S A; 3PFC A; 2PSD A; 3BWX A; 4BZW A; 5C7Y A; 2B61 A; 1HLG A; 3H18 A; 1X2E A; 1WCY A; 2QXT A; 1ISP A; 1LPN A; 1XRQ A; 2HL6 A; 3OC0 A; 4GHW A; 4B83 A; 1UZA A; 5JAT A; 2XUG A; 3F97 A; 3V1M A; 4QDT A; 1U4N A; 4JUI A; 3WIO A; 4ZV9 A; 3R3W A; 4QDX A; 2XQJ A; 2PSH A; 1VXO A; 4FLF A; 3WWC A; 5EHN A; 1MN6 A; 5AIC A; 3HI4 A; 2CZQ A; 1DWP A; 3D7R A; 1TRH A; 4F0J A; 5E4Y A; 3K6K A; 1E5T A; 3HEA A; 1EB9 A; 2H7X A; 3O4H A; 3QPC A; 4FNG A; 1K5P A; 3JYH A; 3M82 A; 1WOM A; 5EHQ A; 5I8P A; 3WWE A; 4JEI A; 4DLN A; 3WK6 A; 2OCI A; 5AIA A; 5EI5 A; 3IVM A; 4EHB A; 1DT3 A; 3OOS A; 5ALK A; 1XZE A; 1ACJ A; 2BFN A; 4CI9 A; 2FX5 A; 1E66 A; 4LKO A; 2WUD A; 3VVL A; 3TEJ A; 2OCL A; 4AO8 A; 1R9M A; 1ZD4 A; 5AI8 A; 2JGL A; 3DDU A; 3I2H A; 3KXP A; 3DYI A; 1TCC A; 5ALM A; 3VJK A; 4L0C A; 3UMJ A; 4FLE A; 1EK2 A; 1XQX A; 1CUB A; 1MAA A; 1T2N A; 3D5E A; 2BJH A; 4G8B A; 4BCB A; 2GZR A; 1QIE A; 2GBF A; 2CJP A; 4XII A; 2QTB A; 2HRQ A; 5ALV A; 4E46 A; 5BWC A; 1EDD A; 1GT6 A; 1W1I A; 4W1Q A; 1G5F A; 1QIG A; 4KYV A; 2FJ0 A; 4DMK A; 3RLI A; 3C43 A; 1QO7 A; 5FRD A; 4H18 A; 2J3Q A; 2PU5 A; 1K8Q A; 4C4Z A; 3H2I A; 3PUH A; 1Y7I A; 3BJM A; 3U1T A; 2Y2V A; 2XQI A; 3QPA A; 1YAS A; 5AM0 A; 3WJ1 A; 2VAT A; 4X7B A; 5DYY A; 1E3Q A; 3RM3 A; 4H7D A; 2HAD A; 2CKM A; 2WFL B; 2HA2 A; 1Q84 A; 1AC5 A; 2LIP A; 2B20 A; 4QWM A; 4C89 A; 2ECF A; 3QIT A; 4LHE A; 1GGV A; 5ALP A; 2PSE A; 3G0G A; 4J0I A; 4DNP A; 1ZJ4 A; 3C6Y A; 4B82 A; 3QMW A; 4FWB A; 4Q05 A; 4OPM A; 3O4G A; 4X6Y A; 5HCU A; 3B5E A; 3ZPX A; 2HFJ A; 2HHA A; 1BEZ A; 4IH4 A; 1RWQ A; 1MX5 A; 2BUB A; 2ONC A; 3KOO A; 4INZ A; 1U8E A; 3IB3 A; 3JW8 A; 3CCB A; 1EIN A; 4OD0 A; 1QTI A; 2UZ0 A; 3V1N A; 4JNC A; 1YS1 X; 5IVI A; 1QLW A; 4H77 A; 2OCG A; 4L3W A; 1IUO A; 3STU A; 4I4C A; 1QIM A; 5C8Z A; 5DWD A; 5FPQ A; 3PIC A; 3DEA A; 2BAG A; 1YS2 X; 2G5T A; 2B4K A; 4FDM A; 3WL6 A; 4CFS A; 3SX4 A; 1EDE A; 3V9A A; 4UHD A; 4Q1V A; 2DR0 A; 1LBT A; 3EQ7 A; 3AIK A; 4M0E A; 5JZ9 A; 1ZOI A; 1AGY A; 2Z5G A; 4M0F A; 3RK4 A; 5AH0 A; 4LIP D; 3STW A; 3D2C A; 4EY4 A; 2GBG A; 2XUJ A; 1EVQ A; 4DYH A; 4U2D A; 3M83 A; 4FLM A; 1XQV A; 4ZRD A; 2EDA A; 3R3Y A; 1QZ3 A; 1GXS A; 2WSL A; 5AK3 A; 3VXK A; 1ODT C; 1VE7 A; 1O6G A; 3B12 A; 2HA3 A; 2W6C X; 5AK5 A; 1EK1 A; 2PBL A; 1GQR A; 3ICW A; 1CUY A; 4MEB A; 1EB8 A; 2NW6 A; 4OU4 A; 2WU4 A; 3N0T A; 3DS8 A; 4KRY A; 2FJP A; 4J0J A; 4FHZ A; 4NMW A; 4AB1 A; 4BCC A; 4WJL A; 1AMN A; 2EDC A; 4IH1 A; 5ALF A; 3H1B A; 2VAV A; 2GYV A; 1DWQ A; 5D3Z A; 3STY A; 3R3Z A; 4DSZ A; 5ALY A; 2XE4 A; 1JKM A; 4LXG A; 1ODC A; 1ZD5 A; 4MYD A; 5DNW A; 2IIT A; 1N1M A; 3LIP A; 1J06 A; 2JGM A; 1XFD A; 3STT A; 5K3F B; 5KWJ A; 3F9C A; 5AIB A; 2P8S A; 1QIH A; 5AK6 A; 3TGL A; 3MUO A; 4BTL A; 4BB0 A; 3G9U A; 5ALX A; 5AOC A; 4PF1 A; 3VDX A; 3G0D A; 1NU8 A; 1PJA A; 3O9M A; 3PE6 A; 4QDZ A; 1UKA A; 5ALQ A; 2Z3Z A; 1E8D A; 1M33 A; 3EQ8 A; 5HQ3 B; 3EIO A; 4O08 A; 3VVM A; 1VZ3 A; 3TJM A; 2PVS A; 1HPL A; 3IUJ A; 3H0C A; 2OPH A; 1YB6 A; 3ESA A; 5LIP A; 4X90 A; 2OGZ A; 4W9R A; 3WIB A; 1UK8 A; 2OLE A; 2Z8X A; 5K5E A; 1CUJ A; 1Y7H A; 2VJA A; 1TCA A; 1GPK A; 4G4I A; 1FJ2 A; 2XUO A; 1TIB A; 2DFP A; 3BE8 A; 3WK8 A; 4UBK A; 4I19 A; 4BE4 A; 1LPO A; 2QUA A; 5JD5 A; 4NS4 A; 1C7I A; 3A2N A; 1MU0 A; 2QT9 A; 1CUH A; 4P08 A; 3Q8W A; 2D0D A; 2Y6V A; 4Y2R A; 1YR2 A; 4UFN A; 3R3U A; 1UK7 A; 3FVT A; 5DYT A; 4PSC A; 5A6V A; 3KWJ A; 2AXE A; 1C7J A; 3FLB A; 4IHA A; 4BP9 A; 3PUI A; 5FKJ A; 1BU8 A; 3I2G A; 1X2B A; 2VQ6 A; 2XQF A; 3QM1 A; 4ZGB A; 3WJ2 A; 3WKB A; 1DX6 A; 2BUC A; 2W22 A; 4F60 A; 4GEC A; 5A62 A; 1W52 X; 4K2A A; 4UUQ A; 4C88 A; 4G9E A; 2WIK A; 4RE6 A; 2VF2 A; 2C0P A; 4MXD A; 4XWG A; 5AKI A; 1ZJ5 A; 1XZG A; 3ESB A; 3DL7 A; 1XZK A; 1UWC A; 1X70 A; 2XMB A; 2WG2 A; 5CH3 A; 1HBJ A; 1SCK A; 6YAS A; 5ALU A; 4J0H A; 2GYU A; 3U0V A; 4WDR A; 1TK3 A; 1I6W A; 1BE0 A; 2QZP A; 3OTQ A; 4B9E A; 2GYW A; 3NGM A; 5JIB A; 1J1I A; 5K3A A; 2ZSH A; 1KU0 A; 2WJ4 A; 4QLA A; 5E4J A; 4MWS A; 4CIB A; 4KE8 A; 1R88 A; 4MWT A; 1G66 A; 3ED1 A; 3MVE A; 5BWB A; 5LZ4 A; 2XUQ A; 1XZA A; 3FNB A; 2DHE A; 4PSU A; 7YAS A; 4YK7 A; 3GZJ A; 3I2J A; 4GBG A; 2C5G A; 3FLE A; 1ZI9 A; 3DKK A; 4DNO A; 4E11 A; 4M0M A; 3W9B A; 1F0P A; 2XB6 A; 4BE9 A; 2CB9 A; 2XMS A; 3L1J A; 5D3K A; 1BEE A; 2WHQ A; 3G7N A; 3B3Q A; 1ZIC A; 2J4C A; 2ORY A; 3EQ9 A; 3TRD A; 1QFM A; 4WY8 A; 2O2H A; 5DTI A; 4EY5 A; 1MNQ A; 3DQZ A; 3AIO A; 2XQK A; 1CUF A; 5I9I A; 2Y6U A; 5CT5 A; 3E1G A; 2HIH A; 1U65 A; 1XRO A; 5A7H A; 1THT A; 1QE3 A; 5EIA A; 4UBI A; 5AHX A; 1L7A A; 2OXE A; 2WID A; 4E15 A; 4MQL A; 3FCY A; 1A8Q A; 1UFO A; 1GKK A; 1UOQ A; 2C4H A; 1CLE A; 3I2F A; 4FKB A; 5KWI A; 3FLA A; 1CVL A; 1GKL A; 3DYV A; 4HXG A; 3KSR A; 4OU5 A; 3WWO A; 4Y2V A; 5E2I A; 3SWW A; 4GLB A; 5FUM A; 4BBZ A; 4K5Q A; 3F96 A; 5HKB A; 3I2I A; 1NU6 A; 3AUK A; 4QE3 A; 5K3B A; 3A70 A; 3VIS A; 4N8D A; 2HU5 A; 1IZ7 A; 3BIX A; 2QVB A; 1P0Q A; 2CMF A; 4B6G A; 4J0D A; 2OAG A; 5K3E A; 4BVJ A; 2ZYR A; 1F0N A; 2H7Y A; 3OG9 A; 3OUR A; 3W05 A; 3WQH A; 2XUK A; 2JGI A; 2WU3 A; 1XRM A; 3ZLT A; 5ALW A; 5AI4 A; 4Q82 A; 4UFO A; 2WM2 A; 3PF9 A; 4GEG A; 2QRU A; 1A88 A; 3WI7 A; 1O6F A; 5JD4 A; 3WYN A; 5AJH A; 4OYY A; 1BN6 A; 1E8N A; 2V9Z A; 5AKL A; 3W9U A; 4Y2S A; 3I28 A; 1P0I A; 1K63 A; 5FPP A; 1JFR A; 2GZS A; 4QEK A; 4OB7 A; 1PFQ A; 5HK8 A; 1QON A; 1DU4 A; 1ZI6 A; 5AI6 A; 4PQE A; 2PUJ A; 4HRY A; 4QA9 A; 3H2J A; 4HZG A; 2VJC A; 1UOP A; 4C87 A; 1R3D A; 5BV7 A; 1L7R A; 2B9V A; 1CUD A; 4A16 A; 4MQM A; 1AKN A; 5HZG A; 1YB7 A; 2EEP A; 1UKC A; 1DT5 A; 3L1H A; 4OB8 A; 5AM2 A; 1JJB A; 2WFZ A; 3L1I A; 3QMM A; 5CH8 A; 1N5R A; 2QM0 A; 3R3X A; 5H3H A; 4G8D A; 1J07 A; 4OCZ A; 2VT7 A; 4BAT A; 1G42 A; 1XKT A; 2ZSI A; 3WLA A; 3STX A; 2RAU A; 4Y2T A; 4K6G A; 5CBK A; 1CUV A; 4LYE A; 4PW0 A; 4AN0 A; 2X8B A; 1MTZ A; 1MX1 A; 4ZRE A; 4P93 A; 3D0K A; 3HSS A; 5JAN A; 3I6Z A; 2OQV A; 1DQZ A; 1L7Q A; 2RHT A; 2Z8Z A; 3DKR A; 4U2C A; 4JH0 A; 2ACE A; 3A2M A; 3WK7 A; 3FWH A; 2DHC A; 3FYT A; 1QTR A; 2HFK A; 1R9N A; 5E4T A; 1UK6 A; 4H7F A; 4TPK A; 4QLO A; 3C6B A; 3KWF A; 5ALZ A; 1EA5 A; 5DLP A; 2HA6 A; 4PO3 X; 4WFK A; 1XZC A; 1XZH A; 2ZYS A; 4GI1 A; 4FOL A; 4C4X A; 1ZGC A; 3QH4 A; 5YAS A; 3G0C A; 5AK4 A; 3QVM A; 1CUE A; 5EHX A; 3LII A; 4H35 A; 1R1D A; 5AKH A; 2JID A; 4DMC A; 4B81 A; 3D4L A; 1FFE A; 1W6R A; 5T4B A; 1YSC A; 4X6X A; 1HDE A; 3O95 A; 5IKX A; 4XVC A; 4G1F A; 2ZJ6 A; 2XI4 A; 1VZ2 A; 5AKG A; 5I7U A; 2VA9 A; 4QNN A; 4UHC A; 3C45 A; 1CV2 A; 4ROT A; 4RE5 A; 4DNF A; 4HS9 A; 4BVK A; 3G0B A; 3H2G A; 4IO0 A; 4BZZ A; 4YPV A; 4L72 A; 1EVE A; 1WPX A; 1JT2 A; 1XZL A; 1UK9 A; 1LPA B; 1MX9 A; 4Y2Q A; 3O9V A; 1IVY A; 1C4X A; 1WHT A; 3ZLV A; 1U2E A; 2BUA A; 5JAD A; 4B0P A; 4AXB A; 1GQS A; 5F4Z A; 1GZ7 A; 4CCY A; 5JAS A; 1QII A; 1XLU A; 4GDM A; 2PU7 A; 1WB5 A; 3FVR A; 4CG1 A; 3BDV A; 1QJ4 A; 1CUC A; 2XUF A; 3K2I A; 5K3C A; 2I78 A; 2RGU A; 1FFC A; 3ICV A; 3ESD A; 2XMQ A; 3ZLU A; 4CCW A; 3VJM A; 2PX6 A; 3C8H A; 5AP9 A; 3GBS A; 5LZ7 A; 3IBT A; 5DNU A; 2V96 A; 2BKL A; 2GBI A; 5IVK A; 1AUR A; 3I1I A; 5LZ9 A; 3HRH A; 4G9G A; 2YAS A; 3QBJ A; 1CRL A; 4HXF B; 3RAR A; 3M81 A; 1CEX A; 3LS2 A; 5A2G A; 4UBL A; 3NUZ A; 5AKY A; 3R0V A; 1OIL A; 1OCE A; 4H0C A; 5DTJ A; 3ANS A; 1UKB A; 4FFW A; 5IVD A; 4RNC A; 1EX9 A; 4BDS A; 4DGQ A; 5KBY A; 4H7I A; 1MNA A; 1WB6 A; 1LZK A; 1OXM A; 2XUI A; 4P92 A; 4W1P A; 4JYM A; 1WM1 A; 3AIL A; 3C6Z A; 5ISM A; 3DOH A; 4X71 A; 3DCN A; #chains in the Genus database with same CATH topology 2FY8 A; 5TSD A; 4I8D A; 5EZY A; 4Q39 A; 4JLW A; 1D7O A; 3S2Z A; 3ZIA A; 3H1E A; 4F3T A; 4R83 A; 3GD5 A; 4G84 A; 3V4V B; 3GBV A; 3W5A A; 4LN1 A; 1M2V B; 2H11 A; 2Q6O A; 2Y0C A; 3MCW A; 4IJU A; 3AUU A; 1XP8 A; 3LWZ A; 2PT7 A; 4IDQ A; 2SKD A; 4ILK A; 2EL7 A; 4XQG A; 1ZOP A; 1MEO A; 3L6R A; 4URH A; 3E4C A; 2J08 A; 4JYC A; 3SHV A; 4E3Z A; 4EX6 A; 3UPT A; 3ANY B; 3QG1 A; 3TS1 A; 3A36 A; 2Z4S A; 3E9N A; 3SZO A; 3AHI A; 4A1F A; 1TIP A; 2ES4 A; 3BYZ A; 3UAT A; 5END A; 1PED A; 3GHH A; 2XMR A; 2XMC A; 1R27 A; 3AR7 A; 2XZD A; 3BZU A; 4KXW A; 1PJR A; 2I8L A; 3LM8 A; 1MP0 A; 4PGF A; 3T61 A; 1DDO A; 4Q3O A; 4RET A; 5E9J A; 5FJI A; 5G4K A; 1P74 A; 2DLN A; 1KPH A; 1PFU A; 4P82 A; 4ZEX A; 4GM4 A; 3TW8 A; 2QVY X; 4RKE A; 4Z6K A; 3A9S A; 7AT1 A; 1T8W A; 4UCX A; 4S1T A; 4BRW A; 4JEM A; 5KDD A; 2DYK A; 1QRA A; 2B34 A; 1CR0 A; 1L8O A; 3GIW A; 2I80 A; 4L28 A; 5D01 A; 4RLQ A; 3DVR X; 2LZ0 A; 1J99 A; 1I1H A; 2OBF A; 2Q3M A; 1TJ4 A; 2I4O A; 4XIF A; 3N8H A; 4J0K A; 1VKZ A; 2G63 A; 3TEF A; 1UDB A; 4LDA A; 4G5F A; 1SVK A; 1MEN A; 1MDF A; 1RXU A; 5ICC A; 2FN8 A; 3KPV A; 2J9Y B; 3OPY I; 1PZH A; 3UVE A; 1EPU A; 4PYK A; 2V5U A; 4CCS A; 1L5F A; 3FCX A; 5B12 A; 1TQD A; 1WD7 A; 2FPI A; 2I0F A; 2XRF A; 1C0E A; 3OHM A; 5ENZ A; 1EP3 B; 1J49 A; 5AVW A; 2LKD A; 1E69 A; 2VYQ A; 1J20 A; 3HXP A; 3VR3 A; 4B5K A; 5JI2 E; 2PAN A; 3DV2 A; 1J90 A; 1FP1 D; 1ZG3 A; 4ZQF A; 1OLS B; 1H0S A; 5CG1 A; 4K40 A; 1X1A A; 1YBM A; 2P8J A; 2C58 A; 1JUD A; 4EGA A; 2G71 A; 4PCL A; 2Y05 A; 2JAN A; 1KZ1 A; 2AV6 A; 4QYI B; 4NCI A; 2C5H A; 5FUV A; 3F3M A; 1Q83 A; 5LKA A; 2OMK A; 1H2Z A; 4BXO B; 1ZMR A; 2HU7 A; 5EMN A; 3OFV A; 1R3U A; 4AWX A; 3VNA A; 4D3X A; 3OA0 A; 4EG4 A; 4W8H A; 1DBQ A; 4Q6B A; 3I3O A; 3QE4 A; 3ZV3 A; 1OX4 A; 5LP6 F; 1L6R A; 1OAA A; 1FVF A; 2GEJ A; 2VZE A; 3GYG A; 3ELB A; 4KO3 S; 4YJF A; 2CC2 A; 1L2T A; 1ORI A; 1NXP A; 1TT7 A; 2DST A; 2F1J A; 2J3D A; 2R8R A; 5K2O A; 1QIK A; 4G97 A; 3FSG D; 1JLM A; 5B3J A; 1AV7 A; 3H9C A; 2H51 A; 3BCS A; 2D7D A; 4D4I A; 4EVR A; 1MB4 A; 3BA6 A; 5KNB D; 3UW3 A; 3T8Y A; 2Q3C A; 3WQS B; 5B2E A; 4A27 A; 2BMW A; 1A9R A; 3LQ4 A; 2XCO A; 2ZAT A; 3QU4 A; 3IE0 A; 5EXE A; 1J2J A; 2CGE A; 1QH8 A; 4UXH A; 4EIT A; 4L4U A; 5DE3 A; 1I2D A; 2AEE A; 2QS9 A; 3HKD B; 3OO0 A; 3SEZ A; 4BVG A; 2RB4 A; 3OQH A; 4Z3K A; 3ILH A; 5IC4 A; 5MF6 A; 3SNH A; 1CG6 A; 4RK5 A; 5FLY A; 5DFA A; 4BGD A; 3KDP A; 4QEL A; 3N2G A; 3QVG A; 1P0C A; 3NGX A; 4X92 A; 1YZK A; 1OH8 A; 4YIK A; 1JPJ A; 1IG0 A; 4JU8 A; 2D6F A; 4FGL A; 1ODS A; 1K08 A; 1Y1P A; 1PR6 A; 4HWS A; 1IY9 A; 5DJ5 A; 4Y67 A; 3O1I C; 5DNC A; 2OOR A; 1RPJ A; 3KKO A; 3LAW A; 1K86 A; 3OZR A; 1S8O A; 2VUT A; 4CV3 A; 1EK5 A; 1Q0P A; 2ZBE A; 3T1O A; 2QN6 A; 3FKS A; 2XIT A; 1QOZ A; 4YP0 A; 3I39 X; 4N5M A; 4OXY A; 4IGI A; 2SIC E; 1KI2 A; 1V11 B; 2WI8 A; 4RM2 A; 8ATC A; 3HDV A; 2DU8 A; 2Q0X A; 1OU4 A; 3DR3 A; 2VDU E; 4YY3 B; 4BTV A; 2HRE A; 2ZVQ X; 3JZF A; 2B37 A; 4KQ2 A; 1N3B A; 2XNK A; 1Z0U A; 3IXQ A; 5EIB D; 1CPY A; 2VAP A; 4J07 A; 3W5V A; 2UYQ A; 3SKX A; 1Q7E A; 3C74 A; 3COW A; 3U7Q B; 4KRG A; 3CCC A; 4MKR A; 3TOC A; 1VE6 A; 4ENP A; 2QLT A; 4C6R A; 721P A; 3OZT A; 1E2I A; 1CR4 A; 3G9Z A; 4YEC A; 4LFC B; 1N2C A; 4BAU A; 2QP4 A; 3S8M A; 1WKV A; 5INT A; 1MRN A; 1N1D A; 2VSQ A; 1ZM9 A; 1A9O A; 1UP6 A; 1ZNW A; 3IVR A; 2C4U A; 4BFW A; 1Y8R A; 4XWO A; 3WWZ A; 1OWP A; 1L4E A; 3GWG A; 3C8D A; 2H4X A; 3RAJ A; 3SIR A; 1I32 A; 4NCN A; 2WLS A; 2IYO A; 1USW A; 1GPU A; 3AHE A; 4JBW A; 3BL8 A; 4Y6Q A; 3FPH A; 3EIH A; 4LMB A; 3REG A; 4GL4 A; 3BHI A; 4MWO A; 4CLX A; 2QMH A; 4HDO B; 2YIN C; 3GFF A; 1FMC A; 2IC5 A; 3HZO A; 4OKN A; 3TSY A; 1JG2 A; 3CLU D; 3ISV A; 5F35 A; 1WZI A; 4AF7 A; 2VWQ A; 2UZ3 A; 3CYF A; 5LN1 A; 5AW5 A; 1LX7 A; 4PSV A; 2D0P B; 3MHT A; 4N0Q A; 2G83 A; 2GHR A; 2BZS A; 3HLY A; 3ZVN A; 1QIF A; 2GEF A; 1BQ3 A; 3GH3 A; 3BJR A; 3LLC A; 1MQ8 B; 2DTX A; 3FPA A; 1CEY A; 1C30 B; 2HXG A; 2GBC A; 1IHD A; 5BSH A; 3J7T A; 3K92 A; 4KE6 A; 5FPY A; 3CBQ A; 3ETC A; 1SR8 A; 5LDG A; 1AXE A; 1BWR A; 2NTY C; 3HVH A; 1W5T A; 1K7E B; 2ZBR A; 1GGK A; 2ZT5 A; 3HQ4 O; 3VHX A; 4JU9 A; 1LVH A; 1O5T A; 2B36 A; 2AN3 A; 4Y6S A; 3WMX A; 1MR3 F; 2Q1X A; 2VA8 A; 1BS1 A; 3AKY A; 3PLN A; 5EUM A; 5TS2 A; 3T52 A; 3VMM A; 4EPY A; 2JGE A; 1Y5N A; 1TUB A; 2Q5Q A; 1YQD A; 5HF8 A; 3ZEA A; 1IM5 A; 1OOE A; 2VPY A; 2QT1 A; 3D4B A; 2D5L A; 2APJ A; 3NEA A; 3DCJ A; 3OU7 A; 4CT0 A; 3QWA A; 1LLC A; 2A7Q A; 5DXF A; 1W0C A; 1YJB A; 3GW8 A; 4D0N A; 4NJO A; 4DV7 B; 2FPS A; 1YT5 A; 4QPJ C; 5C2J B; 4PLY A; 4EA8 A; 2PJR A; 5EIE A; 4LHV A; 3K9B A; 4EB3 A; 4RTQ A; 2C97 A; 3E5H A; 2JLB A; 4F5E A; 5ALO A; 4CLO B; 4QHS A; 1VCI A; 4R8K A; 1AB6 A; 4EB0 A; 2QUI A; 1DCK A; 3NHB A; 2GWH A; 1QB8 A; 1VPE A; 3DE7 X; 3WA6 A; 3C3A A; 3ZZM A; 3ZNN A; 2QB5 A; 5ICF A; 3JQ9 A; 3ZQK A; 4HRA A; 2B1G A; 3EXE B; 4DQU A; 4EF6 A; 2Y1X A; 2HH9 A; 1WJG A; 4HO1 X; 4R8M A; 5LYJ A; 5E3H A; 4CQL A; 1FXW A; 2C42 A; 1CP2 A; 2ZRD A; 5ECG C; 4LJ8 A; 5FIU A; 1KEP A; 2IY7 A; 3B52 X; 1JIU A; 4QBG B; 4WLV A; 5DBH X; 4G6Z A; 4E1A A; 1VFN A; 3R0Q A; 3F71 A; 4PQG A; 1NYQ A; 3BFP A; 3ZH2 A; 3AU9 A; 3RVD A; 4UZF A; 3ZK7 A; 4L51 A; 2G77 B; 3GOQ A; 3UY4 A; 3P2I A; 4F4F A; 2HMV A; 4IQ0 A; 4BFX A; 2I9P A; 1YIO A; 1EI9 A; 1VYU A; 2HZ7 A; 3FNE A; 5FI0 B; 2PWA A; 3PC8 A; 4OFW A; 2OXI A; 3IA2 A; 4DW8 A; 3DZH A; 1UPT A; 2W2X A; 4IVR A; 4R2J A; 1XNR B; 3DR5 A; 1TCB A; 5E8R A; 5C1B A; 4P3Y A; 1UL1 X; 1R18 A; 2Y9P B; 2KHZ A; 3A27 A; 1SUA A; 3PX7 A; 2WTN A; 1FPM A; 2BFW A; 4AG6 A; 1VXR A; 5G1P A; 4FR5 A; 1Q0Z A; 3T9D A; 3UN1 A; 1TKR A; 1I3N A; 4LJ4 A; 1MH9 A; 4DRX B; 2AVD A; 3HNR A; 4ZCS A; 1A4X A; 5LP6 A; 4J1Q A; 5JS1 A; 1YVR A; 3F5M A; 3QOW A; 1GRC A; 5PGM A; 3AQ4 A; 3R41 A; 3UWP A; 2FW0 A; 1K89 A; 2W37 A; 4MI1 A; 5FUX A; 4EUE A; 3JAL B; 1T6X A; 5JEE A; 2D1F A; 3RR2 A; 2R1V A; 2C96 A; 3GFG A; 4QJ4 A; 2GBP A; 3VZS A; 1IE3 A; 2UXC B; 3HO8 A; 3C3B A; 3EDY A; 1AMU A; 1XZB A; 3H1P A; 3C8U A; 3GDF A; 5IBR A; 2CT8 A; 4H7K A; 1YA4 A; 2WUG A; 3V8E A; 3R9W A; 2R4U A; 1BVH A; 3HVI A; 2UXD B; 5LNK 1; 1W0K A; 3CJR A; 3IBC A; 5IC6 A; 3ADK A; 1OLS A; 1TJN A; 2I7D A; 1BX0 A; 3B2E A; 3HJB A; 3OBI A; 1CS4 C; 4L07 A; 5DWQ A; 1P6Q A; 1SGW A; 3DO5 A; 2CUZ A; 3IDA A; 2YOF A; 3MUF A; 4QVK A; 4J16 A; 5FOX A; 2PEZ A; 2VVT A; 5AKJ A; 3WKD A; 3NOQ A; 2LDB A; 3WJ7 A; 4EPV A; 5EHZ A; 1H8H D; 2P2Z A; 2IPO A; 3MCZ A; 1UEJ A; 2B78 A; 1P4R A; 4KVA A; 2D2X A; 4HLD A; 3V42 A; 1DIA A; 3C70 A; 5AXB A; 1P2V A; 1O0C A; 3GG2 A; 3DQP A; 5BSF A; 3UWK A; 2JVJ A; 5DT9 A; 3F6S A; 1WDT A; 3V7F A; 1N0V C; 4YRF A; 1T36 A; 5HFA A; 3IIM A; 3IOC A; 3ZZU A; 4CCY B; 3CEP B; 2GCO A; 3QPW A; 4O63 O; 4AJL A; 3QQ5 A; 1M2H A; 1DB3 A; 5AKZ A; 1YA8 A; 2QL7 A; 5ALI A; 4I5F A; 4ZEW A; 4U6N A; 3G2H A; 1K8Z B; 3H6K A; 1EHY A; 1NBA A; 2GK7 A; 2NWQ A; 5F5N A; 3CPE A; 4R6W A; 3RW9 A; 1RJW A; 1IUP A; 1UAD A; 2NPO A; 3DJF A; 3N9I A; 2I4R A; 4UD6 A; 5T3Y A; 4DMF A; 2F3U A; 4RV9 A; 3G8D A; 3T7S A; 1MIO B; 5JMP A; 2E2W A; 5LZ5 A; 4CIX A; 1OX6 A; 1Z88 A; 2PRI A; 2RIP A; 4FWT A; 4FYI A; 2FEK A; 1WF6 A; 2X2O A; 1LVG A; 1BD3 A; 1JIL A; 1VG0 B; 2YCL A; 3DCI A; 2PX5 A; 5KWS A; 1XXL A; 2HZB A; 1CC1 S; 1SC6 A; 3B1Y A; 4GIB A; 4KN5 A; 4YCC A; 3MGA A; 1T2A A; 2B83 A; 3QHA A; 2PGX A; 4REO A; 4XLZ A; 1KOF A; 1ISI A; 1J33 A; 4I4L A; 4U8H A; 4BCD A; 3PDH A; 1WS6 A; 2Q8Q A; 1MBZ A; 1Y6F A; 4YJ3 B; 3HXM A; 4X60 A; 4N0D A; 1AKQ A; 4X24 A; 3K8Y A; 4C01 A; 4CDN A; 3JWE A; 4OBB A; 1FOE B; 4Q19 A; 3VHR A; 2P77 A; 1EYZ A; 4QUG A; 4C5C A; 4Y6R A; 2WIG A; 3TOX A; 4KN8 A; 1U0J A; 2O2D A; 1XYY A; 3ISJ A; 5I9S A; 2ACK A; 2RJ3 A; 3IHW A; 3S8E A; 3M6I A; 2J9R A; 2WD7 A; 1HQD A; 1ZK2 A; 1QUE A; 2AFV A; 1YRW A; 1QX3 A; 5T41 A; 1G2P A; 1YBH A; 2VSF A; 4TYY A; 3ZBP A; 3CZA A; 4Q34 A; 2CSU A; 1W1U A; 2ZSK A; 1H22 A; 5DP1 A; 2VTB A; 3MTJ A; 1JG4 A; 1LXC A; 1RP1 A; 2J9X B; 2BM9 A; 2GJ9 A; 1IOV A; 1HKH A; 1O95 D; 2CLI B; 3BER A; 1VJ0 A; 3EK2 A; 4DXA A; 5KOJ A; 3HYT A; 1XKX A; 2O2I A; 3PXU A; 1OU0 A; 4LXY A; 1T5S A; 4PZZ A; 1FW8 A; 3K09 C; 3PXD A; 4RIE A; 2AN4 A; 4LF6 B; 1GSI A; 1MC1 A; 2Z1D A; 4NMM A; 1C3O A; 4UHH A; 3R4V A; 2C5A A; 4KQ9 A; 2CNW D; 4TS9 A; 4XYM A; 5IVH A; 4KPY A; 1VLM A; 1RAC A; 3P45 A; 4MV1 A; 5B5S A; 1FFY A; 2CYB A; 1D4O A; 1JMY A; 2JFX A; 1DV2 A; 3DHY A; 3OQB A; 5CXX A; 1AKR A; 4P58 A; 1L3C A; 3MYY A; 4QU5 A; 4RYL A; 5H9V A; 5IU6 A; 4C05 A; 2AX4 A; 4M89 A; 2LPM A; 3FPE A; 3PS9 A; 5A3S A; 3V8P A; 1SIO A; 5CHE A; 4RKQ A; 5ADH A; 5C52 B; 3RTB A; 3EVN A; 3CRQ A; 3RQI A; 3K1A B; 2O7N A; 2ST1 A; 1KQF A; 1R4A A; 2GK4 A; 4F11 A; 4DCJ A; 3EQ2 A; 4DJF C; 3AFM A; 3HTX A; 5M2Q A; 2FCR A; 2MYT A; 3EBL A; 4LYY A; 1SNY A; 4ZWN A; 2X7H A; 1AG9 A; 1TM5 E; 4FIN A; 4L4S A; 3FPK A; 2PGI A; 2Q6V A; 1FFD A; 3GNQ A; 3A1D A; 4NYI R; 2V7P A; 1FVX A; 1YUB A; 3O8C A; 2VRN A; 2NU7 B; 3K20 A; 3GGD A; 1Z7E A; 4CDM A; 1OBC A; 4HAI A; 1KTV A; 1B8P A; 1PT7 A; 5JAO A; 1LBE A; 2Z8Y M; 1JHP A; 2EHJ A; 3R8C A; 4N3G A; 3VSD A; 4ZEM A; 3FKQ A; 4OX9 Y; 5A3R A; 3A4N A; 1V39 A; 2P5S A; 3EDM A; 3NPM A; 4IOM A; 3TLY A; 1V7P C; 3QFT A; 2BEJ A; 3DLT A; 4QAS A; 1V2H E; 1HY3 A; 3FZY A; 4AO7 A; 2FPR A; 3GK3 A; 2XVA A; 2X15 A; 2X5K O; 4JLD A; 1JU4 A; 2CYC A; 2WHP A; 3GAA A; 3CH5 A; 3KQY A; 3ORF A; 3Q88 A; 2MIN A; 2RIR A; 4FKM A; 2GM5 A; 3H3X A; 4WBX C; 3ZWV A; 1U7P A; 4AC9 A; 5AM4 A; 4AUE A; 3PF8 A; 1DS6 A; 4AZ0 B; 1EVY A; 1X1C A; 2FSG A; 2ZEJ A; 3DZ8 A; 1P1K A; 3HXK A; 3SIC E; 4KE9 A; 3URK A; 3ZNP A; 3FHC B; 3O0T A; 4FX2 A; 4B8W A; 3ZOZ A; 1X92 A; 3INR A; 1CY8 A; 1NHW A; 4GSL A; 5DAS A; 1TYB E; 2ALJ A; 3ZS9 A; 1NW7 A; 1UDR A; 1WUH S; 4AS2 A; 4D56 A; 3SXP A; 1L5Z A; 2NU9 A; 3OOY A; 4YXF A; 1O57 A; 2VJK A; 2WSY B; 4GR4 A; 1MA3 A; 4A03 A; 5JD6 A; 1O89 A; 3VJR A; 4GXN A; 4LXH A; 4XX0 B; 1VA4 A; 2QAJ A; 3T12 A; 1V43 A; 3NXK A; 3Q3E A; 1G4A E; 5EIN A; 1AQL A; 4BB5 A; 4R71 A; 1N5I A; 2QTV A; 3X43 A; 1ONP A; 3KHT A; 4B3D A; 2CL7 X; 2HRB A; 4WUT A; 4AJ1 A; 1ZIX A; 3PG5 A; 4TS4 A; 3KYI B; 1KC3 A; 1TYZ A; 2VP4 A; 1AOX A; 2BKV A; 2JJ1 A; 2QD5 A; 1QGE D; 3O82 A; 4MVJ H; 1QO9 A; 2V7B A; 4HQO A; 2XEC A; 3CX7 A; 2PJD A; 1BO6 A; 4IS3 A; 4AIR A; 4F3K A; 2D2F A; 2BKU A; 1D3A A; 1W36 D; 4FEY A; 4RY9 A; 2IVF A; 5KSZ A; 1CUG A; 3D02 A; 4I4T F; 4C0Q C; 1Z36 A; 4GI7 A; 3FGP A; 4CGY A; 4CUK A; 2WSI A; 2XBM A; 1NF9 A; 3ZX4 A; 2ZDQ A; 3FSE A; 4CM1 A; 2JHP A; 3GGJ A; 1JGT A; 1NNI 1; 2DM6 A; 2MOK A; 4LWQ A; 2ZJ7 A; 3TN7 A; 1RNL A; 3BC1 A; 3GT0 A; 4N7M A; 1LI7 A; 2R7L A; 3BB4 A; 3KB2 A; 4KZK A; 5ENV A; 2WEK A; 2IU0 A; 2G8L A; 2DEU A; 4JAV C; 3KSD O; 1D6M A; 3PHI A; 4ESH A; 3QKS B; 3EUW A; 4DJT A; 4KXY A; 1S89 A; 2OB2 A; 4ZYT A; 1DBR A; 4E4R A; 3ZGN A; 1EVK A; 1DG1 G; 2Q6L A; 1FRV A; 1JP7 A; 1L3B A; 1Y34 E; 3RIH A; 5F7X A; 1XNQ B; 4E0B A; 2XTA A; 2DCU A; 4KV7 A; 5BNI A; 5K0E A; 1X3L A; 2O4C A; 4BEV A; 5KQS A; 4YGX B; 1Z34 A; 1ULH A; 3VQT A; 5DBE X; 3L4B A; 4H27 A; 2GH1 A; 4NEG A; 2ECK A; 5DO9 A; 2A92 A; 2WY2 D; 3OQI B; 4DBL E; 3FUX A; 4AY2 A; 3ZN2 A; 2QOE A; 3UUE A; 2O06 A; 3WWP A; 4RKY A; 1VBN A; 1JJI A; 4CNQ A; 3CVO A; 3DT9 A; 2VHJ A; 5JMO A; 4GW3 A; 3WRX C; 3RSH A; 1YB1 A; 3EDC A; 1F4V A; 4EY8 A; 4LZO A; 1XT6 A; 2XWJ I; 3VR0 A; 4DM4 A; 3W04 A; 4TWZ A; 3IUP A; 4CEJ A; 1DRV A; 2RON A; 1E2N A; 1M34 A; 4WOB A; 3VC1 A; 2GN4 A; 4P0T A; 4IXN A; 1ZVK A; 5B30 A; 1UFV A; 1SQG A; 2R9V A; 2VTV A; 1M7H A; 1OX5 A; 4UBM A; 3U5T A; 1QDE A; 4JYP A; 5JAN A; 1TO6 A; 2A9V A; 3B6J A; 2QY6 A; 1A4R A; 3CZ9 A; 2AFH B; 4YJ2 B; 4QXM A; 5FXM A; 4B8Y A; 4UJ3 A; 4D7A A; 2RE8 A; 3CUU A; 4V1C A; 4F96 A; 5IJ9 B; 3QU5 A; 4GU5 A; 4UNP A; 3KKN A; 4Z0N A; 1T6Y A; 5CVJ A; 2RB5 A; 3BXP A; 3CH7 A; 5IAO A; 3WFZ A; 3TXX A; 1XN4 A; 3EOA I; 4HFN A; 2IXA A; 4L0Q A; 5A5F A; 2CXP A; 4UME A; 2D2I A; 1A3C A; 1ZWK A; 3D4O A; 1XIV A; 3QU9 A; 4NQR A; 3RSG A; 1Z5W A; 5ADV A; 3ZV7 A; 2Z83 A; 4KIT B; 3GLI A; 2G0T A; 2LND A; 1ND5 A; 2H3E A; 1IQU A; 1G2I A; 2X76 A; 5CBP B; 4IVN A; 4DDM A; 1TPZ A; 3IEP A; 1ZR4 A; 5MGN A; 4G0N A; 2VHZ A; 5HGX A; 2QR5 A; 3N2E A; 3DE3 X; 1L5O A; 4F06 A; 3VX4 A; 1GU1 A; 1WOY A; 1KOL A; 1CUU A; 1D1S A; 4AZ3 B; 3IUQ A; 5JAH A; 3DTV B; 3SBE A; 3I05 A; 1P91 A; 3CRM A; 1J0C A; 3EVT A; 4FJ1 A; 1X0V A; 3T7K A; 2HPH A; 3FUC A; 4EVS A; 4UEQ A; 3W6O A; 1HDX A; 2QSJ A; 3ZWN A; 2O9J A; 1PWZ A; 2H1O A; 1JHX A; 1Q57 A; 2Q9C A; 5FL7 A; 1JNX X; 5F2K A; 4GUD A; 4JI4 B; 1IHP A; 4V2I A; 5CD1 A; 3S55 A; 4C07 A; 3FWY A; 1OD6 A; 3C7K A; 1F8G A; 2EFY A; 1J8Q A; 3A1W A; 2IXD A; 4K9N A; 2FUK A; 2UYY A; 2C3M A; 3NFR A; 3OFK A; 4I08 A; 1DZ3 A; 1Z0K A; 2YTZ A; 4RSM A; 2OUI A; 2WHX A; 5LQS A; 2PGN A; 3WKA A; 1HNZ B; 3PJR A; 5ALR A; 5IT9 A; 4AEC A; 2AUM A; 3VZR A; 4EY7 A; 4D80 A; 4TTR A; 5D00 A; 4EHL A; 2M6S A; 2UUP A; 1SUR A; 2I65 A; 1P0M A; 4QJI A; 3I5M A; 1RYN A; 3QPB A; 2AJB A; 4WER A; 3V7Y A; 4EKY A; 1TVM A; 1BCR A; 2ZUU A; 5BQP A; 3HJR A; 3L8F A; 2F3Q A; 4OXK A; 4XVQ A; 4LUC A; 3BH7 A; 2AJ8 A; 4L8G A; 4N1A A; 4TLD A; 3WDS A; 5L8V A; 1S7C A; 4EF4 A; 5HG0 A; 1VL0 A; 3C4J A; 4YGY A; 4K9R A; 5LA6 B; 3GMT A; 3AF3 A; 3GA5 A; 1ULB A; 2CXX A; 2VHQ A; 1MVL A; 3MCU A; 4MUK A; 4XYL A; 1VBK A; 2G08 A; 4Y0A A; 1PZN A; 3CXU A; 2OB1 A; 3N6X A; 4KQK A; 1L4Y A; 4AXX A; 1EDB A; 3BQN B; 2ING X; 4MMY B; 3GDG A; 4JX9 A; 2A0X A; 4RPC A; 5CX1 A; 3TLG B; 1YZF A; 4QNR A; 4BEC A; 2WUE A; 1KKM A; 3O94 A; 2GXQ A; 4D46 A; 3AP1 A; 1YS4 A; 1PAU A; 1I30 A; 1YFK A; 3Q5G A; 4A15 A; 2KBE A; 1GPN A; 4M37 A; 5CB8 A; 4NR0 A; 4OJK A; 4EPW A; 2XV1 A; 1GPH 1; 2OV6 A; 2PL5 A; 5DN6 D; 3CLT C; 2I3B A; 4NYM R; 1E2L A; 1KDT A; 4JZK A; 2WZQ A; 2ZUL A; 3GGP A; 3IDZ A; 2ZQZ A; 2PL9 A; 2H4H A; 4HB0 A; 2Y9M B; 3KT3 A; 1Z08 A; 2C2W A; 5HS4 A; 2P4U A; 3HPQ A; 3NHW A; 3HVR A; 2QN2 A; 2M6L A; 2JBA B; 1PZX A; 2E4W A; 2I22 A; 2JLS A; 2RCY A; 2RKW A; 3MXO A; 4PYI A; 2XTM A; 3MWJ A; 4MA0 A; 4D0S A; 2RCN A; 4PHF A; 1YDG A; 4KAC A; 4L0M A; 4O4J F; 2WFL A; 3RKU A; 4CT5 A; 2HQR A; 3BXG A; 1SBC A; 3LGX A; 4WDQ A; 4M8K A; 1PNR A; 4ENW A; 3IP9 A; 4AZ0 A; 4MLC A; 1M1Y A; 1DQY A; 3CBG A; 4YAC A; 1HLD A; 1G38 A; 4WK6 A; 4E5U A; 2XJ9 A; 3KV1 A; 3L93 A; 5KZM B; 4F2D A; 1BAP A; 1KIA A; 4ME6 A; 3BRE A; 1M6N A; 4BNH A; 2WMO B; 2R4T A; 1HDC A; 5JLA A; 3WLF A; 1H2W A; 1JKF A; 3RRX A; 1L5H B; 4WZA E; 4W9M C; 5GQS A; 3GR6 A; 1ENO A; 1XTI A; 3IHL A; 3VOS A; 2ZTM A; 2ZIG A; 2WJY A; 4HDA A; 1SOM A; 1YB5 A; 3K6J A; 4Q4A B; 2PYI A; 2NXN A; 5HE8 A; 4J2T A; 2VPT A; 1YHZ A; 4I4N A; 4BNX A; 4MO9 A; 1SBQ A; 1KIM A; 5TJH A; 1CUA A; 4EY3 A; 5C8D A; 1SBZ A; 4TYP A; 1OKK A; 4IAO A; 1PL8 A; 3HJU A; 2JFU A; 4JLS A; 5EN4 A; 4KV9 A; 4Y2P A; 2YJ3 A; 3UJ6 A; 1PNQ A; 3PWS A; 4HG2 A; 3PWT A; 2JIO A; 2VF4 X; 4AWB A; 3HB5 X; 3MAA C; 2QLJ A; 1OZ0 A; 1LNS A; 2MR5 A; 3C39 A; 4HLU C; 5DPM A; 5SY9 A; 2DKN A; 2NV0 A; 4GM1 A; 2XOD A; 3D6T B; 1GNL A; 2PX3 A; 5DN9 A; 1IM2 A; 3KOY A; 2QLM A; 4I5D A; 2G0N A; 1JQ5 A; 3VU1 A; 2C00 A; 2AFI B; 1IUN A; 2ZI7 A; 5AKX A; 1KEE B; 2CSX A; 1U0L A; 1UAY A; 1D09 A; 2QPA A; 1QYW A; 4JOB A; 5HUQ A; 2CDR A; 1D5W A; 2Q4R A; 5BW6 B; 5CH5 A; 4EI2 A; 4JLA A; 4BO2 A; 1E77 A; 1Y6R A; 1VOT A; 4BO1 A; 4MPT A; 3IDF A; 5B7Q A; 3AR2 A; 4ZI3 A; 1A7U A; 3H9G A; 3Q10 A; 2AA3 A; 5GIP E; 5K20 A; 1S6Y A; 2HBR A; 3QPU A; 1IBM B; 3LDU A; 3OT4 A; 4BEW A; 2CCJ A; 2QMO A; 2ZUV A; 1UP7 A; 3IQH X; 1RI1 A; 3J81 A; 4JLM A; 2FW6 A; 3KJH A; 5KQG A; 1WXW A; 3O91 A; 4NWI A; 2NUA B; 3E7W A; 1H9A A; 4BAZ A; 2IZ1 A; 3L07 A; 1WAM A; 3VNU A; 1KKL A; 3Q0T A; 4QYX A; 1IY1 A; 4E4T A; 4EZE A; 4RWE A; 2GPS A; 4QM7 A; 4CTK A; 3IAM 6; 4NFL A; 4DZZ A; 3KQU A; 2A35 A; 4KAJ A; 4M7W A; 3N2Z B; 2VCY A; 1TO2 E; 2OV8 A; 4RVH A; 3TXA A; 1MTO A; 3AHD A; 3F6R A; 1ZD2 P; 3OKP A; 3OTH A; 4TV9 F; 3DGF C; 4IV5 A; 1HET A; 1EUD B; 4PLH A; 1WTC A; 4A5S A; 1G64 A; 3M9W A; 4LYF A; 4UDY X; 4E5D A; 5KSO A; 4EK9 A; 2NZY A; 3QVL A; 5MN6 A; 1YK1 A; 2BKE A; 5JY1 A; 1KBO A; 3LZC A; 2R6Z A; 1SZD A; 1FS5 A; 1CUZ A; 1QK4 A; 1LPS A; 1R50 A; 5SY6 A; 3V55 A; 3ZGL A; 3CLK A; 1YFZ A; 4R31 A; 1W46 A; 1UDW A; 1UKY A; 2H2A A; 1SOV A; 3UYK A; 3GQB A; 3D3Q A; 3JV7 A; 3UHF A; 4OHU A; 1JPU A; 1LNQ A; 3AUJ B; 4NV3 A; 3C3D A; 5K3S A; 3AHG A; 4DR2 B; 1KO4 A; 2FUG 6; 2SHK A; 3H9E O; 3QLV C; 4R6X A; 3I9K A; 2A33 A; 3AEU A; 3S5U A; 3BB3 A; 4UM9 B; 1YCD A; 1B15 A; 3E0X A; 3O21 A; 4OL0 A; 1CSE E; 2X9Q A; 4LYD A; 1Q7C A; 3VDR A; 5F4B A; 1Z06 A; 2Z2X A; 5FP0 A; 3AET B; 4KK4 A; 1R9G A; 5HJ5 A; 4BB9 A; 1I24 A; 3NRB A; 3GN2 A; 1OFT A; 2BDU A; 2Z9D A; 2FPP A; 3NBZ C; 2YID A; 4CXM A; 3MJV A; 3T7C A; 2HA4 A; 3OZG A; 3P2O A; 3B7G A; 1RI2 A; 5COQ A; 3L7A A; 3BSF A; 3DPS A; 2RI6 A; 4GOM D; 5FO0 A; 5HCI A; 3K4P A; 1G1A A; 3HJN A; 1HNN A; 4ZQH A; 1KYW A; 3GG9 A; 4J43 A; 3V7U A; 4H08 A; 3PXE A; 1RC0 A; 1P1J A; 5EJ8 A; 5JC8 A; 3SYN A; 4L8Y A; 3WA7 A; 2IDZ A; 2GZD A; 3EUE A; 3VFD A; 4EEC A; 1OT3 A; 1E3D A; 2JGF A; 1CNM A; 3PUX A; 4ARB A; 2WKW A; 3CUR A; 1R2Q A; 2G72 A; 3L8C A; 1UPF A; 3BF8 A; 4FQ7 A; 4GD3 Q; 1H6D A; 2XAD A; 4QBF A; 3SLK A; 4JGG A; 2TEC E; 2IX3 A; 3L7O A; 1VJR A; 4KIG A; 2OSV A; 3LRT A; 4LXU A; 4M9R A; 4QPD A; 3MBK A; 5D6N A; 3SG0 A; 3RPE A; 1NW3 A; 3AZP A; 2PKC A; 4RO0 A; 5C81 A; 1RXI A; 3KEL A; 3RTE A; 2FR8 A; 3TE7 A; 1ZUI A; 1A1V A; 1IHU A; 3S1A A; 2PTH A; 3QEL B; 1GGF A; 4YRP A; 3GCX A; 1G4H A; 4D81 A; 3FWZ A; 1ZXL A; 1VCH A; 2Q5J A; 2HLN A; 3X3H A; 1T9G R; 3DEJ A; 4R4J A; 1NR1 A; 2PMB A; 2O3Q A; 1NVF A; 2C92 A; 3FJY A; 1AF7 A; 4LFB B; 4P4N A; 2X8A A; 1TAG A; 4FTW A; 2PZJ A; 1V3U A; 3ORY A; 4OBW A; 1RM4 A; 3GGS A; 4OIF A; 1XDG A; 2WM3 A; 5LA6 A; 2W6J D; 4A8T A; 4KHZ A; 1L5K A; 2X5J O; 1F0Y A; 1QV9 A; 2I87 A; 3HBM A; 4B6R A; 3T1H B; 4KPF C; 4EFL A; 4G0X A; 4WIN A; 4M86 A; 4NBR A; 3W9S A; 2PFK A; 5DHS A; 3QKI A; 2BFC B; 4V0Q A; 5HIJ A; 4JEH A; 2R75 1; 3OWX A; 4GO1 A; 4OHO A; 4X6L A; 2IX9 A; 4GON D; 3T7J A; 1U1G A; 2H5I A; 2LGJ A; 3HB1 A; 4OYR A; 3REO A; 5ALJ A; 2H9Y A; 3IUL A; 1OWL A; 5T4F A; 1EZ4 A; 2OES A; 3OQ1 A; 1SBI A; 1NQO A; 5DGX A; 2Q7X A; 5A02 A; 1AKW A; 1MOZ A; 1SB9 A; 1CRQ A; 5E6R A; 4J0G A; 3F2I A; 4N76 A; 4OUD A; 3COV A; 3KYD A; 3LCV B; 3C87 A; 3CUE F; 2PS9 A; 4KWE A; 4P3X A; 3DO6 A; 4DZT A; 2QKY A; 4G1M B; 2PKQ O; 5AM3 A; 1RHK A; 1XZP A; 3LH5 A; 3MXT A; 3S3T A; 3FZE A; 4O8C A; 1Y48 E; 5FUB A; 1FU7 A; 1ZBQ A; 3Q0E A; 1V8O A; 2JAW A; 2VHV A; 2FRV A; 4X93 A; 3UOW A; 2W4I A; 2RH4 A; 4X1O A; 2XVY A; 2JEY A; 3IVG A; 2HHJ A; 2FJU A; 5GV8 A; 2C4M A; 4AQY B; 1NC3 A; 4G4G A; 2WTE A; 3SDB A; 1Y1K E; 1VIM A; 3GC6 A; 3ZY5 A; 5IAS A; 4PY2 A; 2CHF A; 2H0Q A; 4BII A; 4R5H A; 2PM8 A; 3RRZ A; 3QNM A; 4KO8 A; 3R7D A; 1WDL A; 1ZL2 A; 3D0Q A; 3SDS A; 3IYL W; 3ZQU A; 4ALM A; 5C7O O; 1KP9 A; 3K1J A; 5B55 A; 5JFO A; 3LT2 A; 2OCK A; 4KYI B; 3W6Z A; 1OJL A; 1NBW B; 1OI3 A; 3EA4 A; 3VO1 A; 1SXJ B; 5CDT A; 1V3V A; 2E6K A; 4Z7J A; 2UYO A; 4Q21 A; 2ADM A; 2R7A A; 5TT1 A; 5C7Y A; 3R8F A; 3NWE A; 1ZMT A; 3OC6 A; 3A9R A; 1B7G O; 2B61 A; 3V8B A; 3ZHS A; 5KMG A; 3KJE A; 2P90 A; 4F6L A; 1WY5 A; 1FLD A; 4NC9 A; 3KJQ A; 1YFJ A; 2YJ4 A; 1AN0 A; 4HYY A; 4MQF A; 3H18 A; 3MMP A; 1WXE A; 4ND2 A; 1N78 A; 1WCY A; 2H63 A; 3WDK A; 2QXT A; 1U3G A; 1XD2 B; 3X1M A; 3ZDQ A; 1EWV A; 3U0B A; 1I3K A; 2HEG A; 2NUT B; 3PIT A; 4BE4 A; 5JSU A; 1W5S A; 2FZW A; 3QFF A; 4ENS A; 2GKG A; 2D8A A; 3T94 A; 1T4B A; 2EWV A; 1YQU A; 5EJG A; 3F97 A; 4QGF A; 5G4K B; 4QDT A; 6AT1 A; 3H4I A; 4NKR A; 1NW8 A; 1U4N A; 2RGN C; 3E84 A; 4YRN A; 1UXH A; 3TRJ A; 4BKM A; 3ELD A; 1XEX B; 2W0S A; 2XBU A; 3HBF A; 5SY4 A; 4I46 A; 3AF5 A; 1U7D A; 2JJ8 A; 1VXO A; 3KM0 A; 1G18 A; 2AFI A; 3M6F A; 4YT5 A; 2R6C A; 1TQ8 A; 2MYN A; 1AB5 A; 4LNU B; 3N8I A; 2FWP A; 1QVR A; 2FNA A; 3PR2 B; 3NHU A; 2QYO A; 4WK4 B; 5E4Y A; 4CQN A; 4JHL A; 4WNY A; 1QBK C; 1SUS A; 1XTB A; 1Q7T A; 2AH5 A; 3MYR A; 4L02 A; 1RBY A; 3HEA A; 3NU1 A; 1E0S A; 1QAO A; 1TEH A; 1L7O A; 5E95 A; 5EQF A; 3VPS A; 4BWF B; 4WV3 A; 5B1E A; 4MR9 A; 2PJZ A; 5BPH A; 5FI5 A; 4F2H A; 2B94 A; 2IPX A; 3V5N A; 3RI3 A; 3VPF A; 1G5T A; 2IWR A; 1CIV A; 4HQF A; 1GPD G; 3CUS A; 4ZQI A; 5JSQ A; 4ECE A; 1COW A; 2K4M A; 4AUM A; 1YR1 A; 3KR0 A; 3O4V A; 2H85 A; 1RYF A; 3W3A A; 3TK1 A; 4YV7 A; 3WK6 A; 5EJ5 A; 5E7M A; 2OCI A; 4KQ6 A; 5G0W A; 1PGO A; 4OXQ A; 4QAQ A; 2GBL C; 4MHO A; 2BFD A; 3G0O A; 2E5W A; 3P0H A; 1RHJ A; 2EL9 A; 2VWH A; 2WJW A; 5JDI A; 2D3W A; 3KBO A; 4IEI A; 4CMI A; 2WA2 A; 4BX8 A; 3INY A; 4JP8 A; 4K8R B; 3HDJ A; 4RVD A; 4R0M A; 4YLY A; 5KYW A; 3UJ3 X; 4HD8 A; 2H18 A; 4PYR A; 3U4M A; 2QVG A; 3VVL A; 2EAT A; 5D1N A; 1A50 B; 3IPA A; 1Q16 A; 4EZB A; 2GPA A; 2AML A; 3M8K A; 2OC4 A; 1YGY A; 3KOW A; 1R9M A; 1GUY A; 1ZD4 A; 3NP9 A; 2IYZ A; 1O9G A; 2UZ1 B; 3K0A A; 3R5S A; 3TSW A; 4CM8 A; 2NM0 A; 4JVS B; 3IES A; 4L2I B; 1NI4 B; 2EQB A; 2P1R A; 1XF9 A; 2JRC A; 3A3Y A; 4OQL A; 2BHN A; 4H6W A; 4Z86 A; 3ENW A; 4YS6 A; 3TTW A; 3ZZT A; 3ZW8 A; 3VJK A; 2H31 A; 3K6S A; 4I99 A; 2QE7 D; 5AHW A; 1X7W A; 4IPT A; 3RHZ A; 1UJM A; 2GD0 A; 1M2O B; 2RKO A; 3IX7 A; 4HH1 A; 1UT8 A; 2EF1 A; 2YUT A; 3EIQ A; 1MAA A; 4IOJ A; 2J85 A; 1JR6 A; 3SJU A; 4RGB A; 5DLQ C; 1U3U A; 2I6U A; 4YQY A; 2HU2 A; 1RQR A; 1EFR A; 5KU1 A; 1WU8 A; 4AP6 A; 4BFZ A; 3OF3 A; 3TZK A; 2NO0 A; 4H9G A; 3W79 A; 4MRP A; 4OQX A; 1Q1Q A; 3LQ2 A; 2QW1 A; 1A2O A; 5DSZ A; 3B2N A; 4MLZ A; 3A37 A; 1Z2D A; 2P9H A; 3ZU2 A; 4U5X A; 2H6E A; 2IEG A; 2VXO A; 1ARZ A; 4W1Q A; 2D1Q A; 3JU3 A; 1G21 A; 4DIO A; 1SVN A; 2FJ0 A; 2Q1T A; 4RVS A; 4NIC A; 1I4W A; 2FUE A; 5CUQ A; 1IUL A; 5CA1 B; 2RI0 A; 3U7H B; 1ULS A; 1NJF A; 1KRW A; 4MUJ A; 4MRV A; 2A1T S; 4ZAL A; 3HBL A; 3J81 k; 1NQX A; 4KXL A; 3ZEI A; 4L6A A; 1JLL A; 4F38 A; 4CKC B; 1GU7 A; 4BKR A; 5IUN C; 3SYR A; 3SU8 A; 3UWS A; 3VKU A; 5HIL A; 1MD9 A; 1AHN A; 3DZF A; 3V2I A; 4JLB A; 4KIF A; 1ZK0 A; 3MQ4 A; 3BMQ B; 2GFQ A; 3AUA A; 4C3V A; 2WYW A; 2Y2V A; 1THM A; 3KDS E; 5BN3 B; 1XB2 A; 2UVD A; 2AYY A; 4JV5 B; 1YIA A; 3E9R A; 3F6B X; 3WLK X; 4PIM A; 1W2W B; 3ZWP A; 3UPU A; 1SQ5 A; 2H8K A; 2Q49 A; 2QDK A; 3EAG A; 3F6P A; 5AM0 A; 2VL7 A; 4U8G A; 4AY6 A; 2F8L A; 3S2I A; 1UDA A; 5DYY A; 2NYV A; 2O6H A; 4GKK B; 2IH4 A; 4MOW A; 3KEQ A; 3PEN A; 1KOJ A; 1Z6T A; 3OOJ A; 3OEH D; 4MQ4 A; 4RMH A; 4H7D A; 4RD2 A; 1KNX A; 3OY7 A; 4UVH A; 5E8J A; 4CIV A; 4I4T A; 2CKM A; 2NU6 B; 4Y9D A; 1VLJ A; 3GP8 A; 2EF0 A; 1X80 B; 2XWB F; 2Q0H A; 1OT5 A; 3IEU A; 4TTS A; 2NAD A; 1H6A A; 1LC0 A; 4CRW B; 2WOO A; 4KQG A; 2LIP A; 4U7H A; 3V2G A; 2YWD A; 3O6V A; 2BTO A; 1SVU A; 4RCZ A; 2B8J A; 1B74 A; 3NHJ A; 1E18 A; 4X62 B; 1MG0 A; 1Z2A A; 3IP5 A; 1GGV A; 4LGM A; 2PZE A; 1HK7 A; 4FVT A; 3O92 A; 4O4H B; 2GN2 A; 4PWY A; 2EAU A; 5FLE X; 4YCL A; 1KF0 A; 3AGQ A; 2PUW A; 1ZRM A; 2NLO A; 4L7Q A; 3A0U A; 4QUH A; 5AJT A; 2QVC A; 1GCO A; 4FR2 A; 3QMW A; 4ND4 A; 4QT0 A; 2ISC A; 2Q29 A; 4DAE A; 4Q05 A; 4RFL A; 3HCB A; 1XVV A; 2QD3 A; 3B1X A; 2NP7 A; 3O4G A; 1C0L A; 3ZCX A; 5JIS A; 3PL5 A; 1HQK A; 3C98 A; 3R24 A; 2DR3 A; 3H4M A; 4M60 A; 4F1K A; 2HX1 A; 5CGZ A; 1ZU5 A; 4E1V A; 3PPW A; 2HFJ A; 1SZP A; 1G3R A; 1IQ0 A; 2FGJ A; 2ZR7 A; 4IH4 A; 1U31 A; 1YQG A; 1MX5 A; 2BUB A; 1B0Z A; 2IIB A; 3N7T A; 1GM5 A; 2PUC A; 1VB3 A; 3GX1 A; 1IN8 A; 4FI3 F; 4AZS A; 4QJ3 A; 3BUS A; 1LDG A; 3R09 A; 3RK0 A; 5JHG B; 3CCB A; 4Q4H A; 3IER A; 1QYV A; 2KPO A; 2PO7 A; 3BT7 A; 1FSF A; 1RWW A; 4L3V A; 4QPP A; 1C4W A; 1N0W A; 1GKY A; 2WYV A; 4H77 A; 3BJZ A; 1S1M A; 3OCZ A; 3HJT A; 4IEE A; 4LBY A; 2G88 A; 3M6A A; 5L3Q B; 1P4J A; 4FSD A; 4QOH A; 3KUU A; 3P9K A; 5C8Z A; 5A4F S; 4BND A; 1UEI A; 1WEH A; 5DWD A; 5L3Z A; 2NUT A; 5BQX A; 4V1C I; 1QU3 A; 5FPQ A; 4DHE A; 2ZWM A; 2FEP A; 2IT1 A; 1MO5 A; 1QDU A; 2NUP B; 3PDI A; 3FTF A; 3TQF A; 1JSC A; 1BDH A; 1ZAY A; 1QGA A; 1OZA A; 4DWJ A; 1U9Z A; 2P3Q A; 4K8O A; 3E3L A; 1CZH A; 4CFS A; 1WLS A; 3GLH A; 3LDA A; 3UT5 B; 2QVZ X; 2JNW A; 4OLB A; 3C4V A; 3SX4 A; 4EEL A; 3D4N A; 1AQI A; 2JA2 A; 4BGE A; 4YUZ A; 5KTY A; 3MM4 A; 4S24 A; 4BVA A; 1V1R B; 5TGZ A; 2ERX A; 3E9C A; 4RVN A; 4A91 A; 3IC5 A; 3GRP A; 3CLU C; 4R2W A; 2VTE A; 2W93 A; 2PSS A; 1NXS A; 4MGK R; 3RYF B; 3C41 J; 2R48 A; 5C4I A; 3LKZ A; 1SN7 A; 4XIY A; 5EIO A; 3COJ A; 5FAD A; 2V1X A; 5E7J A; 4KFC A; 4MW1 A; 3I92 A; 2JJ2 A; 2MTB A; 4LS3 A; 3RK4 A; 4UQL A; 3ICQ B; 4OP2 A; 2DGD A; 4XZ3 A; 4FIZ A; 1GPW B; 5D2C A; 3RIM A; 5I8W A; 1EVL A; 3BL5 A; 3VR3 D; 2WJJ A; 3AVP A; 4UU0 A; 2GZM A; 1XNG A; 1DC3 A; 3D2C A; 4LVO A; 2F3T A; 3B9R A; 4RDI A; 1KDZ A; 4C7K A; 2GYY A; 2VSO A; 2XUJ A; 3MQF A; 3QQP A; 1X8K A; 4K1X A; 4PTZ A; 4LRQ A; 4FLM A; 4KGX A; 1J5P A; 4R51 A; 5L3S B; 5LQR A; 1D8A A; 7MHT A; 1VCM A; 2PZM A; 1P1C A; 3RU9 A; 1KJ8 A; 4ZRD A; 3W4I A; 5AK3 A; 1KQG A; 3CLS C; 1GXS A; 1HOR A; 3L43 A; 4NBW A; 2H5L A; 1KY2 A; 1VRC A; 1ECC A; 3BED A; 4RD0 A; 3QVV A; 4L5Y A; 4CF3 A; 2P75 A; 3T5B A; 4M98 A; 1VE7 A; 2C82 A; 3KQL A; 1Z0S A; 3B12 A; 2HA3 A; 2GKS A; 1ZN2 A; 5HA5 A; 2F1K A; 1KY3 A; 2AF4 C; 4REM A; 2QL6 A; 4Y9R A; 1DEH A; 1GQR A; 5LMZ A; 1MV8 A; 1CUY A; 3KYC B; 4MEB A; 4KI0 A; 1TAD A; 3OZV A; 5A4J A; 2NW6 A; 1I3M A; 4OU4 A; 2FXV A; 4TTV A; 1E6C A; 2HN9 A; 3OZX A; 4OHX A; 2QAG C; 3RB8 A; 2C2O A; 5CFQ A; 4Y3U A; 3DAH A; 1YUM A; 1YU9 A; 3NC0 C; 5HDB B; 1NLY A; 4R1Q A; 5AXC A; 2CFS A; 3A25 A; 4BCC A; 1IZ9 A; 3I8S A; 1AMN A; 2I4N A; 2XZL A; 4IH1 A; 3FCS B; 5ALF A; 2ZTL A; 3QKT A; 1CRP A; 3L0S A; 1VSB A; 2XND A; 1ILW A; 5GON F; 3Q8L A; 1NY6 A; 5CW1 A; 4D9C A; 1T24 A; 4RSH A; 1PR0 A; 2Y1O A; 1DWQ A; 2I6F A; 4IRX A; 4ITU A; 1EBB A; 1KOR A; 4P6V F; 1KIF A; 4WUV A; 2WIA A; 1X1B A; 3O1J C; 3BFV A; 1GQO A; 5DHP A; 3E81 A; 3STY A; 1KGD A; 3E9Q A; 4CIY A; 3WRW A; 1T6Z A; 2ICA A; 2V7W A; 3ETJ A; 3G79 A; 3GT4 A; 1RQ2 A; 5A9S B; 4BMZ A; 1NUA A; 2DPY A; 2OHH A; 2YYA A; 1U8C B; 3QVX A; 3MPO A; 4RK6 A; 2EFG A; 5F5X A; 1ODC A; 1Q0H A; 3AEK B; 3NC4 A; 3R2J A; 5DK6 A; 5CJP A; 2VHU A; 1ZD5 A; 2XNS A; 2PVP A; 4MR8 B; 5FHQ A; 4JXK A; 2HLD A; 4JXJ A; 1GPB A; 3UK7 A; 3FKS D; 3SF8 B; 4M9X A; 3K0C A; 2V0N A; 4O4J A; 1V7Q A; 4CMK A; 4P5E A; 5IZ4 A; 4N0M A; 1XFD A; 3TII A; 4B44 A; 1TAQ A; 4RA9 A; 2PL3 A; 3G6W A; 1HT0 A; 5AIB A; 5BXQ C; 3ET4 A; 5BWL A; 3K9C A; 4MR7 B; 1UKV Y; 1YRH A; 2J7K A; 3UJN A; 4U7J A; 1QVA A; 1QE5 A; 3JAT A; 4URV R; 5CA1 A; 3AY7 A; 3AX6 A; 1VM6 A; 2O2S A; 2XCT B; 5C4G B; 2IYF A; 1QGU A; 3CEJ A; 3GMI A; 2OQ2 A; 1F1J A; 4BB0 A; 4BTL A; 3LXU X; 1NOK A; 5ALX A; 4A0G A; 5FLX A; 1UAA A; 1BA2 A; 3K1Z A; 4NK5 A; 1MB3 A; 2WF6 A; 1EDZ A; 1CT9 A; 1FEZ A; 4R9U C; 1UDC A; 4ZU3 A; 3DWE X; 2G82 A; 1SO8 A; 2VDQ B; 2WNS A; 4TRM A; 1UPC A; 4YRB A; 1QUF A; 1NU8 A; 2QN1 A; 4TS3 A; 3L0Y A; 1SU7 A; 1CKE A; 2D0O B; 2OHV A; 3DVL A; 2Z3Z A; 2RK6 A; 1BMQ A; 2KQU A; 3EQ8 A; 3HP4 A; 2XXA B; 4AI6 A; 1MQS A; 2AQK A; 5BRN A; 1BGV A; 4C6T A; 3P52 A; 5C4M A; 3W7B A; 3VVM A; 3ZKA A; 4B6F A; 2C7P A; 1WHS B; 1SA0 A; 4UW9 A; 2Y67 A; 1F2V A; 3BQM B; 5CDG A; 3CSD A; 4EJS B; 4ND1 A; 5IYZ F; 2J8Z A; 5EIX A; 4Q4C A; 4C29 A; 2ZUF A; 2GJK A; 4ARS A; 3TD9 A; 3Q6I A; 5DW0 A; 1EXM A; 2BZ0 A; 4IY1 A; 1E7Y A; 1Z2N X; 1XE3 A; 2P9T A; 3PRY A; 4UAU A; 3WLM A; 1RXV A; 1O3Y A; 3MPU A; 2QBY B; 3ICP A; 1X7Z B; 5I5E A; 3DVA B; 4R1L A; 4DG5 A; 3PQ4 A; 4XSH A; 5AMX A; 2ZBV A; 3D3J A; 4S1N A; 2QG6 A; 4DKX A; 2BH9 A; 3TIX B; 4HT3 B; 2DTD A; 4GZM A; 4EL5 A; 4FT4 A; 2J9F A; 1N2H A; 1EFH A; 4R1T A; 3VO2 A; 3ORQ A; 4P7J A; 2OGZ A; 4IMP A; 4IJ6 A; 2C99 A; 3C16 C; 1CS0 B; 2FH5 B; 2GS9 A; 2VAM A; 1NQU A; 3SHQ A; 2OLE A; 4ZO9 A; 1RFG E; 3SVT A; 2G06 A; 1XCM A; 3EG5 A; 5JEC A; 3EGX A; 3A1F A; 1X1D A; 2QZ9 A; 4B34 A; 2IP4 A; 1JEQ B; 2ETX A; 1C2T A; 3VGP A; 4P86 A; 1B14 A; 3AM3 A; 2WAA A; 2ZJ0 A; 4UKD A; 3IQF A; 2KX7 A; 2XXJ A; 1GUF A; 4HR7 A; 1SUD A; 2Q7O E; 1DO0 A; 2QX8 A; 1FJ2 A; 4L4X A; 3O84 A; 3BGS A; 2DFP A; 1W2G A; 4GLO A; 3ZJT A; 4TV9 A; 1CR2 A; 1Z4Q A; 3RUD A; 1AGN A; 4WKO A; 1MFZ A; 2JGA A; 2GKO A; 3L7J A; 5ICS A; 2E37 A; 1FUI A; 2FUC A; 1YBF A; 1QFJ A; 4OHB A; 1QQB A; 2BPO A; 2WWH A; 2QUA A; 3D3W A; 4DUY B; 5H3C A; 1KSF X; 2PUT A; 3B6M A; 4Q18 A; 4QDJ A; 3DMP A; 3HB9 A; 4GX0 A; 4BUJ A; 1ZA2 A; 1LKZ A; 1Y6J A; 2NXE A; 3TZB A; 1YHN A; 1QKK A; 1T0U A; 3LYL A; 1VKJ A; 4ZMR A; 2PE5 A; 2NUP A; 3ZGO A; 5JH7 B; 2WMD A; 3ER9 A; 5FEM A; 4W7S A; 3LHF A; 1N5D A; 1BW9 A; 3PQ6 A; 4F6R B; 4WE3 A; 4Y2R A; 1YR2 A; 4TSF D; 4K3U A; 5JQG B; 3K0B A; 1PDO A; 3DP7 A; 4PRZ A; 1NLM A; 4C0L A; 1SUF A; 2N75 A; 5DGT A; 3R3U A; 5ABP A; 3FVT A; 3IGF A; 4NL4 H; 1GPA A; 4Q9K A; 5DYT A; 5A6V A; 2ZJO A; 1QB7 A; 2OYC A; 4OKS A; 4BNL A; 5JJB A; 5GIN E; 4R7Z A; 4OSO A; 3Q8M A; 4E21 A; 3FLB A; 1AY0 A; 1ORH A; 3OMX A; 2NXC A; 1V9S A; 2GZ1 A; 3IKL A; 1M6Y A; 4YAL A; 4XLL A; 4A8J A; 2VH5 R; 2PMW B; 3D81 A; 4ZAF A; 4BO4 A; 1E0A A; 2R7M A; 4NCJ A; 1NRJ B; 2SCU A; 4PGG A; 2VQ6 A; 1W7A A; 1OFU X; 3QM1 A; 1K98 A; 4J29 A; 3AU8 A; 3R5X A; 4RK0 A; 1C1X A; 1N3Y A; 1XAJ A; 2DKF A; 1Z7W A; 4ORE X; 3NS7 A; 4LJL A; 4CHG A; 1OH7 A; 3KPW A; 2AG6 A; 3D3E A; 3ZQ4 A; 5L4N A; 4KNY A; 3P94 A; 4EMS A; 3QLU A; 4R81 A; 1OH5 A; 3UXM A; 1BQE A; 1W0K D; 1W52 X; 2VSX A; 2W11 A; 3DNF A; 2X3F A; 4Q1E A; 4GSY A; 5FYD A; 4UUQ A; 1L1F A; 4D3Z A; 4KL8 S; 1GLG A; 3A9V A; 1Y6G A; 3G5T A; 2FFR A; 3KBB A; 4XUE A; 1R8S A; 5CLH A; 4HHE A; 5FOU A; 4DCC A; 2PO5 A; 2X4G A; 3I04 A; 3LCJ A; 3LYH A; 4MXD A; 3LNC A; 3B3G A; 2J0U A; 5T7B A; 3NB0 A; 4XWG A; 3K70 C; 2OK7 A; 4ABT A; 5KUT A; 2Z2I A; 4PHH A; 5JZO A; 1D6S A; 3DV0 A; 2DFV A; 5MHT A; 1XZG A; 2I4L A; 1I2B A; 1D1P A; 1XZK A; 2G4V A; 3NVA A; 2Z57 A; 4Q9D A; 5K4H A; 2FRD C; 2WWY A; 4HYT A; 3H6G A; 4IUC S; 1F38 A; 3OZU A; 2H8G A; 2AB0 A; 5E6U B; 2C7R A; 2GJ8 A; 2R8O A; 3G89 A; 1T9D A; 3CPH A; 1C4K A; 4JD5 A; 1I1Q B; 3AEQ A; 4BFT A; 3GEE A; 1GEE A; 1I4D D; 2ZJ2 A; 3F1K A; 4HMO A; 3VA7 A; 3KJZ A; 1VIY A; 5LU8 A; 5BOS A; 4ZD6 A; 2GYU A; 2GN8 A; 2VZ9 A; 3N74 A; 1KWK A; 1Q6Z A; 4JJE A; 1VGR A; 3CXR A; 4L7V A; 4MGZ R; 4B3I A; 4D92 A; 3U6B A; 4NHE A; 5BST A; 3R6G A; 3VTA A; 1VDZ A; 3GED A; 4WDR A; 3V6M A; 4FW8 A; 3D6J A; 3ZDR A; 1ESZ A; 1I6W A; 1MLD A; 4MIY A; 4A82 A; 1M2F A; 1NAH A; 1ZPD A; 2B4Q A; 1P77 A; 4D9M A; 3H2B A; 2BON A; 1XPO A; 4Q16 A; 3C7T A; 3NGM A; 1FW6 A; 2YMM A; 2L17 A; 5IUL C; 2ENW A; 3M2P A; 2ZRH A; 1ZDM A; 1EK6 A; 1W5A A; 1PBN A; 4OAQ A; 5K3A A; 3WFI A; 2XCS B; 2FSV C; 1AV6 A; 1KU0 A; 3W0L B; 5A4I S; 4QLA A; 5DSX A; 1E1D A; 3ZJU A; 4ZAX A; 3TJZ A; 3CIN A; 2FXI A; 1ITZ A; 1X0X A; 4HX5 A; 4MWS A; 3T5G A; 1NUR A; 1P60 A; 2I2A A; 4LSZ A; 4KE8 A; 3NLL A; 4MCH A; 5IAR A; 1B42 A; 5C46 F; 2IHB A; 3NHK A; 1X14 A; 3QOX A; 4IIJ F; 3DKI A; 3ABR B; 4BLT A; 4EJ6 A; 5CNK A; 4L5A A; 5A1J A; 1GIA A; 2P9J A; 4YMG A; 3RU3 A; 1M2E A; 3ED1 A; 5EIF A; 4DAB A; 1LTK A; 3L0O A; 3WQQ A; 5BWB A; 1QH9 A; 1GAW A; 2OLJ A; 3T38 A; 4NCH A; 1XZA A; 2NNL D; 4U3J A; 5K9Z A; 1TF2 A; 4KUE A; 4LOA A; 1JDB C; 1KO7 A; 1GP2 A; 3PA8 A; 4URE A; 2AMJ A; 1Y1Q A; 1B6S A; 1BOF A; 1G0N A; 1VRV A; 2DHE A; 3ANL A; 4K9D A; 2COK A; 4JX8 A; 2FPU A; 4PSU A; 4YFV A; 2EJV A; 4YK7 A; 1RBQ A; 1SKB A; 2JH8 A; 3EY9 A; 3NHR A; 3ZHY A; 2W6Z A; 3NJR A; 1BFU A; 4EMU A; 5A4M S; 2JK0 A; 1P2G A; 5BMV F; 2QTH A; 2BFF B; 4NCB A; 3EG9 A; 4C7V A; 1J8M F; 2KB0 A; 1KET A; 3AEZ A; 4E11 A; 2FWR A; 1GSA A; 3W9B A; 2Z6Q A; 3D8S A; 4ZT2 A; 2WJG A; 5IM4 F; 3LCU A; 3FCA A; 1T8Y A; 3LCM A; 5CB4 F; 3T7A A; 2CB9 A; 3L1J A; 4DQ0 A; 5IM5 A; 4CKB B; 1NXX A; 2WHQ A; 5A0T A; 4YMU A; 1J0X O; 2V7V A; 2R6J A; 3EIS A; 4H15 A; 5EWM B; 4CKX A; 1BYU A; 5JXG A; 3AB7 A; 3G17 A; 3Q1K A; 3VO8 A; 5JMT A; 2DBQ A; 1NDH A; 1C8B A; 2ORY A; 3WGI A; 1H2A S; 1BKD R; 3A6L A; 4NI5 A; 4TRJ A; 3MFQ A; 1GNQ A; 2PXX A; 3GU3 A; 3ZXX A; 2DEC A; 1E7S A; 3F2V A; 2O2J A; 3SIW A; 3VZB A; 1Q7G A; 2BM8 A; 3QGM A; 1QH0 A; 5DI9 A; 3L2O B; 4EY5 A; 4YQF A; 2OYS A; 3AR6 A; 1HT1 E; 4AYW A; 1TVC A; 3IAM 1; 2XQK A; 1CUF A; 4PLW A; 4TL6 A; 5I9I A; 5U9C A; 4DV3 B; 2Y6U A; 5JJL A; 5CT5 A; 3FCE A; 4P54 A; 4FYW A; 5CVV A; 1T91 A; 3HYR A; 1DAG A; 1U65 A; 3TLA A; 4S1O A; 1B26 A; 3ECA A; 4F4J A; 3QKR B; 3KHK A; 3OIF A; 4CF6 A; 2JFB A; 4WLN A; 4CN8 A; 3W90 A; 4B45 A; 2POC A; 3B1K A; 3CZC A; 2Z95 A; 3FTP A; 4PG5 A; 2GN6 A; 4UBI A; 5E2B A; 3DVS X; 4CTJ A; 2DH5 A; 1FTG A; 2QAI A; 5HX7 A; 3FCY A; 3N2I A; 3F6E X; 3IQG X; 3OFJ A; 1UPX A; 3TEM A; 2FF7 A; 1AQU A; 1I01 A; 2OR3 A; 3WHL A; 1CLE A; 4JOS A; 3HB7 A; 4IOK A; 3FRJ A; 1KKU A; 2ZZN A; 1KK2 A; 2HK6 A; 3RK1 A; 5EXD A; 3W5H A; 1MFP A; 3BBL A; 2FUG 1; 1PUI A; 2YHI A; 4ARC A; 1F2D A; 1KZ6 A; 1M76 A; 2Z2Z A; 4KQE A; 1J2U A; 3PPY A; 2XE7 A; 5AHK A; 3DYV A; 3HUE A; 4IIF A; 3ACB A; 4WZB B; 3RC2 A; 2VII A; 3B70 A; 3CNN A; 4UTR A; 1UBR S; 4BLR A; 2OZE A; 4MYT A; 2WF8 A; 4B6E A; 1SEP A; 1WWK A; 5JAM A; 4TV8 B; 1JQB A; 1K8Y B; 1NWH A; 1OFV A; 1WXD A; 3CG0 A; 3LO8 A; 4MUL A; 4P4L A; 3ZY2 A; 4OM7 A; 3ONE A; 1LF5 A; 4AYT A; 4EB6 B; 5WRF A; 2BFA A; 3AUK A; 3FHO A; 3VVC A; 5AXD A; 3O38 A; 4QE3 A; 1OW3 B; 5K3B A; 3T9A A; 2NTE A; 5DIF A; 4FD2 A; 3FCU B; 1Q1Z A; 5FTK A; 2O3S A; 2JZO A; 4JR0 A; 4F6R A; 7REQ B; 2HCR A; 4TL8 A; 2HUN A; 2HJG A; 2VUS A; 1UMK A; 4WD3 A; 1TKB A; 2HU5 A; 2ZB8 A; 3VYW A; 3V7N A; 1P0Q A; 3AEY A; 1P8J A; 4LY0 A; 4ODI A; 1F6B A; 4Y6P A; 3FHJ A; 1SEL A; 2VK4 A; 4J0D A; 4W9N C; 4ZVV A; 4NRD A; 1KDP A; 1USK A; 2HNA A; 2OAG A; 1QG2 A; 3F7N A; 5CX4 A; 5FBH A; 4UNS A; 1F0N A; 1E3J A; 1L5G B; 4H17 A; 2VOJ A; 3IPY A; 3OLZ A; 2YN4 A; 4H6P A; 1XBT A; 1HX7 A; 2WU3 A; 2P2G A; 1P81 A; 4NYJ Q; 2VXB A; 3ZIU A; 1ADY A; 1C3X A; 2UZA A; 1QF9 A; 1M5U A; 1VG1 A; 2DLC X; 1P33 A; 4HN4 B; 3C8Z A; 2R7K A; 4Q82 A; 2O7S A; 4EKN B; 1UPU A; 3O26 A; 3OPX A; 3P0R A; 3PF9 A; 3IV6 A; 3R5J A; 4GEG A; 4NYJ R; 4QGG A; 5EJA A; 1A88 A; 5AJZ A; 1U9C A; 4DCV A; 1JSX A; 3WSV A; 3BRS A; 3WYN A; 1N2C E; 1O8B A; 5ESD A; 3OQM A; 3GH1 A; 2B0C A; 2DY0 A; 3NWP A; 3PE3 A; 1GYQ A; 1H6B A; 3LUC A; 1E4Y A; 1Y3H A; 2AR7 A; 4N9W A; 3HL4 A; 4W6Q A; 3W9U A; 4DLL A; 2EFC B; 2RJN A; 3PR3 A; 4MH0 R; 1YRB A; 3VKH A; 4X1Y B; 1V1M B; 1P0I A; 1U1C A; 3P9C A; 1JTE A; 5L3R A; 1KMM A; 4YJH A; 3WGG A; 3GJ7 A; 3I9J A; 4F7W A; 5J63 A; 5DW3 A; 1W87 A; 1W5B A; 3ZE7 A; 5EMJ A; 4D43 A; 4ND8 A; 1FJX A; 1LJL A; 2PK3 A; 2Q2W A; 1CY0 A; 2OOS A; 3MJR A; 3QYA A; 1B4D A; 2FPX A; 1UBT S; 2OPB A; 2P35 A; 3PDJ A; 4NE7 A; 4TWA A; 1DU4 A; 1LS1 A; 3OIG A; 5JSK A; 4E07 A; 4G1E B; 4QA9 A; 4MVJ A; 4HZG A; 5JD3 A; 9LDT A; 5J4K A; 1H5N A; 1ZH6 A; 2Y1F A; 1ZM2 A; 4HN9 A; 1QM5 A; 3KT0 A; 3I5Y A; 5GP1 A; 4OVK A; 1TZB A; 3EGE A; 4FQ5 A; 2IW3 A; 2DFL A; 1OLU B; 2B9V A; 4AWA A; 1CUD A; 3BM5 A; 3WME A; 2OCD A; 2VHX A; 4MQM A; 3T3E A; 2G80 A; 3ANM A; 1AKN A; 2ZXE A; 5F7O A; 5HZG A; 4PAM A; 4AUR A; 4EUN A; 2ZJ5 A; 2ZBF A; 3PL1 A; 5JG7 A; 1DJM A; 1UKC A; 2OTC A; 3E9Y A; 3GLQ A; 4ENQ A; 3L1H A; 3HA7 A; 4QUK A; 4GI5 A; 1TQ4 A; 4BQO A; 5AM2 A; 2GZA A; 1WIW A; 3IAX A; 4RPG A; 4ERX A; 1KK1 A; 4AAY A; 4QKQ A; 1Q1N A; 3UK0 A; 3H7A A; 2IQ5 A; 1UBC A; 3QMM A; 5H3H A; 1SD2 A; 2CFC A; 2QRL A; 3AOE C; 3WOH A; 4JGB A; 3VYT B; 4JXX A; 1RJ9 A; 3CRW 1; 1UBF A; 4WZZ A; 1D4Z A; 4JP5 A; 1XKT A; 4N6O A; 4BAT A; 1IUK A; 1Z7G A; 1WA5 A; 3JAR B; 4LJ6 A; 3B9Q A; 4BQN A; 3EMD A; 1B6T A; 5IKM A; 1Z47 A; 2CV0 A; 3THO A; 3ZGV A; 621P A; 1TJY A; 3KFL A; 2GCI A; 5AJW A; 1JZS A; 4XUD A; 3WVN A; 2J46 A; 1CUV A; 2YR0 A; 4Q7E A; 1Q15 A; 1V9L A; 3UUG A; 1HMY A; 3KSM A; 3ZU4 A; 4DLU A; 4AN0 A; 3F6C A; 4GAE A; 2GT1 A; 3CUY A; 4BV9 A; 1MJG M; 2V54 A; 1F8Y A; 3HKE A; 1MTZ A; 1MX1 A; 3A8T A; 3C85 A; 3D3K A; 4WN9 A; 4C22 A; 5FBW B; 1RRF A; 2WHE A; 3VNQ A; 1PTJ A; 2JCY A; 4KTT E; 1IAT A; 1VHJ A; 2D74 A; 2HJ9 A; 3SHF A; 4KR7 A; 1BXG A; 3L41 A; 4RTP A; 3U4Q B; 2Z2K A; 4FR0 A; 3CB4 A; 1GDD A; 3IUS A; 4IIY A; 2ZYU X; 1DQZ A; 4UVI A; 1DC5 A; 1EAM A; 2C2B A; 3QEM A; 2BHL A; 4PYO A; 1QQ5 A; 5TQJ A; 3T1Q A; 3IR7 A; 3PBK A; 4KUH A; 1PVO A; 1GQF A; 2BV3 A; 1VI5 A; 2CXU A; 1VME A; 4KSB A; 3ZTW A; 4GPD 1; 3TEC E; 4XR4 A; 4BKU A; 4U0K A; 5GGX A; 4E5S A; 3JAS B; 2W2P A; 3IOE A; 2ACE A; 3WK7 A; 4EHD A; 3FWH A; 1UP4 A; 3OZC A; 3OCY A; 4XWA A; 3MT0 A; 2VTK A; 2A5D A; 4MA4 A; 1U2G C; 1QTR A; 1UMD A; 2HFK A; 3NQ4 1; 1EX7 A; 1SU4 A; 1SQ0 A; 3QRW A; 1JED A; 3A7D A; 4AW9 A; 3QLJ A; 3SVZ A; 1QHY A; 3AR5 A; 3HE3 A; 3P4Y A; 4PC3 A; 4RHC A; 4ZK2 A; 4MYC A; 4YUA A; 5J4L A; 2O8C A; 4UM5 A; 2ZYG A; 1Z5V A; 2JI7 A; 2VPP A; 1WXG A; 2RGD A; 4TPK A; 1ZIP A; 4XD7 A; 1WEK A; 3KAK A; 2QN8 A; 3BGW A; 4AP9 A; 1I2M A; 2PYZ A; 1AS3 A; 3TVY A; 2ZRM A; 5LQU A; 1SXP A; 5TJ9 A; 3G9Q A; 1YVE I; 4PO3 X; 3BE5 A; 1IBR A; 4WFK A; 3I62 A; 1PI3 A; 2EZU A; 4ZYU A; 1KR5 A; 1II8 A; 1E3T A; 5IDW A; 3U60 B; 2W2K B; 3HA5 A; 1JHR A; 1KA9 H; 1SWW A; 1VPV A; 1SY7 A; 2VP0 A; 2W4M A; 3PC3 A; 3QF4 A; 1MHT A; 1R37 A; 2PEY A; 2VI5 A; 2C3O A; 5L9A A; 1YVP A; 3QH4 A; 1ES9 A; 2NZ8 A; 4E09 A; 4JBH A; 4E2F A; 5BYK A; 3RBA A; 3UC9 A; 1CUE A; 4PS0 A; 1EEX B; 4G25 A; 1U9J A; 5JOQ A; 2RGW A; 1C7Z A; 5LDH A; 1GFW A; 3I8O A; 2C2Y A; 3I9F A; 1P5J A; 1PR2 A; 3LII A; 3SBO A; 4XEU A; 1R1D A; 4M53 A; 5L6L B; 5F2N A; 3TAU A; 3CTP A; 1QQ6 A; 3KKZ A; 2UKD A; 3KK0 A; 4AMV A; 3IKT A; 4WZB A; 2PYD A; 1RXS A; 2WTB A; 5TC4 A; 4QOI A; 2HNK A; 3F7M A; 4E5K A; 2PFS A; 3MJD A; 1DWU A; 1DOA A; 1PGN A; 1TA0 A; 1XVU A; 4O6V A; 2CFI A; 3CF6 R; 2I71 A; 4GX1 A; 3JAW B; 2FDC A; 3H5L A; 2IV7 A; 3I5U A; 4P2G A; 4KMO A; 4Q37 A; 3U49 A; 5AVV A; 4LTN A; 3FDZ B; 2JHC A; 2Y71 A; 1U8Z A; 2XI4 A; 1VMA A; 4CHT A; 5THK A; 3L06 A; 2E5F A; 2ZR0 A; 3DON A; 3ASZ A; 4D79 A; 3Q2I A; 3QXX A; 5IJZ A; 3QNW A; 2P4Q A; 1T3G A; 1U3C A; 2I66 A; 4LYA A; 5J23 A; 5F9H A; 1OKK D; 4HS9 A; 1UBU S; 1UPB A; 3G0B A; 4N6G A; 1J8V A; 4IO0 A; 4BZZ A; 1WPX A; 3VU9 A; 1JT2 A; 2BOV A; 5GON A; 1YCF A; 4GMD A; 1TQ6 A; 2DPH A; 3HZH A; 3MMZ A; 4Q1B A; 5T8K C; 2JGD B; 1M5R A; 1O97 D; 4D8W A; 4K05 A; 1WHT A; 2YWF A; 2HYI C; 4FDV A; 3LHD A; 2UZI R; 3RAS A; 1SWV A; 1VMK A; 4N82 A; 2BUA A; 4HXY A; 4UE6 A; 3T54 A; 4AL4 A; 3F5S A; 4G09 A; 1ISM A; 3H1T A; 4ILE A; 2XYH A; 4RLF A; 1DC4 A; 3NP7 A; 4NH4 A; 4PFS A; 3PFG A; 5TNX A; 4DRE A; 1L4F A; 1MA0 A; 5K8L A; 3SS8 A; 5F4Z A; 4BV2 A; 1GZ7 A; 4BGI A; 4JC4 A; 4LOH A; 1QZT A; 2XW6 A; 4QNM A; 1J39 A; 4BMN A; 1OFG A; 2DB3 A; 4D6X A; 5JNW A; 3TMA A; 4CG1 A; 1H8E A; 1CRZ A; 4TKV B; 3SZM A; 3RE1 A; 1PWX A; 3BDV A; 3B3J A; 3NRC A; 1CUC A; 1SQF A; 3CZ4 A; 4ENV A; 2G7Z A; 2XJ4 A; 1AZT A; 3ERC A; 1E7W A; 2DKO A; 3PGV A; 3TTU A; 4BQS A; 4IJ5 A; 2BGW A; 1E62 A; 3EVA A; 4D41 A; 3GRF A; 1T9A A; 1LLU A; 3LO5 A; 2E21 A; 3DE0 X; 4I50 B; 1BXK A; 2HBY A; 2H1R A; 2X1H A; 1E99 A; 2Z1N A; 3NT5 A; 1HC7 A; 1DP4 A; 3PEU A; 1MX3 A; 5CYF A; 4ASU A; 5JFG A; 3VX3 A; 4CCW A; 3EAS A; 4N3N A; 1KDV A; 3Q2O A; 1NQV A; 3Q7I A; 5DNE A; 2O14 A; 3ROT A; 1YBV A; 4PBR A; 1PL6 A; 1VIV A; 2I5P O; 5DNU A; 1A8H A; 1UMB B; 3L6U A; 2YV1 A; 1HG1 A; 3SLR A; 9LDB A; 1YKS A; 1H4T A; 3V8Q A; 4WKB A; 4UVE A; 1AKE A; 1AUR A; 1UBL S; 2WF5 A; 5ARD A; 3GO4 A; 3M4J A; 3O8N A; 4MRS A; 4CV1 A; 5LZ9 A; 5JCP A; 1EES A; 3NHM A; 1Y8Q B; 3CZP A; 4G9G A; 4GQI A; 5IYZ A; 4B3S B; 2FPL A; 2QRZ A; 4H0N A; 1VI2 A; 3S81 A; 4G1C A; 1JFF A; 3KHS A; 1ZKM A; 2VP5 A; 3KXK A; 2V7Q D; 5F1O A; 1SUL A; 1VMD A; 3NYR A; 3NUZ A; 3FOZ A; 3DBO B; 4US0 R; 5DHF A; 2FHS A; 1S3G A; 3BGO S; 4TXM A; 2ZYA A; 5DEI A; 4R8S A; 4J3N A; 4BIM A; 2K6G A; 3U3J A; 1XVL A; 3MRT A; 4FU0 A; 1KYQ A; 1NVE A; 4V0N A; 3M0E A; 5AJU A; 2IOH A; 3ZXU B; 5DTJ A; 1Z8D A; 4ZOL B; 2EHD A; 1JDN A; 1X87 A; 2DQS A; 3VO9 A; 4UE2 A; 1KAQ A; 3IQI X; 1M0T A; 5ELL A; 4CMC A; 3D8R A; 2O1X A; 2BMD A; 4PZ0 A; 2EP5 A; 3ITD A; 3KSZ O; 3PUZ A; 4C3Z A; 3DMN A; 1NI5 A; 1V35 A; 4BLQ A; 2ZUB A; 1LZK A; 5JBI A; 3KFX A; 3TUI C; 1J2R A; 1FUK A; 1N1G A; 1NON A; 1SUB A; 4TRN A; 3ORH A; 1V96 A; 3C6Z A; 1Z39 A; 4HNS A; 4TTT S; 4XVR A; 5EAI A; 4CME A; 1VI6 A; 3D6U A; 3GT7 A; 1HYH A; 2BC9 A; 3TNL A; 1Z4P X; 4N3A A; 3EMB A; 4BO6 A; 2P6P A; 1E58 A; 2XYQ A; 4FHF A; 3PD1 A; 4YBR A; 1H5U A; 3ETD A; 3B1J A; 2X1V A; 3I52 A; 1QFZ A; 1SC3 A; 4FOP A; 4A2Q A; 4NBV A; 1O94 A; 2E1R A; 2VNS A; 4OA5 A; 4KAX A; 4ZPJ A; 4QAJ A; 2WJ6 A; 1GGJ A; 1PYD A; 1UOD A; 2XE6 A; 2PH5 A; 2EEZ A; 3FYU A; 4V0O A; 5JAR A; 1DTW B; 4BV3 A; 3QXG A; 4WX2 B; 2NP6 A; 3ETL A; 5EER A; 3KQH A; 5FWX B; 2Q3B A; 3B9P A; 3LLM A; 4OI2 A; 4G5Q A; 4TWB A; 4A36 A; 1I4O A; 4DG8 A; 4TLL B; 1RBM A; 1Y1T A; 3TWO A; 2BR3 A; 4ALL A; 3BOS A; 5H7X A; 2PPL A; 4ZT6 A; 4DLR A; 3DTT A; 3QUQ A; 1Q44 A; 4JWT A; 4NFS A; 1FFA A; 2HJH A; 1NIP A; 4P4G A; 3RIG A; 5FI3 A; 1N3I A; 1R5O A; 3SM9 A; 3E3N A; 1S2L A; 4GYK A; 1FX1 A; 4HJR A; 4HX9 A; 2OY0 A; 3KT8 A; 3NND A; 4YRL A; 2CE7 A; 4XX1 A; 5DX1 A; 1UM8 A; 5FR2 A; 3UW1 A; 2YRX A; 4DYD A; 4CVN A; 3RF6 A; 2H7C A; 4HAD A; 4BAG A; 1YL6 A; 4GXT A; 3HDH A; 3O0D A; 1M8F A; 1T5B A; 1RTK A; 3C3W A; 3FTC A; 1BS2 A; 3GPI A; 3M7I A; 1SCQ A; 4MRM B; 4UHF A; 5EA2 A; 4ALN A; 4R7T A; 1LLF A; 1P6X A; 2FVY A; 3OCX A; 4D9J A; 5BT8 A; 3VZC A; 1T9Z A; 2WIB A; 3EXI A; 3GQV A; 4ZZ1 A; 5I7W A; 5JS6 A; 3DLH A; 3JQF A; 4D0R A; 1ZI8 A; 2GVD C; 5JZB A; 2QQ5 A; 2YXB A; 2JVI A; 1R4M B; 1XMQ B; 2CHV A; 2W2K A; 2O4U X; 3MIZ A; 3CF0 A; 5EWJ B; 3H8W A; 1DTS A; 1VL1 A; 3PPV A; 3L6C A; 3SZG A; 3WNC A; 4M1Z A; 1VKH A; 3AHH A; 2ZG6 A; 5JSH A; 2WTM A; 3WIC A; 3TL2 A; 5I45 A; 3DH0 A; 1G5H A; 1L7N A; 3N3A C; 4X3P A; 3U31 A; 2B69 A; 4UU8 A; 3UOG A; 2HF2 A; 1PV4 A; 3JQB C; 1P29 A; 3PJE A; 5BN4 A; 1WV1 A; 2RHH A; 3Q7Q A; 1M34 E; 1FS4 A; 4P80 A; 4KEA A; 2OFX A; 2NSH A; 1X9H A; 3MRV A; 4EVQ A; 2O2Z A; 1DHR A; 1NPY A; 3GXO A; 1U8U A; 1SVO A; 5ED4 A; 1GCG A; 3LCC A; 2C59 A; 1RWO A; 2H2F A; 3DUW A; 4L57 A; 5F5L A; 3UUW A; 3WLE A; 1PQU A; 2UVP A; 4HPJ B; 1X7Y B; 4NJM B; 3U7E B; 4BNJ A; 1XL0 A; 1J97 A; 3PXA A; 1RYD A; 1EWK A; 4R9N A; 1FRF S; 1AQN A; 1QS0 B; 4JR2 A; 2XWP A; 3RYI A; 1KTI A; 2ANB A; 3WR5 A; 1Z6P A; 3I8X A; 3LTN C; 1ULT A; 4M9Q A; 5I01 A; 3OM8 A; 3ER8 A; 2E7Z A; 1WX1 A; 4JBX A; 4F8Y A; 3H4T A; 5F23 A; 3GLI E; 3EGW A; 4K28 A; 4YA6 A; 1UM9 B; 2DUM A; 2JAX A; 3N0X A; 3FXA A; 5BOR A; 3S7Z A; 4X95 A; 2G04 A; 4ERN A; 1XRS B; 3LCR A; 4YWJ A; 2C88 A; 4BNU A; 3RO7 A; 4BZX A; 2JI9 A; 5TF0 A; 1HR0 B; 1KO8 A; 3QMV A; 5TAR A; 2O8B A; 1KC1 A; 2FU5 C; 2OCX A; 2VK2 A; 1PU2 A; 1IWO A; 2H07 A; 1P1B A; 2BF7 A; 2XCU A; 2YVA A; 2ZCU A; 1QP0 A; 2B7L A; 3OZD A; 1JB9 A; 1WTA A; 3OJO A; 2YRF A; 2IPL A; 3MUN A; 3RFT A; 5KC8 A; 4K6M A; 1NVU Q; 4HB2 A; 1XZ8 A; 2X6T A; 4CMB A; 5FUL A; 1PK7 A; 4TKU A; 1AQY A; 1GGE A; 1A8I A; 1AUV A; 1DAP A; 2BGR A; 2EKB A; 4DZR A; 2VXY A; 3T5D A; 1BH2 A; 2YXP X; 4YRG A; 1DJQ A; 3O8Q A; 3HKB A; 3PGM A; 2SNI E; 1ISS A; 4M9S A; 1T0B A; 1V4N A; 1L4U A; 3HV0 A; 4MAM A; 2XUA A; 5EFZ A; 3JWJ A; 1A2B A; 1B1C A; 4WQM A; 2HAE A; 4JUF A; 4IIJ A; 3DLB A; 3SBX A; 3HAC A; 4D3Y A; 1V41 E; 2RG7 A; 3U61 B; 4OXR A; 3FOW A; 4RII A; 1E4E B; 1XD9 A; 3LF2 A; 5G3Y A; 3DB1 A; 2IK8 A; 2PLF A; 1HM5 A; 3SC6 A; 4N7R A; 1E2J A; 1K9S A; 1Y33 E; 2A9P A; 2VHW A; 3CKM A; 4KQC A; 4OJQ A; 2YMQ A; 2PRS A; 4BRZ A; 4C20 A; 4DQX A; 5HYV A; 4LJK A; 3WZL A; 4DAR A; 4MW9 A; 4K9O A; 3W3A D; 2AR9 A; 4GS5 A; 1ZON A; 3NYW A; 4X84 A; 4CZ2 A; 5EJ6 A; 1T26 A; 3E70 C; 2W6M A; 2RGC A; 1WMD A; 1XLW A; 2X0J A; 1VC2 A; 1W75 A; 1RAH A; 1AUQ A; 2HMU A; 2ZBD A; 4O2B B; 4UOQ A; 1IIR A; 1P4G A; 2R8Y A; 5IMB A; 2WDZ A; 1U7Z A; 4S12 A; 3TLJ A; 4Z2A A; 2QSU A; 1ZZE A; 2X2E A; 3OBB A; 4IDG A; 4WNA B; 5DIZ A; 3RYH B; 2HDW A; 4CFY A; 5FTJ A; 4POO A; 1JIX A; 3PSA A; 5AR1 A; 4EYG A; 4ZOQ I; 1A7A A; 1UMB A; 1UJ6 A; 5IM4 A; 1E98 A; 3AVY A; 2AJL I; 3LEC A; 1FZT A; 1ACL A; 5ALT A; 4C7O A; 3WND A; 4FE7 A; 2FFH A; 1Y9E A; 2Z1M A; 1Y1L A; 4BQQ A; 4K1L A; 3KXL A; 2V4U A; 3WB0 A; 3SZL A; 5LC1 A; 4FIL A; 2IDM A; 3ZD8 A; 1RWX A; 1T9H A; 3EVB A; 3BZW A; 4U6Q A; 3U80 A; 2OIL A; 3USC S; 2C5L A; 3DLI A; 4GMK A; 3E15 A; 1OV6 A; 1XZJ A; 2VDK B; 1Z6A A; 1DXY A; 2NSD A; 3DWF A; 4LRW A; 4U2E A; 3IQZ A; 4JJ8 A; 1O5I A; 3UAZ A; 3A26 A; 3GYB A; 4GP6 A; 2EG9 A; 2PDA A; 1KEZ A; 3G1W A; 2C2M A; 4BQF A; 1DKM A; 2OO7 A; 3N7A A; 4OUK X; 4B9A A; 3UGJ A; 1QO0 A; 4D8V A; 1JBQ A; 4G37 A; 5ILQ A; 1F2T B; 3PNP A; 2WFM A; 5M7E A; 4L9S A; 4YDH B; 2YN0 A; 1DPG A; 2O0L A; 3HBA A; 5HF5 A; 4WCF B; 1G3I A; 5C8Y B; 3TI9 A; 1XZI A; 4KLZ A; 1M1Y E; 1K6E A; 4MKG A; 2JDI A; 2UVQ A; 2YKG A; 1SC9 A; 1ZYU A; 3K0K A; 1PT5 A; 2ZIE A; 4OGL A; 1XMX A; 2ROQ A; 3VU0 A; 4M46 A; 1FFB A; 4DB4 A; 3OF2 A; 3SC2 A; 2PMD A; 2NTJ A; 4KH0 A; 2R96 A; 3L7K A; 5C1P A; 3DZK A; 2ADO A; 3X1Y A; 2ZUE A; 1ZCF A; 2EBU A; 4QZV A; 2GGS A; 3UMV A; 2Y76 A; 4DC0 A; 2XB4 A; 1DAR A; 4EFD A; 4LXQ A; 4PC2 A; 1LRJ A; 1QC5 A; 2EW8 A; 2OK8 A; 3AFO A; 4HGP A; 4PXS A; 1J7J A; 4K86 A; 2B4Y A; 3P2M A; 4YHB A; 4DBZ A; 2FZV A; 1AHH A; 3HAB A; 4XLT A; 5B4V A; 4FQ8 A; 3MGK A; 4J75 A; 4ANF A; 1F7U A; 1V8Z A; 1IBL B; 2NGR A; 3ESZ A; 2ATC A; 2EW2 A; 3LA6 A; 1ND6 A; 3NR2 A; 2E3J A; 4GYW A; 4NBT A; 3L05 A; 5H8W A; 2FKA A; 1DAD A; 2WMY A; 4HQ0 A; 1H05 A; 3QHQ A; 3TGW A; 2HV9 A; 3RVM A; 4X0E A; 4B32 A; 5LQD A; 1OFH A; 2Z9C A; 3NM5 A; 2BI7 A; 3OKA A; 2PV7 A; 3VTF A; 4KPE C; 2JEZ A; 1A98 A; 4TGL A; 1SBT A; 2FN3 A; 4LNH A; 4CG3 A; 4OQZ A; 3B5I A; 2IS2 A; 3FY4 A; 4G0Q A; 1TW2 A; 1P6U A; 2GF9 A; 3DOI A; 3AO0 B; 5AOV A; 4RJT A; 2GVZ C; 3GRY A; 3EC2 A; 4ACB A; 4D44 A; 3PD7 A; 3FMI A; 5HQT A; 3AYZ B; 5JAL A; 4KRF A; 3H17 A; 3OE4 A; 2AG1 A; 2RAP A; 1DJO A; 2DBV O; 2J5X A; 4GWK A; 4WGF A; 4WRR A; 4BLS A; 5ALG A; 2WC1 A; 1NL3 A; 2WG0 A; 2RI1 A; 4N3B A; 3T4E A; 3WLS A; 5FAK A; 5I51 A; 1AK1 A; 4KDF A; 2PUH A; 4E6E A; 3P2E A; 5BT9 A; 1EZZ A; 1KEU A; 4DXD A; 1NP6 A; 1B93 A; 1MGO A; 2NUB A; 3FIU A; 2Q5I A; 1JT9 A; 2DPM A; 2E87 A; 1GTJ 1; 1K4K A; 4DLX A; 4H3V A; 3JYN A; 1SQS A; 3TSC A; 4XGK A; 1E1N A; 4GRA A; 5LOV F; 4EGC B; 4X20 B; 4J4H A; 1Y9A A; 4J1T A; 2CXS A; 5L7T A; 5JBQ A; 3H5I A; 1TKC A; 1R0Y A; 2WYO A; 4LCJ A; 2O0J A; 1U0O C; 2VS1 A; 4X45 A; 4AMY A; 4MWD A; 4C0W A; 3EJW A; 1K7X B; 3L7Y A; 1WXI A; 5J47 A; 3V48 A; 4MUY A; 3KLN A; 3OKF A; 3OJX A; 1E1L A; 3FJO A; 3QEA Z; 1UF9 A; 5B36 A; 3JD3 A; 2C9Y A; 3SFV A; 4EAB A; 4I1U A; 5TS3 A; 3JQR A; 2ZRA A; 4FS3 A; 3LXM A; 3OX4 A; 3FSG A; 1HYB A; 2JW5 A; 4F2W A; 3K94 A; 3JVV A; 1JEA A; 3UUF A; 3V4F A; 2WWF A; 3ZVL A; 1CYD A; 1BRT A; 1QH1 A; 1J42 A; 2M32 A; 1RR6 A; 3LTE A; 3AVW A; 1YDF A; 4G4S O; 5BN5 A; 1WSW A; 1L9X A; 4HXE B; 3K9F C; 1LBS A; 2OBN A; 3L40 A; 4P3N A; 5JNV A; 2AFH E; 5K3D A; 4B4O A; 3G9X A; 1SC1 A; 2GAF A; 3VNR A; 3ZD9 A; 1TUE A; 3PNS A; 4US2 R; 2E55 A; 2YXN A; 2OD2 A; 4RF4 A; 4L50 A; 2VDM B; 4EUS A; 2P78 A; 4IXW A; 2KLH A; 2GM4 A; 4F8E A; 3PEF A; 2WTU A; 4PNP A; 3M1L A; 4XQC A; 2W2O A; 1DVR A; 1ZD3 A; 2B4T O; 1IG3 A; 4CFZ A; 5T3U A; 3RUE A; 2JIP A; 3DBV O; 2VQF B; 5FR1 A; 3FOF C; 1EXW A; 4WXH A; 3AT1 A; 1FB5 A; 4C1B A; 2IZO A; 1KZY C; 4PSK A; 1LS6 A; 1EWR A; 5EP1 A; 2C4T A; 4ZCT A; 4F46 A; 1ECB A; 2OSB A; 4H4E A; 2CJY A; 2P2L A; 4MS1 B; 3TQH A; 3TTV A; 2FET A; 3Q3I A; 3V8N A; 2XOE A; 1LI5 A; 1AO0 A; 5FCD A; 1A5Z A; 4JCC A; 1RWB A; 3RV4 A; 4MAF A; 4ZCL A; 1T0P A; 3CHY A; 1C50 A; 5EGN A; 3RVK A; 2WJP A; 2B5V A; 2FWJ A; 1Q95 A; 4G1B A; 4X5O A; 1KYZ A; 3MMX A; 4GVL A; 1H2Y A; 1Y5R A; 1OGY A; 2H06 A; 4Z3O A; 5IZC A; 1KRX A; 4MUN A; 3LMK A; 5KCZ A; 2HMW A; 4ESV A; 1LY1 A; 4DV4 B; 1K5D A; 4P8R A; 5A71 A; 4LF4 B; 3KQN A; 2JJ2 D; 4RA6 A; 4FXP A; 2VRB A; 3T7E A; 5EXT A; 4PWZ A; 1S02 A; 4QEZ A; 1HUR A; 1UQR A; 4QPC A; 4YBZ A; 3BXF A; 3VC3 A; 4EG1 A; 1S0U A; 3VZP A; 1JAL A; 2H92 A; 3KOU A; 5BMV A; 3KR1 A; 2R1T A; 4OQE A; 4HDM B; 1T4M A; 5K8V A; 1ZA1 A; 2CXR A; 5KYX A; 1HNW B; 1U99 A; 5CEE A; 1ZD8 A; 2J1L A; 1POX A; 2O7V A; 2X9V A; 1ZU4 A; 4FGU A; 4L9A A; 3R9K A; 4I3F A; 1X15 A; 1IEQ A; 5KYY B; 3JVI A; 4QU8 A; 2ZAO A; 3L18 A; 3ITN A; 4AKE A; 1WBD A; 3B38 A; 2ZUD A; 4R1M A; 5DBI A; 5EXP A; 3TAX A; 1N9G A; 3NID B; 5AVQ A; 2VDJ A; 4FFN A; 5K0T A; 4LSM A; 3LGS A; 3RJZ A; 2FMX A; 4MYU A; 4DIE A; 5ED0 A; 3WLI A; 3FR7 A; 2PHZ A; 2C9O A; 2FP4 B; 3LUP A; 3SSN A; 2D1T A; 3AHF A; 3D8T A; 3BMN B; 3DAF A; 3JQ9 C; 4JB2 A; 4XAR A; 1AUP A; 1V7Z A; 2VNJ A; 2HHC A; 4DQK A; 3RS8 A; 4LJ7 A; 3OLW A; 4BQP A; 3PU6 A; 1Z4I A; 1MEJ A; 4C4S A; 2YV5 A; 4J1N A; 3K9G A; 1E2M A; 1PZF A; 1I2C A; 3UA4 A; 4WCV A; 4KR0 A; 4TVS A; 5K6A A; 4FNA A; 5JWH A; 3MCA A; 3OOW A; 4PV7 A; 2NZW A; 1ORW A; 1H2X A; 1WU7 A; 2GNO A; 4JP2 A; 1C0K A; 4HYQ A; 4PZ9 A; 4XB2 A; 4A0S A; 5BW4 A; 8ADH A; 5HOQ A; 4W5Q A; 2QL5 A; 2XND D; 4GOO D; 2AOU A; 1C81 A; 2HWY A; 3PC8 C; 4LQ4 A; 4V0R A; 3W8D A; 2PLN A; 1PCX A; 4IHJ B; 1FYW A; 4RJJ A; 4F5Y A; 3HYS A; 2GLX A; 1F2T A; 5LPN A; 1F17 A; 3NXS A; 5AO9 A; 1G5P A; 5BNF A; 4PIP A; 4R3U C; 4OFZ A; 2FF5 A; 2UYG A; 4FDA A; 4ZGW A; 3FAZ A; 4Q66 C; 2EKD A; 3VDM A; 4YAS A; 3BK7 A; 4NE9 A; 2XLB A; 3UMR A; 2GET A; 2F55 A; 3FDF A; 2HPA A; 1WMB A; 2OC9 A; 2PS3 A; 3HR8 A; 2XOX A; 4A23 A; 2HLD D; 4C7M A; 4CWA A; 3OE1 A; 5J5A A; 3UGK A; 1Q0T A; 3D7M A; 5KU7 A; 1ZW1 A; 1KHH A; 2R5N A; 4GZ6 A; 1RLU A; 3FYC A; 4HWO A; 4L3O A; 1US8 A; 4H1H A; 1P2S A; 5LUB A; 2IW1 A; 3DVQ X; 1I7D A; 1TMK A; 1Z5N A; 2OXF A; 4C0J A; 4INP A; 1XDD A; 2E4A A; 2F6R A; 2OBX A; 2WKQ A; 2CFR A; 2WWR A; 3D5T A; 4FHC A; 4Q93 A; 4W8O A; 4AT1 A; 4BVH A; 1BVY F; 1E4O A; 2VTD A; 3EPR A; 1I5O A; 2FMT A; 5CPB A; 1LSJ A; 2XED A; 2ZT8 A; 1ISF A; 3R5H A; 3WIS A; 3KKK A; 4LBZ A; 4CR8 A; 2HWW A; 1H1L B; 1AN9 A; 2R8Z A; 2WB4 A; 2PCJ A; 2XUD A; 5JZS A; 2W6O A; 1WXQ A; 3L7M A; 1K68 A; 1FQJ A; 5CTJ A; 3RYC A; 4RSG A; 1E0D A; 3M3H A; 4S1W A; 2QTC A; 4J16 C; 3CEA A; 3H86 A; 5I9N A; 3T7I A; 5DYG A; 3B6K A; 2FQX A; 4I1Y A; 4N03 A; 5HDQ A; 3GA0 A; 3TBK A; 4JI0 B; 1W5E A; 3OND A; 3HGQ A; 3KVR A; 4Y7C A; 5AW1 A; 3LD9 A; 1D6N A; 1CTQ A; 1TND A; 2PMF A; 2FZL A; 3OZS A; 4O33 A; 5LLS A; 4ZI2 A; 4J7A A; 4RW0 A; 2F1R A; 3ETG A; 5HF6 A; 1X8J A; 1ETU A; 1IAQ A; 4D6P A; 4MW2 A; 2ZAN A; 4GE0 A; 4OMC A; 2AFI E; 2PRZ A; 1FZQ A; 5FNV F; 2FZC A; 3VU3 A; 1R16 A; 1R6T A; 4TTQ A; 3UAG A; 4A3S A; 1XRN A; 1CZN A; 2GAJ A; 1E2F A; 1R5J A; 2FX3 A; 2IF0 A; 2Q3D A; 3MD9 A; 2VT6 A; 4E2X A; 2CXH A; 3A9T A; 5DZS A; 3VPA A; 1GTS A; 3AIN A; 1CUX A; 4LF9 B; 4QOD A; 4HWR A; 1L4K A; 2QZS A; 1CJS A; 5TXG A; 1N5M A; 2EJB A; 4G41 A; 5IBC A; 2IVZ A; 3TQI A; 1QYX A; 1QE0 A; 1YB2 A; 4CLR A; 4OK6 A; 1FBT A; 2XGJ A; 3JQC A; 1XQW A; 2JIR A; 1EHC A; 5LQA A; 4S3M A; 1E7M A; 1N2X A; 4YJK A; 1E1K A; 1H8H A; 3CA8 A; 3N58 A; 1J23 A; 4OSE A; 4RDH A; 4IIC A; 1Z68 A; 4HEH A; 4BJ3 A; 1ZSV A; 2V55 B; 5JJ5 A; 1CY6 A; 4KX5 A; 3VV2 A; 2BGN A; 5KEI A; 4BVL A; 5FYQ A; 5C7I O; 5C51 B; 2I99 A; 1E92 A; 3AIM A; 3BXH A; 5AVO A; 1NKV A; 4ZDK A; 4GKV A; 2OP0 A; 2V8B A; 2OAP 1; 4H3K B; 2ZB3 A; 1EJB A; 3MN1 A; 4DM7 A; 3LC0 A; 5CT9 A; 2QPL A; 4C21 A; 4RTS A; 3CH6 A; 2UUA B; 5CJ2 A; 4D26 A; 1KMI Y; 3K9N A; 3GUU A; 1K7S N; 2FSI A; 3LQ1 A; 3EIE A; 3M6X A; 1BXR A; 4H60 A; 3OZW A; 5FAX A; 5T4H A; 5JHH B; 3E22 B; 4ZHS A; 4LJR A; 4ZOD A; 1YJX A; 2HF7 A; 3K8Z A; 4WZA A; 4CT4 B; 3ITE A; 1XKL A; 2DTE A; 3JQQ A; 4RJK A; 5F3D A; 1PKX A; 2FX2 A; 1A76 A; 3P2K A; 4K6F A; 3DFM A; 5BSE A; 2RG1 A; 1WB9 A; 2A0F A; 5EY5 B; 2EWM A; 5B2F A; 2JIM A; 3BA1 A; 3B9B A; 4UNQ A; 2XIJ A; 1UH5 A; 2WWW A; 4P84 A; 4CKE B; 2B4R O; 4LJS A; 3TKA A; 4R1U A; 2H1L A; 1P5Z B; 1UXI A; 2C3P A; 4KAA A; 2IA5 A; 4L27 A; 1JA0 A; 3ZWY A; 1G55 A; 4JJ7 A; 5AVZ A; 4MJL A; 1WZU A; 3L1V A; 3ETH A; 3AC0 A; 3Q2K A; 1F6W A; 3GNS A; 1EZ1 A; 1H2R S; 1K6J A; 1MJH A; 3DO8 A; 1E64 A; 1NVM B; 1R6A A; 1UBN A; 2VJP A; 2XVM A; 3FMO B; 3LPC A; 4L6C A; 4OLJ A; 4DXH A; 4Y1R B; 1QR2 A; 4G56 A; 3ZK9 A; 5BVG B; 2NPI A; 4CM9 D; 4OV6 B; 2XX3 A; 5CYP A; 4GKA A; 4FNM A; 3GZT B; 2VJB A; 3N86 A; 3P9I A; 5LZ8 A; 4DIU A; 3OTI A; 4PGH A; 1RZR A; 2QRN A; 1JU3 A; 4N3C A; 1XK7 A; 1W0J D; 2OGT A; 1RAA A; 4E32 A; 5CFL A; 4JJZ A; 1CQI A; 1TF7 A; 1CNE A; 3A3K A; 3D54 D; 2XQG A; 2AJC A; 2V3C C; 2NIP A; 5I7R A; 3I5X A; 2G1K A; 5H16 A; 1YZQ A; 1DWO A; 1M5T A; 2GES A; 4ARA A; 1ZM3 A; 4EYE A; 4ENT A; 1O95 A; 3LT0 A; 2P9F A; 4OP1 A; 4WFJ A; 3AL0 C; 3CE9 A; 4KAF A; 4D97 A; 4RIG A; 4OI0 A; 3KAL A; 2B5O A; 2FPG B; 1XTU A; 5KOH B; 4IIH A; 5GJC A; 3ZE1 B; 3ITK A; 3THY B; 5BV0 A; 2J4D A; 4PRY A; 5IDV A; 4RZH A; 2NO1 A; 2Z04 A; 2CNW A; 4L5B A; 4LXI A; 3IO5 A; 3QNU A; 2UV8 A; 3R20 A; 3NDB B; 3KKL A; 4MVW A; 1ZJ6 A; 2EBF X; 2R65 A; 3QUA A; 1EBF A; 3R3S A; 4M99 A; 3WO1 A; 3FOE C; 3CLR D; 3L8H A; 1ECP A; 4GUA A; 3A6Z A; 4N40 A; 2G9N A; 3EWA A; 1HMP A; 5KYY A; 1NFR A; 1UXO A; 4EUH A; 3D0O A; 4EPT A; 3I1Y A; 3H19 A; 5T5Q A; 4U7W A; 4R2X A; 3RUF A; 4A73 A; 3MTD A; 4J0E A; 1AXR A; 3JYP A; 3G1T A; 4C4Y A; 3UV6 A; 1E3E A; 1PEA A; 4LHZ A; 1JFT A; 4A26 A; 1JS1 X; 4YR7 A; 1KU6 A; 4ZJZ A; 3EFE A; 5LVA A; 4FDT A; 1N32 B; 3ELU A; 4ZT7 A; 3PDC A; 1EIW A; 1E9A A; 1RYE A; 4MKH A; 3IHK A; 2ONJ A; 1T4C A; 4V02 A; 3V33 A; 4FMP A; 2EC5 A; 5I9M A; 2QTG A; 3PXB A; 3RG2 A; 3QU7 A; 1KK0 A; 4QYW A; 5KKC A; 1QIJ A; 1NUQ A; 1DUS A; 3QBE A; 4XFK A; 5LYY A; 1DRW A; 4B4V A; 2LCF A; 4ZYY A; 2F4N A; 3VR6 H; 5TMP A; 1N57 A; 1JSL A; 1QKI A; 2P40 A; 1B3B A; 4L73 A; 4MW4 A; 2VF8 A; 4BFY A; 1BGX T; 4BJ1 A; 3G82 C; 2FE1 A; 1RKB A; 5FB3 A; 4OIM A; 1WPR A; 5JCO C; 1P39 A; 3MS2 A; 5CW2 A; 1PJ8 A; 2GR9 A; 1IZY A; 1NLU A; 3CF4 A; 5EXS A; 2Q28 A; 1D2A A; 2ZSD A; 3BCU A; 5LB2 A; 4UDO A; 4HQK A; 1B0U A; 1G20 B; 2QUB A; 5FFN A; 2PX8 A; 4ZRM A; 1VGQ A; 3GPD G; 3IIJ A; 1ZMO A; 3AVO A; 4GWG A; 3TJR A; 2XXB A; 5U4Q A; 3UJ9 A; 1JR2 A; 1QCD A; 2C4N A; 5CFM A; 4RTR A; 2V3V A; 4LGC A; 1LV8 A; 4K0K B; 1R0X A; 3EZX A; 1FRQ A; 3FNI A; 1FX0 B; 1U02 A; 3NHS A; 2BDE A; 2X58 A; 1OA0 A; 4JA0 A; 2X23 A; 1U98 A; 121P A; 1KJQ A; 3O4I A; 3Q9U A; 3V46 A; 3PIR A; 3PYM A; 1SIB E; 1ZGH A; 1V48 A; 2Z3W A; 2GPB A; 2C9C A; 4DWV A; 4BO3 A; 4YMT A; 4HAX A; 2ZW9 A; 3DE2 X; 1UT6 A; 4MEJ A; 2WKS A; 4JML A; 2IOF K; 3WL7 A; 1TTP B; 1SCI A; 3DNM A; 5JTU A; 2G40 A; 4XEH A; 2BCE A; 3CRR A; 3ED5 A; 5JUW A; 1UZL A; 3RTG A; 3W2F A; 2GPJ A; 2IZZ A; 5AI9 A; 1BDI A; 1O87 A; 1E3I A; 1ONS A; 3A6F A; 3HCF A; 2V45 A; 4MYS A; 4I2V A; 4JVN A; 4Z9F A; 1RMY A; 3WTC A; 2J48 A; 1UBK S; 4JJK A; 3L4B C; 4YTE A; 4KR9 A; 1OGI A; 5H0I A; 1EM8 A; 1KFC B; 1RYB A; 1M2K A; 3VPH A; 3PCT A; 1R4Z A; 2B3T A; 1Z75 A; 4G8C A; 4MW0 A; 1YL5 A; 3AM1 A; 4J03 A; 1VMI A; 3NV9 A; 3QJG A; 3PX3 A; 1HL7 A; 1AA9 A; 1OXX K; 3WGU A; 4E12 A; 5LP1 A; 2QMZ A; 2P62 A; 4IJM A; 3VKG A; 5BSW A; 3KFV A; 3E4D A; 2V6M A; 2C7O A; 4KHP B; 1G8Y A; 1XQ9 A; 4YO7 A; 2EWW A; 1KXZ A; 2HHL A; 1ZID A; 3OCU A; 2FW8 A; 3RS9 A; 1OTH A; 2PSF A; 4CQM A; 1GR1 A; 1G5R A; 1ZXB A; 1NFQ A; 4GI3 A; 4ITV A; 1I6K A; 4RQT A; 1R4N A; 4RZX A; 3JQG A; 2F59 A; 3TUZ C; 1T2F A; 4MHS A; 4JB0 A; 3ROX A; 2VBF A; 1GTG 1; 4HJX A; 3EIU A; 3QVU A; 1IWB B; 1ZRH A; 5TGD A; 3I0M A; 4QDO A; 3WGZ A; 3TTY A; 4O4L F; 4IIB A; 1DKL A; 1IAV A; 4NZ9 A; 2UUO A; 3HH8 A; 2X4D A; 3AF4 A; 3PGA 1; 1PJ4 A; 4J4T A; 2QJW A; 1BCS B; 4CM4 A; 4ZI7 B; 2R2A A; 1EXN A; 3I3S R; 4P33 A; 2KSQ A; 4RJ2 A; 4DLB A; 1HYG A; 2JFH A; 1M54 A; 4ZT4 A; 1I1N A; 3FOB A; 2RH5 A; 3T1N A; 4A0Q A; 3MIN B; 1CUL C; 4B2P A; 1Z7Y A; 4PPY A; 5ILG A; 4DCN A; 3WKC A; 1D2H A; 3SAJ A; 1ULU A; 3ZWA A; 3AG5 A; 3F98 A; 2A3R A; 3F78 A; 3EXH C; 3MT7 A; 5CTA A; 5EHG A; 5IDQ A; 3GVH A; 1KER A; 3PKZ A; 3QOY A; 3U56 A; 2OFF A; 5JO0 A; 2OCY C; 3A1V A; 4RGY A; 3BD9 A; 1PWH A; 2JG3 A; 3QX3 A; 1KFB B; 4C43 A; 3RAN A; 4L76 A; 1Z5P A; 4F5D A; 1C9Y A; 3M5C A; 2X0N A; 3P32 A; 2VHY A; 2BY4 A; 2ACV A; 2DOK A; 5IAG A; 3HC7 A; 4YAK A; 2DUU A; 1LFA A; 4IQ8 A; 5FFF A; 1J5X A; 2BP7 B; 3JD4 A; 3MOI A; 4LQL A; 3ROM A; 1Z15 A; 1O4W A; 3IDQ A; 3KB6 A; 4NVR A; 3BH6 A; 3Q9L A; 1VA5 A; 2YX1 A; 2IRW A; 3UMC A; 3RS7 A; 4QG3 A; 1EGV B; 4G89 A; 4CKE A; 1RLO A; 1Z4N A; 4AZV A; 2WKP A; 2GZH A; 1Z3C A; 1ZHH A; 1N0U A; 1C29 B; 1B8O A; 2P41 A; 3B36 A; 4L9W A; 4QWW A; 4R0V A; 3TLB A; 1XAH A; 4M9Z A; 4OP3 A; 1ANK A; 2VKT A; 2WQZ A; 1TM7 E; 2ZRB A; 4HAW A; 4WEQ A; 2CK3 D; 1ZCZ A; 4QOS A; 3U3O A; 5U4X A; 1F6D A; 4M1E A; 3DMT C; 4FOT A; 4KDE A; 5C1O A; 4CEI A; 4MTC A; 4R9V A; 1MXH A; 2I1L A; 3JRX A; 3NBF A; 2BLN A; 1VE3 A; 5JFT A; 1Q3K A; 3AJA A; 3R40 A; 4Y0N A; 4LG1 A; 5MGB A; 1T0J B; 4YAF A; 1ZUH A; 4IDB A; 4TLM B; 5GJV C; 3EPL A; 3KPJ A; 4Q03 A; 5REQ A; 4BJ6 A; 4CU2 A; 3R3V A; 4FFR A; 3IAE A; 5F52 A; 1B23 P; 2FSU A; 3I9O A; 4ABY A; 5ALD A; 1Z4M A; 5IN5 A; 5KYK A; 1PJQ A; 1E9S A; 5LJL A; 4R40 A; 3ASU A; 3QEJ A; 3GAR A; 3IMC A; 5AW9 A; 2ODW A; 3UWD A; 4MUE A; 3F8W A; 2DER A; 4CPD A; 1XQY A; 4UM8 B; 3PE4 A; 2ORW A; 2WWI A; 3NHZ A; 1SKQ A; 2IE8 A; 521P A; 4P4U A; 4ZHQ B; 1SAY A; 3VOB A; 5DK4 A; 3ZWK A; 1IPF A; 3T9C A; 4BO7 A; 4TQA A; 5FOR A; 1WMA A; 1MDB A; 3D1L A; 5EEG A; 3HVU A; 3WYE A; 4M1S A; 3OWH A; 3EEQ A; 3HXT A; 1C7E A; 3JZM A; 4HZU A; 5B3L A; 3LL7 A; 2RGR A; 4DJJ A; 3NT2 A; 4OOF A; 2GQ0 A; 5EUC A; 4ZZ3 A; 2B99 A; 2OHI A; 4X97 A; 1FE8 A; 1W9M A; 1FYX A; 2I6S A; 4X85 A; 1RWP A; 2ATX A; 3B48 A; 2W8B B; 5ELM A; 4HV4 A; 2QTM A; 4JBG A; 3REQ A; 3T0I A; 5HML A; 4PBS A; 2FOI A; 4F1W A; 5I6D A; 2X77 A; 2RSL A; 2FG7 C; 3DEI A; 3NWO A; 2ZSE A; 1QP7 A; 3D59 A; 4Q1D A; 1OYY A; 1R0W A; 1ZM4 A; 3QEO A; 1L8X A; 2XMD A; 1Z12 A; 3AER A; 3UJP A; 1JSR A; 4QGA A; 3EAR A; 2JGJ A; 3GJY A; 3NVM B; 4M4S A; 1LVU A; 2F3P A; 3NTL A; 2ZBQ A; 4GNK A; 1IY0 A; 2DHD A; 4R1O A; 4CDR A; 3GEL A; 5HZK A; 1ZMB A; 1HYE A; 3BE6 A; 3KYB A; 3S70 A; 4AJI A; 4MMZ B; 1ZCJ A; 3E9D A; 2Q5O A; 1M8P A; 3X2S A; 2QN7 A; 3EEI A; 4MVJ D; 2PNJ A; 4L5C A; 4CF4 A; 4D6Y A; 2OAE A; 2G6B A; 2R8B A; 4QUA A; 2VDW B; 3AES B; 1O08 A; 3NMU F; 1XKQ A; 3MQ2 A; 5BSR A; 1YOV A; 4GLL A; 2Q1Y A; 5JU6 A; 1EUC B; 4IMR A; 2DQZ A; 4REQ B; 1LH0 A; 4EW2 A; 3EFO A; 2RAR A; 5FBV B; 2OM6 A; 4I2N A; 4QQR A; 1AGX A; 1P5H A; 5K3F A; 3BFK A; 4W63 A; 3WKE A; 4KKV A; 3MRX A; 1ZKD A; 5K0N A; 5H70 A; 3R7W B; 3TX2 A; 2VDN B; 1ZK3 A; 3TW6 A; 2JLQ A; 3C6K A; 4KR6 A; 1XD8 A; 1ZLX A; 2FR0 A; 1EFT A; 1W93 A; 3M1Y A; 4U7T B; 4IOL A; 4ZCP A; 4A4D A; 4OQN A; 2HTE A; 1CEE A; 3IP3 A; 3JXE A; 2XT0 A; 5LZ2 A; 4KFT A; 5LOV A; 3L7I A; 2J6H A; 1DTE A; 4DVJ A; 3NHL A; 3AZQ A; 3THR A; 3A1U A; 1ZIT A; 1GZV A; 3M6U A; 2W8Z A; 3NEJ A; 1XFA A; 3EYA A; 4GIC A; 4WES B; 4K6C A; 4BFL A; 4XQ0 A; 3SQX A; 3T3O A; 1IEX A; 1G6O A; 3COS A; 5ESR A; 3RD5 A; 1CZR A; 1DBS A; 4HSG A; 4XQ9 A; 1G63 A; 3CU5 A; 5LBT A; 2EVW X; 5C3U A; 3ZZ0 A; 3FNG A; 2XVZ A; 5KB3 A; 5FDF A; 4D9G A; 1QTQ A; 3R0J A; 3MCT A; 2O1S A; 1GYP A; 4X6U A; 4B80 A; 5BN1 A; 1V26 A; 3QVK A; 4E88 A; 1UFQ A; 3GPB A; 3QXC A; 3F5D A; 2I9K A; 1NME A; 4ID9 A; 1Z0Z A; 3NAL A; 1QGZ A; 1IZ8 A; 1EUZ A; 4RZQ A; 1ZJJ A; 1EE1 A; 3C6Q A; 2FLK A; 3H1G A; 4KQI A; 4LI3 X; 4EBF A; 2W6Q A; 1OMO A; 2YVL A; 5EJF A; 1OY1 A; 2V7U A; 4W6Z A; 5TQ2 B; 3UOA B; 4E14 A; 4EVI A; 3HP1 A; 4ESE A; 4FGZ A; 2LKC A; 2PUA A; 2G09 A; 2QD4 A; 1GRQ A; 3KT6 A; 2XWS A; 4QRI A; 2IZ7 A; 3PFC A; 1L4G A; 3S44 A; 4OKI A; 2P27 A; 1WQ3 A; 4WAD A; 3RS3 A; 1Z22 A; 1YC2 A; 5L95 A; 3LZ6 A; 3JR3 A; 4KXX A; 1FP2 A; 2EFE B; 3N6V A; 4RPL A; 2DWC A; 1RKX A; 4CLE A; 4I9H A; 4NXM B; 4RFV A; 4ZYL A; 3FBG A; 4FYV A; 3N0R A; 1WCV 1; 1X2E A; 3NVD A; 4YMI A; 4EAQ A; 4EG6 A; 2O8F B; 2PBR A; 5EZQ A; 4IFT A; 1N6N A; 3L5K A; 1XRQ A; 1LPN A; 3UM9 A; 2HL6 A; 2G5G X; 1O03 A; 5JVD B; 2KLB A; 1V5F A; 1XJ0 A; 3HB0 A; 4QD3 A; 2YVQ A; 3P1J A; 1U3F A; 2IOQ A; 1CET A; 1VYT A; 1UZA A; 2MMG A; 3I01 A; 2XUG A; 4RH0 A; 3UZU A; 2ET6 A; 4D9K A; 3V1M A; 5TQV A; 1DI0 A; 5GIO E; 4YWH A; 1VIA A; 4F5W A; 4JLU A; 2AZM A; 3FD8 A; 5EC8 A; 3WIO A; 3IVC A; 3M9X A; 3QLT A; 4ZI7 A; 1PNT A; 3OT1 B; 2EGH A; 3A3O A; 4MQE B; 4QDX A; 1N2B A; 4ZV9 A; 1ULZ A; 2PSH A; 5ITS A; 2DRI A; 4FLF A; 1YHY A; 4F2G A; 3WWC A; 5EHN A; 2CLH B; 2HJS A; 1QHX A; 1QRS A; 3C2Q A; 1MN6 A; 2F9F A; 3LUH A; 1I10 A; 1XTV A; 3HI4 A; 2MMC A; 2CZQ A; 4KH1 A; 4NDZ A; 4BBA A; 2KJQ A; 4JVL A; 3RUC A; 1UCF A; 4AFN A; 4TMZ A; 3D7R A; 5BYB A; 1TRH A; 1UIR A; 3B51 X; 2YYZ A; 3EC7 A; 3K6K A; 4MP6 A; 4XJS A; 3CVW A; 3L8G A; 2PUF A; 3GUY A; 2F4V B; 2RBG A; 3HM9 A; 4MUI A; 2ZYV X; 4OHZ A; 2CHQ A; 1NVV Q; 2K7F A; 3KEF A; 4GI2 A; 1EVZ A; 4TXH A; 4IX9 A; 4DA0 A; 4OH1 A; 2H4J A; 1JA9 A; 1N6O A; 1ZHF A; 4HAU A; 2WSS D; 4TMV A; 2FGK A; 4DQD A; 1R6X A; 5EZ5 A; 4EHN A; 1VL6 A; 1WOM A; 1KFK B; 1RLM A; 5I8P A; 1SVL A; 1WXH A; 1CF4 A; 3WWE A; 1IN5 A; 2J0E A; 3R3H A; 1NVV R; 3TD6 A; 3H0E A; 4D4M A; 4DLN A; 2B7B A; 2ODA A; 1WUK S; 2GCP A; 3OH8 A; 1J2Y A; 2BEK A; 1L7P A; 5AIA A; 5ER9 A; 3G59 A; 3IVM A; 1YZL A; 3ZNO A; 1N5J A; 2VHC A; 4BZP A; 1Y4D E; 2ODN A; 3HKC B; 4M0K A; 3OOS A; 5KTR A; 5F9F A; 1AM4 D; 2AEF A; 4OGF A; 3SXJ A; 1XZE A; 5ULL A; 1JKJ A; 1ACJ A; 1QHS A; 2F9L A; 4LHX A; 4EHA A; 3PC6 A; 4F84 A; 2BFN A; 2JVK A; 2PX0 A; 3QBV A; 3RRF A; 3N59 A; 3DGE C; 3K0F A; 4KBY A; 5BRJ A; 3C1U A; 5FPH A; 3WB9 A; 4BO5 A; 5M3B A; 4AMB A; 4UCW A; 3EUA A; 2EBW A; 4JHX A; 4LKO A; 1JEC A; 2ORV A; 2B1R A; 2PWP A; 3TEJ A; 3X1J A; 3IMK A; 4L5I A; 2MII A; 1LA2 A; 1M1N A; 4Q0M A; 4A92 A; 1O0B A; 5DNY B; 3OJW A; 4AJ4 A; 2VDO B; 4AO8 A; 1VID A; 3DSR A; 2RAF A; 1HZZ C; 2WPN A; 3QKU A; 5MDH A; 1AVT A; 2WYU A; 4M6W A; 2BEV B; 2Q8P A; 4YRM A; 3Q4G A; 3MSC A; 1PSW A; 4JFR A; 3OJF A; 1F2U B; 2IZ0 A; 2VNH A; 1KYY A; 3VBC A; 4C08 A; 2L42 A; 3B13 B; 4YAW A; 4QGS A; 4E33 A; 1JNY A; 1T29 A; 4JLJ A; 4JI2 B; 3KTA B; 1OYO A; 3UMJ A; 3M5D A; 2WU8 A; 1QZX A; 1Z35 A; 1XQX A; 1CUB A; 3LYU A; 3ZHC A; 2ZTV A; 3C3C A; 1T2N A; 3WP1 B; 3N05 A; 3D5E A; 3KB8 A; 1HQY E; 1H8E D; 3DZB A; 4A8H A; 2CBX A; 3BCR A; 2RGE A; 3ES9 A; 4ND7 A; 1I9C A; 3MZ0 A; 5TEW A; 1DIB A; 3TNJ A; 2KYR A; 4PRL A; 5GVR A; 2GZR A; 2H2I A; 1U8Y A; 1O6Z A; 2Y77 A; 4LZP A; 1HT3 A; 4XII A; 1JHM A; 2GRA A; 2QTB A; 4JL5 A; 4EA9 A; 1GD1 O; 5DI3 A; 4BWG A; 3ULK A; 1Z5Z A; 5ALV A; 4RD1 A; 4P35 A; 1UFK A; 2GJS A; 3P97 A; 5MN4 A; 1O0S A; 4QM6 A; 2A84 A; 2NVW A; 1N25 A; 2R84 A; 2YV4 A; 2W6J A; 3D8N A; 1G5F A; 2GM9 A; 5KSW B; 3JQ7 B; 1QIG A; 3MNF A; 4GQB A; 1RCU A; 3N2N A; 4QUD A; 5K4A A; 3SF0 A; 4ASU D; 3MCV A; 1D1Q A; 4W5R A; 4M8L A; 1E63 A; 5TEV A; 3GP3 A; 1C3V A; 1Z29 A; 3DG9 A; 2JD0 A; 3O1L A; 4GBE D; 1QHH B; 3BMX A; 4R2I A; 3WRY C; 4KOD A; 3VR2 A; 2GEB A; 3IS3 A; 3IA7 A; 1I2Z A; 3MJF A; 2FTK E; 4OG2 A; 4PLT A; 4IKO A; 1B73 A; 5FRD A; 1G29 1; 1B8N A; 2O2C A; 1PV2 A; 2I7V A; 1TZK A; 2PKE A; 3EHD A; 4H18 A; 4QYT A; 4RTN A; 4DUG A; 2PU5 A; 2BVN A; 3V9P A; 2J6I A; 3PQ2 A; 5IJ0 A; 1IYZ A; 4NOJ A; 4BMX A; 2Z4R A; 5FNV A; 3VSA A; 5BWM A; 4MVC A; 1JYF A; 5TMD A; 3SSO A; 3FSR A; 5TDH A; 2EXX A; 3COZ A; 4QC2 A; 3A1E A; 1D6J A; 2W2L D; 3SMQ A; 1KEV A; 4B40 A; 1NMY A; 3UQY S; 1SD5 A; 1WD5 A; 3GLK A; 3NTO A; 1GRV A; 3F4K A; 4ZQC B; 1REQ B; 5LXT B; 3I45 A; 1USG A; 2WBL C; 4X7B A; 1KQN A; 2ZJ3 A; 5JW6 A; 5DYI A; 5AW7 A; 3TL6 A; 3OLV A; 1NPD A; 4TKZ A; 4TQ9 A; 1PJ3 A; 2DHR A; 2QF7 A; 4X41 A; 4BLO A; 3W6P A; 4HEJ A; 1V4B A; 1W85 B; 4BU0 A; 2FZE A; 2QYT A; 4LOI A; 4M0C A; 4O3M A; 1IM8 A; 1E6E A; 4M7P A; 5TPW A; 2GNP A; 1JIJ A; 1Q3T A; 3O3C A; 4YJD A; 2B20 A; 2G3Y A; 4QWM A; 1U8X X; 2Q6T A; 4FFB A; 1NVB A; 5CLJ A; 2FKN A; 2Y0P A; 1C9E A; 1NNS A; 4FMD B; 4OL9 A; 5D85 A; 3DZI A; 3JYL A; 2QN9 A; 3KUC A; 3ROG A; 4JJA A; 4DIL A; 2ZXU A; 2YHA A; 4LHE A; 1XTS A; 2QGN A; 2PSE A; 4MGH A; 1ESN A; 3DC7 A; 3NHA A; 1XU9 A; 5FBZ A; 4FGW A; 3E9E A; 3PA6 A; 3RC8 A; 3FKJ A; 3KZK A; 3LDF A; 5AD1 A; 3GAM A; 1COZ A; 2MSE B; 5J43 A; 2DMR A; 4FWB A; 3G2M A; 1U3W A; 5HZH A; 5ARC A; 4LF5 B; 1UXJ A; 2JA1 A; 4X6Y A; 1R5B A; 4B17 A; 1U90 A; 3FGO A; 1JZQ A; 3B5E A; 5K99 A; 4WOC A; 4FYK A; 5E6S A; 4IX1 A; 4D45 A; 2V03 A; 2HHA A; 3AGP A; 3THW B; 1RWQ A; 1JHY A; 1JI0 A; 1UJ5 A; 3UK6 A; 4FBG A; 3O2Q B; 3KOO A; 4P32 A; 4PIC A; 4INZ A; 1IJB A; 2OQR A; 3V4P B; 3IB3 A; 1YNO A; 1FU4 A; 2NQ2 C; 3JW8 A; 2J0S A; 3ZCV A; 1EK0 A; 1W2H A; 1W88 B; 4OD0 A; 2WF7 A; 1YOB A; 4JNC A; 4ZVR A; 5JDI C; 5IVI A; 2BSA A; 4QPZ A; 3Q3H A; 4JJI A; 2X3Y A; 2XGO A; 1ZUW A; 3BH0 A; 3O1H B; 5K6T A; 4WJK B; 2ZRQ A; 4I4C A; 1BSV A; 5IMS A; 1PEY A; 1RWK A; 3EXA A; 4JB3 A; 3FPJ A; 2DBZ A; 2IFT A; 1JQE A; 3LY8 A; 1XV5 A; 3DXZ A; 4MS3 A; 3DEA A; 2OHJ A; 2IXE A; 3FF1 A; 2OVB A; 3SN6 A; 4FW9 A; 4AKH A; 2OFO A; 2H4W A; 3SLG A; 4FDM A; 1QZU A; 3EZ6 A; 1YZY A; 2Q5E A; 1SVW A; 1IJF A; 4HDK A; 1G41 A; 4QTU B; 4EAR A; 3SX2 A; 2RC5 A; 4KMM A; 1C1D A; 1ZIS A; 2TMY A; 4JIG A; 1P3Y 1; 4UHD A; 3OT1 A; 2F7S A; 2GTH A; 3PQ8 A; 2I1Q A; 4Q1V A; 2XGS A; 5FTM A; 5LU9 A; 5BY2 A; 3E1S A; 1Y3C E; 1KMH B; 1QNL A; 2G6T A; 1DBP A; 1UR5 A; 2D4A A; 4HDN B; 2IDK A; 4GSK A; 4H31 A; 1FFX B; 2ZOS A; 4V39 A; 4FFD A; 4Q1F A; 5E1O A; 3WMG A; 2BFM A; 4PQF A; 1P2B A; 3R7B A; 4ZYV A; 1EJ6 A; 5H68 A; 1NR7 A; 2JRL A; 3AON B; 1ZOI A; 4G0O A; 4F3Y A; 2BCJ Q; 3THX A; 4ZDJ A; 5GY7 A; 3Q1L A; 1GVH A; 3CWZ A; 2TS1 A; 4M0F A; 4EUK A; 1CP3 A; 3UAV A; 1Y0B A; 5AH0 A; 3PGX A; 4FFO A; 3HUU A; 2Q2Q A; 3STW A; 2JFN A; 2QTL A; 4HLC A; 1UJC A; 3DW1 X; 1Z6Q A; 4L95 C; 4MVJ E; 4XYB A; 1Z8F A; 2LWI A; 3ZE8 A; 1CRW G; 4HTF B; 2NAF A; 1P31 A; 3HU1 A; 4EY4 A; 4JR1 A; 1GZS A; 2O0H A; 5J4V A; 1EVQ A; 3FVV A; 4HY3 A; 1Z62 A; 1P1R A; 4RTK A; 1YNS A; 1UMC A; 3M83 A; 4B74 A; 1EHI A; 1XQV A; 3CW2 A; 3RJT A; 1F9A A; 4XR9 A; 3ZI0 A; 3ADD A; 3G5L A; 2EG5 A; 3KOZ A; 3P9Q A; 4K9Q A; 3R3Y A; 1QZ3 A; 5HMM A; 2WSL A; 2WZC A; 1NMX A; 3BHM A; 3NFY A; 1BHJ A; 3ECS A; 4WTO A; 2NVU A; 2BFB B; 2QE7 A; 2ZCV A; 1O6G A; 1GG2 A; 1JR3 D; 4RF2 A; 2CJX A; 5I7V A; 4AM8 A; 2Y1E A; 3HG7 A; 2QRG A; 1YQX A; 3OX3 A; 2GO1 A; 3AI7 A; 2FFJ A; 4OGG A; 2OKG A; 1J1Z A; 4G9B A; 2IWH A; 3SCZ A; 1EVI A; 3BZB A; 4C5A A; 4PZY A; 4L85 A; 1A5B B; 2A5Y B; 3KCQ A; 4OJT A; 3W78 A; 1EJ0 A; 4DV2 B; 1GTL 1; 3G2L A; 2J30 A; 1OBH A; 4Q15 A; 4ENR A; 1GDH A; 2N42 A; 1Z6X A; 2O2X A; 5B1S A; 2Y99 A; 3CZR A; 2PH7 A; 3L02 A; 1HDZ A; 4J0J A; 4JOD A; 3LT4 A; 2YWE A; 3PB3 A; 4R16 A; 1F2U A; 2A4K A; 1HTB A; 1CJB A; 2Z4T A; 3Q9N A; 3HZZ A; 3DFZ A; 2F1H A; 2VAV A; 1JG7 A; 1DRJ A; 1LBI A; 1KAE A; 2GX6 A; 2RDM A; 3HUD A; 2GYV A; 3OM0 A; 4WCF C; 3UGQ A; 2IHU A; 3DBH B; 5DYN A; 1J7K A; 1TD1 A; 1VL5 A; 2ZJA A; 3KJG A; 1EM6 A; 4J3F A; 1YE3 A; 2A1U B; 4U9H S; 1XWW A; 1QG4 A; 4RXM A; 2O1W A; 3R3Z A; 4ZAR A; 3IP6 A; 4LWP A; 2P5Y A; 5U1P A; 1D1T A; 4RK7 A; 2XE4 A; 4F4C A; 2VT3 A; 1JKM A; 3AKE A; 2CVP A; 2VK8 A; 2NMV A; 3DQQ A; 4Z4E A; 1Q0L A; 4XSG A; 5F33 A; 3U3K A; 4RK4 A; 1P8A A; 1C7F A; 3NAZ A; 4HF7 A; 5EWL B; 1CK4 A; 3PQ7 A; 5NUL A; 2E4Y A; 3WX0 A; 1N1M A; 3LIP A; 1J06 A; 4I3H A; 1IOL A; 3ESX A; 3T0G A; 3MWY W; 4YYE A; 2GVN A; 2GEK A; 4XYG A; 1HTW A; 4KGD A; 1SHK A; 2YJ6 A; 3LQQ A; 1NMZ A; 1GV1 A; 4CGZ A; 1Q1A A; 4KNV A; 5KWJ A; 2AL7 A; 1YZV A; 3F9C A; 1K8X B; 1Z5U A; 3NTQ A; 3O8L A; 3C14 C; 4IDD A; 4X1I B; 1OBV A; 1FXS A; 1U2D C; 3OEH A; 2W2M A; 2IYT A; 1QIH A; 3LOP A; 5AK6 A; 4UMV A; 3UHA A; 4YDR A; 3TGL A; 5D86 A; 3VH3 A; 2PD6 A; 3QLV A; 4Z4G A; 3TBF A; 2QPT A; 3A7E A; 3ET5 A; 5J1T B; 3RFU A; 5CCB A; 4UU7 A; 5AOC A; 4PF1 A; 1YZH A; 1AGP A; 2FOL A; 4S1P A; 3WNZ A; 3AF1 A; 2AWN A; 3KSA C; 4G0U A; 3HY4 A; 2PID A; 4YJ3 F; 1GLN A; 3QXJ A; 4RTO A; 4N41 A; 4QDZ A; 3MWD A; 1YJA A; 2P6M A; 1AK2 A; 2QRV B; 1JII A; 3CUW A; 4U6S A; 4BFS A; 1XXH A; 5HQ3 B; 5LXT A; 5I7S A; 2HXS A; 2W9X A; 3EIO A; 4O08 A; 6REQ B; 4DV1 B; 3U44 A; 4G5R A; 4FE3 A; 2J0Q A; 3ILX A; 4RZI A; 3CUT A; 1VZ3 A; 1Z33 A; 5BQF A; 3TJM A; 5EXX A; 3W2I A; 1QO0 D; 2V6F A; 5BWO A; 1LD3 A; 1Y42 X; 3A04 A; 1P2D A; 1H1D A; 2BZ1 A; 2C57 A; 3KU4 A; 2C04 A; 4AJ9 A; 1C14 A; 4DIF A; 3IP7 A; 3WHK A; 4OK3 A; 4EWL A; 3IUJ A; 1MVO A; 1Z17 A; 4MS4 B; 4R2K A; 3I53 A; 1C0I A; 1NF2 A; 3DL2 A; 3TRF A; 2B98 A; 3W4K A; 4WL0 A; 5FQO A; 1QPB A; 3B53 X; 1KE2 A; 5K09 A; 4UC0 A; 2JDI D; 3BTV A; 4UPV A; 3ST7 A; 3ESA A; 3ZQJ A; 3CVV A; 2XDQ A; 2R44 A; 5ITZ B; 4JRO A; 4WZB E; 4W9R A; 3WIB A; 3LC7 O; 1RCT E; 5GZ1 A; 1TGY A; 4GE3 A; 1NA5 A; 3PHC A; 1L4H A; 1UK8 A; 4RAD A; 1OA1 A; 1UC9 A; 1ADF A; 1HH4 A; 2F00 A; 3E8M A; 4NU8 X; 3B6E A; 1TU4 A; 3MLB A; 3EUF A; 3V32 A; 1H42 A; 3IEV A; 2JFV A; 5KVV A; 1Y1R A; 3JAK B; 3W34 A; 2XIQ A; 3SJB A; 4NAU A; 4QBK A; 3WLP A; 5KJ6 A; 3SJ7 A; 2BGM A; 4DY4 A; 3LGA A; 4DEV A; 4IV8 A; 1NMS A; 4NZO A; 3UHP A; 3B5J A; 3THW A; 4MAA A; 4D9I A; 1T5D X; 4I8X A; 1V5E A; 4TQU S; 1T43 A; 3D6N B; 2XUO A; 3E5M A; 3UWO A; 3DF9 A; 1I0I A; 4DC1 A; 1W88 A; 4I19 A; 1EFP A; 3IOY A; 1UXG A; 1U2Z A; 2JB9 A; 2Q99 A; 5DXA A; 3GFO A; 3ETN A; 5KQP A; 1KXJ A; 2Z08 A; 1DJL A; 3ELY A; 3E1K A; 1C7I A; 1J8D A; 3BAZ A; 3A2N A; 4NEN A; 3F4L A; 1MU0 A; 2BFE B; 2QT9 A; 3CLH A; 1CUH A; 2PXC A; 4P08 A; 4P3O A; 3EDQ A; 4LBV A; 4YI5 A; 1D7A A; 4ER6 A; 2Y6V A; 1N75 A; 2XYV A; 3SYM A; 1KFE B; 3BMO A; 4W5O A; 1I1O A; 4AII A; 2R6R 1; 3RC9 A; 1I0L A; 1A0O A; 1DG3 A; 3FNQ A; 2HUP A; 3LZD A; 1DEL A; 4ZKD A; 4Z3D A; 1RE1 A; 3KWJ A; 4UUL A; 5CAE A; 2WSB A; 5FWY A; 5FBQ B; 3BQ9 A; 3U81 A; 3EGC A; 5C80 A; 4W82 A; 1FY2 A; 3PUI A; 5FKJ A; 1MOR A; 3F6Y A; 3D6M A; 3G2O A; 2H16 A; 3V8V A; 5HP4 A; 3I2G A; 3G2J A; 4DQL A; 1JH9 A; 2AW3 A; 5F2E A; 3S2G A; 4PBT A; 1ECL A; 2AEJ A; 4ZGB A; 5BMO A; 1A71 A; 1N1E A; 3WJ2 A; 2FE4 A; 3BWE A; 3N8N A; 5IFL A; 3L4J A; 3WKB A; 1EKX A; 4HNH A; 5KKN B; 3QV1 A; 3RSZ A; 2HWX A; 1BX1 A; 3NPA A; 3QUC A; 5E1M A; 1XUO A; 4B7X A; 5F7Z A; 2IHZ A; 5CIQ A; 1DKQ A; 1CDE A; 2W22 A; 4MPR A; 3VV3 A; 3LUD A; 5A62 A; 2QDX A; 4K2A A; 5ICM A; 4XJT A; 1T8T A; 5JAZ A; 4ZIR B; 3ME5 A; 3LPN A; 3TPF A; 4D4L A; 2VG8 A; 3OT5 A; 4PLF A; 4G9E A; 4PZP A; 4BAS A; 4LPS A; 2HDH A; 1U28 A; 4KXU A; 4M7R A; 2P3L A; 2WR8 A; 3RWM B; 1USV A; 2H65 A; 2I6T A; 3A1T A; 3LPM A; 4DSU A; 2RGN A; 2NU8 B; 3FVW A; 1XZQ A; 1TLT A; 1P9W A; 1P7P A; 2HJW A; 2IJG X; 2C98 A; 1PFK A; 2AE1 A; 3N6R A; 3NYQ A; 1GGH A; 4LRX A; 3ZE0 B; 5LMU B; 1T1R A; 3CNQ S; 1JVN A; 3CM0 A; 3DL7 A; 3JSA A; 5D84 A; 2AC4 A; 5DM4 A; 3B78 A; 3LKB A; 4HB3 A; 1PGJ A; 4IDN A; 1FK8 A; 2FWN A; 1VSQ A; 2OO5 C; 1VJ1 A; 1X70 A; 3G8A A; 3AVT A; 2VP6 A; 1YDW A; 1A7K A; 1EFL A; 2MSW A; 1DFI A; 2O3U A; 5FOS A; 3ZY4 A; 1U5B A; 5T67 A; 4AKS A; 4B3T B; 3HWS A; 2I29 A; 3UFX B; 5KX5 B; 3BPS A; 1OLX A; 1UBE A; 4HSE A; 4JKY A; 4R1S A; 5CM2 Z; 3VKV A; 4D96 A; 4JBN A; 1D0I A; 1SCK A; 1Y3A A; 1KJZ A; 2C4W A; 2FMH A; 3EDR A; 5J5V A; 3ZY3 A; 3DP9 A; 4RQB A; 2HCB A; 3GDH A; 3ZCQ A; 4YV1 A; 2G1P A; 1G3Q A; 2L19 A; 3JQ8 A; 4F71 A; 3DBH A; 1TK3 A; 2AMF A; 4Y6L A; 3IP8 A; 4FJ0 A; 4IDF A; 4OHW A; 5GZ3 A; 2REQ B; 4WIA A; 4U00 A; 4G85 A; 5FBR B; 3O75 A; 2QZP A; 3MEL A; 1GCU A; 3NBY C; 1XVI A; 3OTQ A; 4N12 A; 3RS2 A; 2PQ6 A; 1XU7 A; 3GCD A; 1HLP A; 4NF2 A; 3BGV A; 4DDT A; 5IWA B; 2OCB A; 2GIL A; 3GOH A; 2ZS8 A; 3TD2 A; 3N7U A; 1U6K A; 4ZV4 A; 5IAN A; 5JI3 E; 2ZYD A; 5EL0 A; 5HIM A; 5IS2 A; 2IBT A; 2GHQ A; 4IFE A; 1MOP A; 4D74 A; 2WJ4 A; 4JOQ A; 2Z56 A; 1BG6 A; 2DLD A; 3F90 A; 1INL A; 1G8H A; 1FRN A; 4HRV A; 1CY1 A; 3V4S A; 3GLG E; 1K1E A; 4B8Z A; 4A2W A; 4CIB A; 1ZY2 A; 4K88 A; 5JC1 A; 4PHC A; 5HQJ A; 4A0F A; 1NDU A; 4YRT A; 3CPS A; 3SP1 A; 3NEP X; 2FQ1 A; 4NOK A; 3FR8 A; 2Z0M A; 2QAG B; 4YJ2 F; 4CCA A; 4DT1 A; 1KAO A; 2ZI3 A; 3MVE A; 4FZJ A; 1G8A A; 4OO3 A; 1KH2 A; 4PX9 A; 1QFY A; 1UIM A; 1CQJ B; 4YP6 A; 3GPL A; 4L8F A; 4V03 A; 3CEM A; 3CKL A; 3MJC A; 5F64 A; 1WCI A; 4KPU B; 3VUE A; 4LHW A; 2ZXF A; 3AHC A; 3R8X A; 3MJE A; 1OAS A; 4YHH B; 1SIU A; 1PW7 A; 3AEX A; 1NYT A; 1V25 A; 1C1Y A; 4RVG A; 3DKK A; 1RJG A; 2PS1 A; 4LE2 A; 3EGD B; 5B64 A; 4BU1 A; 4OF4 A; 1IEW A; 1OYW A; 1ODL A; 2ZRJ A; 4DNO A; 2OWE A; 2R00 C; 4DV6 B; 1E1Y A; 1S3S A; 4DPK A; 2JAU A; 4B79 A; 1HE1 C; 1F8X A; 1JJV A; 5GJB A; 3NUQ A; 5B7P A; 1FMJ A; 2X41 A; 3G23 A; 2XB6 A; 1PG2 A; 3GNL A; 3DTN A; 2WWV D; 1OBF O; 4GVP A; 1Y82 A; 2ZU0 C; 4FCV A; 4NBN A; 5D3K A; 4PE5 B; 3ZWO A; 4RL4 A; 1FNS A; 5JED A; 1H2F A; 4FWI B; 4UHK A; 2VKQ A; 4DQV A; 1SHJ A; 2XPC A; 3A4I A; 3IPR A; 3QU2 A; 2G5C A; 3TTX A; 5I5G A; 5F51 A; 5GWX A; 3DZJ A; 1SUI A; 2QUK A; 3R7N A; 1NBE A; 4XIG S; 4HEA 6; 3AR8 A; 1L9K A; 5EP4 A; 5KTM A; 5JAI A; 2D54 A; 3E61 A; 1QFM A; 1RE0 A; 1R6V A; 1M2V A; 1VHD A; 2O2H A; 1TFU A; 3DPU A; 4H4D A; 3NTR A; 1MNQ A; 1OJ7 A; 1W8D A; 2QNB A; 1QOP B; 2XTN A; 3ZNQ A; 3DTY A; 2G1A A; 5LQK A; 1IHY A; 4A8J C; 2I8C A; 1IN4 A; 5AJX A; 3WSW A; 2O3T A; 4ARZ B; 2BDT A; 1J00 A; 3ZUG A; 3QY9 A; 5ITZ A; 1PG0 A; 4EJ0 A; 4G0Z A; 3B6I A; 3SQW A; 2HGS A; 3D7N A; 5ERX A; 3USE S; 5A7H A; 4QRE A; 1THT A; 5IAK A; 1MEE A; 1QE3 A; 2HI0 A; 4MUF A; 2CMH A; 1Y5L A; 1T1E A; 2PGD A; 3LUB A; 3OFN A; 4BBJ A; 4CLH A; 1L7A A; 1XCJ A; 2OXE A; 2WID A; 3CRN A; 3QLU C; 1G4U R; 3JAK A; 5HGJ A; 1S2M A; 3T5M A; 3TQO A; 3Q12 A; 1JR3 E; 3GVX A; 1UC8 A; 1UFO A; 2FMU A; 4RJI A; 1E61 A; 3H16 A; 4TSF A; 5AXA A; 4O5O A; 5LA6 F; 2V59 A; 1UOQ A; 3MGC A; 3TCX B; 1AZL A; 3QY0 A; 3I2F A; 2PFG A; 5FTN A; 3MF4 A; 1KH3 A; 3QVW A; 4AJK A; 2JIZ A; 2Q07 A; 2D5B A; 3QA9 A; 4FKB A; 1Z6Z A; 4N5N A; 5F78 A; 5JYE A; 5KWI A; 1DPF A; 5F7J A; 2XNJ A; 4MP3 A; 4BO8 A; 2XMP A; 1CVL A; 4ACA A; 4ZAW A; 5EUB A; 5C0O E; 1TTQ B; 2Y66 A; 2ZUS A; 4Q3A A; 1N6R A; 4HH0 A; 5A5V A; 4HXG A; 2YYY A; 4RZS A; 3RAP R; 4OU5 A; 4LDN A; 4KKX B; 3LRX A; 4BS9 A; 4Y2V A; 1XOI A; 3AWI A; 4TXJ A; 1QU2 A; 1NXW A; 2EFJ A; 3Q7E A; 5KNC A; 1P9N A; 3BKX A; 3VC7 A; 5LUA A; 1ZQ1 A; 4K5Q A; 3F96 A; 4R05 A; 1GTM A; 3I2I A; 4F12 A; 1N3L A; 4FFS A; 1T5I A; 1IZZ A; 3A70 A; 5HJE A; 4IGA A; 1LLQ A; 4FPX A; 1YE5 A; 4GMG A; 4Z0P A; 5IJA A; 1A4U A; 4N8D A; 2E18 A; 4MQ5 A; 3VNS A; 1W19 A; 2C1X A; 1RWM A; 1IZ7 A; 4RY0 A; 3FFA A; 4D0G A; 2ZBP A; 3AY6 A; 2CMF A; 1JR4 A; 5KTN A; 1YRY E; 1N7J A; 1NT4 A; 1SIW A; 2OZP A; 1NUL A; 2YOH A; 3RTD A; 4D0L B; 1GIT A; 5C3M A; 2JD1 A; 3PFF A; 1ONO A; 1E6U A; 4QYI A; 3RVJ A; 5K3E A; 3F5Q A; 3I6B A; 4GPE A; 3OOE A; 2WT8 A; 1NFF A; 3BD8 A; 2EKL A; 3OG9 A; 3OUR A; 3PWK A; 4MCA A; 4Q3N A; 2QK4 A; 5FBK A; 3LFT A; 3QOF A; 1ZGZ A; 2CFO A; 2JGI A; 1U2P A; 1L5N A; 5HW0 A; 5F98 A; 3ZLT A; 3RS5 A; 5ALW A; 3OE5 A; 5G0S A; 2IH5 A; 1J9G A; 5JS2 A; 3L6D A; 1TJ3 A; 1Z38 A; 3DEZ A; 1XSE A; 3FWP A; 4OTZ A; 3DAO A; 1YJQ A; 4F85 A; 3AK4 A; 2QMY A; 4KXN A; 4LSW A; 2GRU A; 4DUP A; 1QWO A; 3MLE A; 4ML3 A; 4F04 A; 2X40 A; 4IGF A; 3HFR A; 4RAC A; 3CJQ A; 1KY4 A; 3TND A; 4DAD A; 1RVD A; 3RLF A; 4AQ7 A; 2G07 A; 5AJH A; 1PG5 A; 2QX4 A; 4MDF A; 2PUV A; 2ZR9 A; 4II7 A; 1BN6 A; 3SC3 A; 1XAI A; 2NX2 A; 5H4H A; 5GOT A; 4X7M A; 2IBS A; 3NOO A; 1F51 E; 1HOT A; 5KTT A; 3DT8 A; 2OL4 A; 1OGJ A; 3FTD A; 3KD3 A; 1E6M A; 3FDX A; 3HKD A; 2LD4 A; 3I28 A; 4QHY A; 5DM2 A; 1K63 A; 5DCL A; 3KEG A; 1DIR A; 1JFR A; 3C3K A; 3H5A A; 4HDC A; 5FPP A; 3D4P A; 3W7A A; 1PFQ A; 3D7K A; 5DIS B; 2P30 A; 1A82 A; 2V58 A; 1JQ3 A; 5BNZ A; 5BR8 B; 3A9U A; 1P80 A; 1QON A; 4YO8 A; 1Y3F E; 2B8N A; 3H2S A; 1ZZG A; 4UMD A; 5LM3 A; 4PQE A; 1RWN A; 2PUJ A; 4MWG A; 3WMW A; 2KML A; 4AN4 A; 4TYN A; 5NLL A; 2R6H A; 1UOP A; 4ZOB A; 1UBM S; 2W83 A; 1SBY A; 3ANN A; 5BV7 A; 1ABB A; 2A5G A; 1W1W A; 1L7R A; 1TI7 A; 1JAZ A; 3FDJ A; 1RZR D; 5G2O A; 1DAH A; 1AD2 A; 3JY6 A; 1V11 A; 1TGV A; 1EQ2 A; 3E2P A; 4TT3 A; 2EEP A; 1JDZ A; 3EXF B; 1ZNY A; 2DPX A; 3JZD A; 4YLG A; 2VPZ A; 3FOA A; 4Z1M D; 2GER A; 2VDP B; 4OB8 A; 4BNF A; 3ZBU A; 4E08 A; 5AVY A; 1KRH A; 2RH9 B; 3CVJ A; 3N2K B; 1HEU A; 4MYR A; 4WKC A; 5CH8 A; 3R3X A; 3WYF A; 4PNE A; 2OA0 A; 4M8I A; 1Z4O A; 3U7Q A; 4EX7 A; 2GO7 A; 2ZDH A; 4I5G A; 4KQF A; 5A5G A; 1BWP A; 1OI4 A; 2QUH A; 2ZSI A; 3WLA A; 3K70 B; 3PID A; 1OVG A; 4NJE B; 1NUH A; 2H5E A; 3KEO A; 3LU1 A; 4TZ0 A; 3TX6 A; 2GNA A; 4GC9 A; 1JXQ A; 1M6V B; 3S27 A; 4Y2T A; 1Q91 A; 3T35 A; 1UJB A; 3U7I A; 4WXP A; 4II3 A; 1RQX A; 4F7D A; 2ZIX B; 4PU0 A; 2VN8 A; 2DAP A; 4F6C A; 5A0V A; 2X6Q A; 3E23 A; 2JIQ A; 3NHF A; 4LYE A; 4PW0 A; 1TYE B; 1ZVQ A; 2RK4 A; 1H0R A; 4JMP A; 3RQS A; 1FU8 A; 2D62 A; 1FLV A; 2X8B A; 1I5R A; 2ZBG A; 3NI2 A; 4Y6O A; 2ZR8 A; 1ZQ9 A; 2HSW A; 4OM8 A; 4P93 A; 1MAV A; 4V3B A; 3RDP A; 3WLT A; 3HSS A; 1ONN A; 1VCN A; 1CZK A; 3DW3 X; 5ITV A; 1AXG A; 1BZY A; 1CE8 B; 2QVX X; 2BLL A; 2OZK A; 4DSO A; 3VOT A; 1D4F A; 5CIY A; 2JFO A; 3GRR A; 2NO6 A; 2NYA A; 4IIE A; 2R5W A; 1LTH R; 1C30 A; 1ZK1 A; 4QSH A; 1DEK A; 2O57 A; 4RYD A; 4EF5 A; 2RHT A; 4QTT B; 3NG1 A; 4B6S A; 1MRS A; 1OTY A; 4C03 A; 3IAH A; 2FFQ A; 4CMG A; 4G0P A; 3RIY A; 3N5C A; 2C7V A; 2IYN A; 3FPL A; 2O8D A; 3D6V A; 2P5T B; 1PIW A; 4P83 A; 4UTZ A; 2PA0 A; 3A2M A; 3GZ4 A; 1A9S A; 4WKG A; 4X1K B; 3U7R A; 1VHQ A; 4YRZ A; 2DHC A; 4OQM A; 4MI3 A; 1JQL B; 3FYT A; 4I36 A; 1F7V A; 4D5G A; 2Y27 A; 4FCW A; 3DA8 A; 3C5E A; 1DBV O; 5BO5 A; 1I3C A; 5ANR B; 3UFB A; 2Y93 A; 3IEK A; 1SHT X; 4ONE A; 3RC7 A; 2PT9 A; 3CNH A; 4AUK A; 1JMV A; 4CLO A; 3FFB A; 1E9D A; 3G1U A; 5D5H A; 5KAM A; 3AFH A; 4AYX A; 3CVU A; 3JZA A; 4QLO A; 3RKR A; 5H82 A; 1PSD A; 3EW7 A; 3KWF A; 4X9X A; 1KSG A; 2BKX A; 1JDB B; 2ANC A; 3JVU A; 2IZ6 A; 5LQM A; 2I2W A; 4D1Y A; 3EXE A; 1EA5 A; 1ZN7 A; 3KQV A; 3DV9 A; 1PYO A; 4S35 A; 1UXK A; 3WGO A; 5DLP A; 3I6Q A; 1GN8 A; 2HA6 A; 3ORR A; 5LSA A; 3GT3 A; 4M1W A; 1XZC A; 3AJ8 A; 2HSG A; 2HYD A; 2ZYS A; 3IUB A; 2BMJ A; 1FDV A; 1L5S A; 1HDY A; 3L7D A; 4IIU A; 2A3N A; 2GDZ A; 3TZQ A; 4FDL A; 3ZQ6 A; 4LV8 B; 1HF3 A; 4FS8 A; 1I0Z A; 4RPJ A; 4LZZ A; 1GG8 A; 2IYS A; 2Z2Y A; 2YJT D; 3P7M A; 2Q5H A; 3LST A; 1A9X B; 3M33 A; 1NF3 A; 3QFH A; 4NPH A; 4TUY B; 5EPE A; 4YRC A; 2Q5C A; 1WZ2 A; 2ZB4 A; 2Q21 A; 3UG7 A; 5YAS A; 4K9L A; 5BRA A; 5P21 A; 5HKK D; 2VUI B; 3KOX A; 4MQB A; 5AK4 A; 2JLZ A; 3KE8 A; 1ZHO A; 1LWO A; 2I67 A; 4FYH A; 5I3C A; 3JRW A; 2WYI A; 5EKS A; 4XWT A; 3IO3 A; 3A1N A; 3KQW A; 3IR5 A; 4UPE A; 5EHX A; 2ZVJ A; 3EW9 A; 3JYQ A; 3SSA A; 1V6S A; 1W44 A; 4ITR C; 4O8Z A; 3TCN A; 4DRX A; 3M2T A; 4DMC A; 1L7B A; 2MSC B; 1MO4 A; 3S2Y A; 2DQ4 A; 3VR6 A; 4AIC A; 3CIS A; 3RAD C; 2NZX A; 3R76 A; 4I8I A; 3GLG A; 2AZX A; 1LNZ A; 2L0X A; 2LUO A; 4II2 A; 4C78 A; 1N8N A; 4DLK A; 3IB6 A; 2QGZ A; 4CQM D; 4EUF A; 4KOE C; 5IKX A; 4B3H A; 1V16 B; 4XVC A; 4LRU A; 3JAL A; 1VE9 A; 5HRA A; 3LQK A; 1VZ2 A; 1R0K A; 1THZ A; 1YXM A; 5AKG A; 1YGP A; 3BWC A; 2PKR A; 4C4P A; 1HKU A; 1G8K A; 4I6E A; 4TWR A; 3E05 A; 2HWU A; 2VA9 A; 3FMF A; 4ROT A; 3GLF B; 4M36 A; 4I55 B; 4YRJ A; 1Z9B A; 2PXA A; 3FTQ A; 4X8L A; 1U1D A; 2WA1 A; 2ZRP A; 3H2G A; 4U1R A; 2X8J B; 5JDC C; 1JL3 A; 4L72 A; 4E5Y A; 1CJT C; 2CUK A; 1XZL A; 3OWO A; 1LPA B; 4YCD A; 3DHE A; 2ZWA A; 4Y2Q A; 2YL2 A; 5DXJ A; 1H94 A; 2P69 A; 2PH3 A; 2QQG A; 3H11 B; 1ORO A; 3O9V A; 1C4X A; 1OC2 A; 3OPY B; 4EO7 A; 2NYR A; 4ZCM A; 3NHP A; 3B1Z A; 4KN6 A; 1YH3 A; 2AYX A; 2VYN A; 3INN A; 4CIW A; 3T5A A; 4B0P A; 3FCO A; 2CXN A; 1JIV A; 3HMK A; 3L5O A; 3IRV A; 2XY4 A; 1NVX Q; 4WVL A; 1YZ3 A; 4DJA A; 2DUL A; 4ZTZ B; 2BEW B; 4CCY A; 2KE5 A; 2ZI6 A; 1Q0S A; 3K1T A; 3WAG A; 1KAG A; 4GDM A; 3C3P A; 2QNR A; 3FTN A; 1YFL A; 3VVB A; 1R6B X; 1S7G A; 1TA4 A; 2A9R A; 1B76 A; 2YIM A; 1EGM B; 3EWN A; 1URP A; 3DBR B; 1W94 A; 4DCO A; 5DST A; 1IZ0 A; 1KMQ A; 1QJ4 A; 2GN0 A; 2GZ3 A; 2AG0 A; 3BXO A; 3FMG A; 3IAL A; 3TW4 A; 3B60 A; 3K2I A; 4GEK A; 4LJY A; 1NZD A; 4JI7 B; 1ZD9 A; 1V5I A; 1MIO A; 3SEA A; 1J5E B; 1FA9 A; 1I7Q B; 4TLA A; 1WP9 A; 3HRK A; 4IJK A; 1PTJ C; 1XN1 A; 1L1E A; 1BFD A; 3QW4 B; 1LW6 E; 5CIT A; 3HXL A; 1N9K A; 4DDH A; 3WGY A; 2P2C A; 4ZGG A; 3EEY A; 5IT5 A; 5AP9 A; 1EFV B; 3GBS A; 4MHT A; 3IXM A; 4X44 A; 3DMD A; 4KIW A; 3DAY A; 4YJ3 A; 3PR1 A; 1R0L A; 1U1E A; 4KUG A; 3DEY X; 4B3R B; 4DRZ A; 2H9A A; 2QQE A; 4XDH A; 2AXP A; 2JGN A; 1KI1 A; 4YRD A; 5GTD A; 2GBI A; 1KVR A; 5CIW A; 3GFT A; 1V2G A; 2J3L A; 5IHP A; 3KSU A; 5JBN A; 1T1G A; 3S28 A; 3HRH A; 2GFI A; 3O03 A; 5IVP A; 2NVB A; 2HIA A; 2X13 A; 3QBJ A; 1CRL A; 3ORS A; 2IYU A; 1MUU A; 4I1R A; 5KOI A; 5A3J A; 3O8R A; 2WLT A; 3EXF A; 4IUD S; 2BE9 A; 4ZVQ A; 5I4I A; 1CEX A; 2RGA A; 4KO4 S; 2Q4T A; 5AVS A; 1E9V A; 2AYZ A; 1C9J A; 5A2G A; 1E2B A; 1OUM A; 2C07 A; 2NOA A; 4HUQ B; 3H5N A; 3F4B A; 5I47 A; 1PVV A; 3BSS A; 4JC0 A; 1P19 A; 3OEY A; 5IF9 A; 5CA0 B; 3O90 A; 1KYI A; 2RGY A; 3M4X A; 3MT9 A; 3NBQ A; 2DP4 E; 4IMJ B; 4QOE A; 3S2S A; 3ZG6 A; 2I2X B; 1NBM D; 4AVY A; 4JYZ A; 1DAE A; 4FSH A; 1QXS A; 4FID A; 2YML A; 2C9Z A; 2FRD A; 4MJ7 A; 5A4A A; 1B2L A; 5KND D; 5A4D A; 4DGQ A; 4ZT3 A; 3ZWB A; 3II6 X; 3F7O A; 4MV0 A; 5GM1 A; 5I7H A; 3ZXS A; 1N7I A; 4H7I A; 1U0T A; 4BMD A; 4X64 B; 4CUZ A; 1Y3D E; 3PTL A; 1ISR A; 1D8T A; 3PC2 A; 4P92 A; 4K46 A; 4W1P A; 4ZCR A; 1X7X B; 4TYM A; 9ATC A; 3AIL A; 3BWM A; 3K1A A; 1L5L A; 3DOH A; 1HRK A; 5ECY A; 4DL9 A; 3AG6 A; 2BHR A; 2YS7 A; 2PTG A; 4UUP A; 2CNK A; 5KDO A; 3LPA A; 3G5A A; 1ABF A; 2OZV A; 3PKJ A; 3IXL A; 5D7N A; 2AG5 A; 4Q00 A; 3Q7S A; 4RJL A; 5EE0 A; 1PUJ A; 4YC7 A; 2OKC A; 3PEY A; 2SKE A; 3KXD A; 4IWX A; 4B7Z A; 2COE A; 3NJ4 A; 3QK7 A; 3QVO A; 3T9E A; 2W58 A; 4MVJ G; 4AJO A; 4UM7 A; 4NZZ A; 5D6O A; 5ERY A; 2LTA A; 2NU7 A; 2YFQ A; 4LHY A; 4TSR A; 1FTY A; 2GEU A; 3ZOK A; 4CKW B; 1H9B A; 2LVB A; 2QW8 A; 4MV5 A; 3BHJ A; 2O05 A; 1E9B A; 2W00 A; 5DM6 0; 1MT3 A; 1TU3 A; 2YFY A; 4CLD A; 1NKS A; 2XTZ A; 4JC5 A; 1MAB B; 2FSV A; 1DC6 A; 3CB2 A; 4I9N A; 4OQP A; 1SPX A; 1ADC A; 1GUD A; 2EX0 A; 1HE8 B; 1NV9 A; 2NZ2 A; 4UQO A; 4ZOA A; 1NJ1 A; 1CW2 B; 3LL4 A; 4UFP A; 1ODJ A; 4E70 A; 4H16 A; 4B2Y A; 4XPI B; 5IX8 A; 5JAP A; 5F3W B; 1TJ5 A; 1KOG A; 3VHL B; 1ETH A; 1GPM A; 1WXF A; 2JGK A; 4MKI A; 4CYM A; 4A8P A; 1TVE A; 2FQY A; 4Q4J B; 4WK2 B; 1TYC A; 1SHU X; 2XKK A; 3LRP A; 3NDP A; 4NES A; 1UUF A; 3UO8 B; 3FJ1 A; 1XRR A; 3AXT A; 3DFK A; 3L82 B; 2QGF A; 3MHP A; 2FPW A; 3HFG A; 3SQO A; 1ZOS A; 2FMI A; 3HV2 A; 5J40 A; 3AV0 B; 1CLU A; 1YDH A; 3ELW A; 4BNN A; 2PZF A; 2XMZ A; 4TKL A; 5CA1 F; 1M8J A; 4K9K A; 5BR4 A; 3KRT A; 5KHO C; 4KRE A; 4ZXF A; 3AB8 A; 3L9W A; 3MWK A; 2JH3 A; 1NDQ A; 3HN6 A; 3VYR B; 4YCS A; 4P81 A; 1W4A A; 3R7A A; 5IXY A; 2H29 A; 4G3X A; 4GRD A; 1V7C A; 4Q48 A; 1S2O A; 2BSQ A; 3NTX A; 1S01 A; 4OJM X; 4XX0 A; 3S0J A; 3HCD A; 1ZIO A; 2JLV A; 5EYP B; 4D5E A; 1RAD A; 1E89 A; 1DKP A; 1VLH A; 2D13 A; 3HVK A; 1A9X A; 1JJF A; 4H3H B; 3WLR A; 1G5Q A; 3F73 A; 4A1O A; 5DTM A; 1EQU A; 5D3M B; 2ADF A; 1V6C A; 4FZ3 A; 4E6P A; 3FUW A; 1J22 A; 5C8V A; 1C3L A; 2CBG A; 3GGO A; 3V7W A; 4EI7 A; 3LC1 O; 3W5D A; 1IXY A; 4X8O A; 1K97 A; 3QSY A; 1DRU A; 1L8A A; 5K1Z A; 4AMU A; 3OLQ A; 4RZ3 A; 3WGJ A; 8GPB A; 3EGO A; 4HN2 A; 1ENZ A; 4R8V A; 3VPG A; 1D5C A; 5FBL B; 5BPF A; 4LQD A; 1E2H A; 4PGM A; 4IMI B; 4J2H A; 4JC8 A; 1BA3 A; 2CMD A; 2Z5L A; 4X94 A; 4KJX A; 4ZP1 A; 1XMO B; 4DVG A; 2YH2 A; 4NCK A; 2BFO A; 1WKB A; 4CJX A; 4ZO6 A; 3D2A A; 3ZHZ A; 2MCF A; 3DDH A; 4DG9 A; 1LCI A; 1TB4 A; 3UJA A; 4JJ6 A; 4JLK A; 4ZI5 A; 2ODE A; 3RVB A; 3ZCR A; 3OUT A; 1TYA E; 5AJY A; 2C5B A; 3QKS A; 5KVC A; 2WVH A; 1U94 A; 4O4H F; 3A4M A; 3DPI A; 3FET A; 2V6B A; 4K26 A; 5HFN A; 1TXG A; 1UZM A; 4A2C A; 4IQG A; 2Z6V A; 1E1Q A; 3QYF A; 3IL2 A; 3C4Q A; 3AZO A; 1NRW A; 3PRJ A; 4S04 A; 2XNH A; 1JA1 A; 3CT4 A; 1CF9 A; 1DFH A; 3EH1 A; 4PNH A; 1CD5 A; 1J0D A; 5DOZ A; 1U9Y A; 3BWB A; 5F73 A; 1CIJ A; 4DMU B; 1BH6 A; 3RSS A; 3UJC A; 3OQI A; 2JLW A; 1DFG A; 3HS3 A; 3PNK A; 2QIP A; 4FHE A; 5HF9 A; 2ZJY A; 4B85 A; 4C4O A; 2BIF A; 1QG0 A; 2VDV E; 4E1B A; 1ISG A; 3JRN A; 1RKV A; 5EKD A; 1N0I A; 1XWF A; 3PX2 A; 3QP9 A; 4X66 B; 3SC4 A; 1YVU A; 2H59 A; 1J0E A; 4URU R; 3NDI A; 1FJH A; 4YN3 A; 2ZUT A; 3IUE A; 4IO1 A; 5IWC A; 4MPD A; 4JS0 A; 3RCB A; 1R8Y A; 3WAK A; 2R87 A; 1C8V B; 4RMJ A; 3H9U A; 3OUZ A; 4GOL D; 3DBL B; 4Q7L A; 1TM1 E; 1XHL A; 3VMF A; 1F4L A; 2WIF A; 2F1I A; 2RHM A; 4YJ2 A; 2AFH A; 3WL4 A; 4NU5 A; 1D2C A; 4RX1 A; 5KAP A; 1QF6 A; 2JBW A; 5IJ9 A; 2WU1 A; 3TW0 A; 2Y2U A; 2VBG A; 4M21 A; 421P A; 1GZH B; 1K4M A; 3F3S A; 3WWY A; 2IYQ A; 1C41 A; 5IKO A; 4CME C; 2E6U X; 2ZSA A; 4MV9 A; 2Q45 A; 1O77 A; 2V7Q A; 2WZB A; 2ZSB A; 5J2U B; 4Q9A A; 4QU9 A; 2A6P A; 2Q1W A; 2QRQ A; 3L4E A; 4PZE A; 2WD8 A; 4ZQE A; 1OTX A; 4W9N A; 1C8K A; 4PCV A; 4XCX A; 4X61 A; 5K0C A; 4X7P A; 1WP4 A; 4WNC A; 3TF5 A; 1EFV A; 1SOA A; 3CNO A; 3RUH A; 1O94 C; 3VH1 A; 1RMT A; 1DO2 A; 2CLK B; 4FBL A; 3AJE A; 2ZET A; 3PNY A; 2HA5 A; 4FHD A; 1IFX A; 1U2S A; 4XUG B; 3KL4 A; 4FYY A; 3FYU C; 4K0R A; 5COB A; 4ZVP A; 5IXJ A; 3I6Y A; 1K9V F; 1NMQ A; 2ZJB A; 4G23 A; 2BBT A; 2W98 A; 4DR4 B; 2WTX A; 4F56 A; 4OEH A; 3DBI A; 3K9L A; 4NZP A; 2C03 A; 3PQC A; 3NKV A; 4AZ3 A; 4C4T A; 3PLA E; 2NTN A; 1AE1 A; 1GY8 A; 3DTV A; 2P79 A; 4TX1 A; 1SD1 A; 1PHP A; 4ZRN A; 4ZN6 A; 2EOA A; 1N0H A; 1BU5 A; 1DX4 A; 3LUG A; 2C61 A; 3WG9 A; 2WAB A; 1DJN A; 1F0K A; 3NH6 A; 3AOG A; 3N07 A; 1A04 A; 2WV9 A; 3KIP A; 1GZ4 A; 1JJA A; 2BKA A; 3PFB A; 2H1V A; 3QF7 A; 3W8N A; 2GN9 A; 3U17 A; 3WYC A; 4D8X A; 1IMJ A; 5IXS A; 2R1Z A; 4W4T A; 1CJV C; 2F1O A; 3O4J A; 1PQW A; 3BFX A; 3KQQ A; 3BDA A; 9MHT A; 2ZSF A; 3OJC A; 5EZ6 B; 4IXD A; 4HPW A; 1JDV A; 3SJA A; 1LPB B; 5BUZ A; 5KYG A; 2P6R A; 3G3E A; 3VAZ A; 1CXZ A; 3B8H A; 1XP5 A; 2BM0 A; 3WMR A; 3JQF C; 3PFN A; 3P8Z A; 1H4S A; 4IEF B; 2A58 A; 2GPW A; 2G5P A; 4WH0 A; 3SR3 A; 4HNA B; 4NAV A; 1U9I C; 4LE1 A; 1Z5O A; 3F0T A; 3TRK A; 5J2T B; 1FYD A; 1HI9 A; 3TVD A; 1U63 A; 2J6C A; 2ZRF A; 4PSA A; 4NU0 A; 2D4H A; 5IJX A; 5L4L A; 3W5B A; 3IHT A; 821P A; 1WNF A; 1JI3 A; 3Q5E A; 3WLH A; 2OM2 A; 1FND A; 2Z6U A; 4RMN A; 5A9T A; 3WRV A; 1M6E X; 2IPA B; 4OQF A; 3TLE A; 1OBO A; 3ND6 A; 5B4T A; 1XCP A; 4Y98 A; 1HL3 A; 3E7D A; 1S8N A; 4EH1 A; 2FYT A; 3TLG A; 1QAM A; 1G9T A; 2IYP A; 4HQN A; 4TL9 A; 1DOZ A; 1E8M A; 4P7K A; 4Y7D A; 5A6S B; 3VR2 D; 3K2E A; 3OZA A; 1VL2 A; 2HPZ A; 3LBA A; 4HAO A; 1KSH A; 4D3D A; 5C7L O; 2VYF A; 10MH A; 4E2Y A; 1W34 A; 1WE2 A; 1KH1 A; 4DUG D; 1NOJ A; 1VPT A; 2GA9 A; 4FMC B; 2HU8 A; 3GJR A; 1RD4 A; 2PRY A; 3SHO A; 4CL8 A; 1PX2 A; 1T3L A; 1S5P A; 2AJT A; 3TO5 A; 2WAM A; 4J9V A; 1Z0D A; 2GXS A; 3KZV A; 2OHX A; 3ZKW B; 1BMF A; 2P6U A; 1MKY A; 4YRO A; 4H2D A; 1FUY B; 4GRI A; 1W76 A; 4HWT A; 4YX9 A; 2PQ0 A; 2JBA A; 5JYD A; 3HU3 A; 3NIF B; 3WBJ A; 2Z6A A; 3KD2 A; 3ZVM A; 3BIW A; 4NFH A; 3C0K A; 5CXU A; 1E2E A; 4KLA A; 4HDS B; 3PXC X; 1GL3 A; 5HCN A; 1ORV A; 3HM2 A; 1Y7L A; 4JYA B; 1I7P A; 3E03 A; 4HTA A; 1TPY A; 4BKO A; 4X3C A; 4PG4 A; 2P7I A; 3GRK A; 2H1I A; 3P4U A; 1Y6Q A; 1YZG A; 1W4L A; 3KKE A; 2B4A A; 4TVB A; 3DHN A; 1E0K A; 3I12 A; 1HXH A; 2PD3 A; 4MJ3 A; 4C4X A; 1GA4 A; 4WBS A; 2JHG A; 5B7N A; 1M75 A; 1KWG A; 4COD B; 4HAV A; 3TI7 A; 1DLJ A; 1RU3 A; 2G1J A; 3F46 A; 3C7D B; 3H5R A; 4H7H A; 3P5S A; 4MIN A; 1F5S A; 3NDR A; 4CF5 A; 1L5H A; 1EUQ A; 2DFD A; 5AVR A; 3QYP A; 1H93 A; 2F43 B; 4PGA A; 3ZVK A; 1M1X B; 3SIP A; 5C1I A; 3H13 A; 2A2Z A; 1HFJ A; 2FMK A; 2X8W A; 2XZT A; 2YNM C; 1RYY A; 5DX8 A; 5HII A; 4OHV A; 4AX4 A; 4Q4A A; 2A0U A; 2H4Y A; 1XTZ A; 3DZD A; 1L1Q A; 5G41 A; 4OQ4 A; 5C35 A; 1XRL A; 3MUE A; 1RPA A; 3U37 A; 2Q3E A; 1T8R A; 2GLU A; 5HM6 A; 1CR6 A; 3T5P A; 1D2E A; 2AN5 A; 2X14 A; 4W1S A; 3LBY A; 2R25 B; 3L6O O; 1T5O A; 5FWN A; 4H1V A; 4DKJ A; 3JX9 A; 4GZ3 A; 4MV4 A; 1FNC A; 2AKE A; 1KO1 A; 4CKZ A; 2QMQ A; 5D51 S; 4AUN A; 5EOV A; 2ZVD A; 3UCX A; 2VED A; 2F90 A; 2WAX A; 5DGI A; 3JCZ A; 5BXY A; 1D2R A; 1Y3I A; 4Q9J A; 4C7X A; 4IXT A; 2PUB A; 4G67 A; 2F48 A; 1QYC A; 2ID7 A; 3SYT A; 1TT5 B; 2V9C A; 2C7B A; 1KEE A; 1VLV A; 1XLT C; 2FV8 A; 3EF0 A; 4QU0 A; 5T6B A; 3DEH A; 2VJM B; 2BW0 A; 4KLR A; 1RFL A; 1SVM A; 3HGM A; 1K2W A; 3L0P A; 2FPK A; 3CLT D; 4YJG B; 3UMF A; 1DQP A; 1QYI A; 3RAF C; 3PVS A; 4N8E A; 1CJU C; 3CGW A; 5I8Z A; 3KWL A; 1PFW A; 3JR7 A; 1J8F A; 1S9D A; 1OQA A; 1WZC A; 3D3N A; 3LSY A; 3HO1 A; 1YS6 A; 1YWG O; 1M3S A; 3ZHU A; 2G8N A; 3AVU A; 4HC4 A; 4MK4 A; 3LKV A; 2BMA A; 5A9R A; 3FFW A; 5ALN A; 1NBM E; 1E96 A; 2NUA A; 3IUN A; 3I83 A; 3FG9 A; 1JEY B; 4HLQ B; 4Y9S A; 3MJS A; 1RHU A; 2C4K A; 1E9F A; 2VNK A; 3EPP A; 1KZ7 B; 1BXZ A; 4IR7 A; 4Z9B A; 4D4K A; 2YY7 A; 3A4T A; 1NX9 A; 1ZGJ A; 1AZW A; 2JCS A; 4JNK A; 4R8L A; 3S40 A; 2DWU A; 2PR7 A; 3K35 A; 2O20 A; 2PN1 A; 2E5L B; 5E0K B; 1KI6 B; 1T9K A; 4BP3 A; 5HPY B; 2C8L A; 5EE1 A; 1EUD A; 1QFS A; 2G9V A; 2H5J A; 2ATV A; 4FT2 A; 4NML A; 5IAE A; 4UHB A; 4ZV7 A; 3T3G A; 5KQM A; 1WDK A; 1AA6 A; 1X1S A; 2Y0B A; 5EEH A; 1D2G A; 5CDY A; 1KVS A; 1C1H A; 2PGL A; 1U7U A; 2Q3L A; 1TC2 A; 1DUV G; 4GLJ A; 4WY5 A; 2VWG A; 1O96 B; 3U9T A; 4WKN A; 1BMD A; 2FEM A; 4LC9 A; 5ESW A; 1V47 A; 1Y37 A; 2YQZ A; 2QL9 A; 3ZHR A; 5JH7 F; 1RW4 A; 3GZN B; 2F64 A; 4LZJ A; 1J2E A; 3R6D A; 4QHT A; 3TPC A; 3OJL A; 3TR5 A; 1N5O X; 3IAU A; 4EY6 A; 3IEM A; 5JQG F; 1WUL S; 2EJT A; 2AG8 A; 4KVG A; 3H1A A; 2OBM A; 4LPK A; 1G3M A; 4QOJ A; 1NSY A; 2QNI A; 2X9G A; 3MTI A; 2RHL A; 3WBB A; 2DCM A; 3AET A; 2QVL A; 4NZM A; 5T8K A; 4BP8 A; 4YRQ A; 2ENU A; 5FPT A; 4RTL A; 1XJW A; 5TQ0 B; 1I4T D; 1Z28 A; 4J0Q A; 1FNM A; 2HK8 A; 2P9E A; 3CS3 A; 1DJP A; 4HEA 1; 3WLO A; 1BDJ A; 1P4A A; 1WW2 A; 4ETM A; 5B1H A; 1MO6 A; 3DXY A; 1MV5 A; 5F1P A; 1G20 E; 4E3W A; 3TFQ A; 4YCM A; 4Z0H O; 2R47 A; 4I1I A; 2UUC B; 1QCZ A; 4C3U A; 4WBN B; 1SU6 A; 3B20 A; 5KKA A; 4QTX A; 3FHL A; 2P5U A; 3O2S B; 2JAH A; 2P7H A; 4JSY A; 2D7C A; 1TEZ A; 3Q7P A; 1QHI A; 3UB3 A; 1VE1 A; 2Q9A A; 4CTM A; 3KSB C; 3KG2 A; 1D4A A; 4UCF A; 4P0Q B; 4N8S A; 1UDX A; 1VST A; 1N7H A; 1ZH0 A; 2GFH A; 2WFA A; 1T13 A; 1S2I A; 3B7P A; 3LWN A; 2ZIZ A; 3KUK A; 3R5K A; 3VDQ A; 4WAC A; 2BYT A; 1GGM A; 4GPZ A; 4Y2J A; 5DYW A; 2JI8 A; 2IED A; 2GF0 A; 3O93 A; 4HB4 A; 1EH5 A; 4B84 A; 3VR4 H; 1QV6 A; 4DR7 B; 4KXF B; 3PUY A; 2C0Q A; 3TSZ A; 2C20 A; 6LDH A; 5EES A; 2CDC A; 2ZE7 A; 4HNG A; 5F3R A; 1SCU B; 2REX B; 3QEL A; 3GB9 A; 4RC9 A; 3HCR A; 1IXZ A; 4WSO A; 1QLJ A; 3A3N A; 3IBS A; 3III A; 3CTM A; 1GGN A; 4L78 A; 4X00 A; 5DM0 A; 2J7P A; 2VUH B; 1IVS A; 5BVH B; 3TQT A; 3MJ4 A; 2B8T A; 3NAQ A; 3VH2 A; 4YV2 A; 5EZY B; 3OEE D; 5LKR A; 1U4S A; 3MEQ A; 4ETW A; 2Z09 A; 3GN1 A; 4P2B A; 3C3J A; 5JZV A; 1PUX A; 4HNA A; 4J1T C; 3WK9 A; 3C8M A; 1OSN A; 4MB6 A; 1SKY B; 2II6 A; 5JRO A; 2C9B A; 1CUW A; 4JDU A; 3T1T A; 4MVJ F; 3HWR A; 4JBL A; 4EGF A; 4PCA A; 1NJ2 A; 1FXU A; 3AP3 A; 4URF A; 3WBK A; 3RJM A; 3IBY A; 3TSO A; 4QQ8 A; 2E48 A; 1ZUN B; 2NSL A; 2VJQ A; 2RBE A; 4JT8 A; 4LVU A; 2XVX A; 4FUT A; 4MLD A; 2Q4E A; 4HZI A; 3SI7 A; 4EW3 A; 3J80 A; 3UTC A; 1JW9 B; 2XCQ A; 3MC1 A; 1WNT A; 1EU1 A; 1YE8 A; 3QZU A; 5HFJ A; 3M3D A; 2BFC A; 1SQ6 A; 2H7V A; 3R77 A; 5CYO A; 3G13 A; 3EBO A; 3QUS A; 1G6H A; 3BSY A; 2GTI A; 3LUK A; 3LVQ E; 4FZV A; 4N6N A; 4J7U A; 5C5E A; 3W06 A; 1CZL A; 2Z58 A; 2ORE D; 3DL0 A; 4MA5 A; 4EGS A; 1JDT A; 1ESD A; 3TW8 B; 2FAX A; 3EE6 A; 5JYC A; 1NM5 C; 2QX9 A; 2Y1W A; 4UBJ A; 5KVA A; 2VH1 A; 1GNV A; 3E7X A; 1AS2 A; 2G4L A; 2Y1C A; 1TU0 A; 3NM4 A; 2HRC A; 4FBM A; 1SRR A; 3I9M A; 3K2Q A; 3GEH A; 4DF3 A; 3FPC A; 3OZ7 A; 4NZN A; 4H1W A; 3UHO A; 1GG9 A; 1H23 A; 1TEC E; 2AXQ A; 4AJP A; 3DOO A; 1NVA A; 1QPG A; 2JPD A; 2XD9 A; 2HRZ A; 4JYB A; 2IUE A; 4O4I B; 4GQA A; 2VAX A; 2GED A; 1A8U A; 1OC4 A; 1XLV A; 4PNZ A; 5F77 A; 1FUE A; 3BOR A; 5AI0 A; 2JLR A; 1RCF A; 1GZ5 A; 2FPI B; 1RLJ A; 1SXJ A; 2H1W A; 2Q5L A; 4GOJ A; 1U3D A; 4P7G A; 3VZD A; 5KJF A; 3PCR B; 2J9Z B; 3U3X A; 3RNL A; 5KO2 A; 4Q3K A; 5SV2 A; 4UAV A; 2ZM6 B; 4K3V A; 1ILE A; 5KYV A; 1LPP A; 1UWL A; 5CM9 C; 2XUH A; 3AI3 A; 1Z45 A; 5EIB C; 5FUQ A; 2C3Y A; 3IUR A; 4JP1 A; 5EIH A; 5J4H A; 5KYU B; 2BWJ A; 2E49 A; 4Y2X A; 3CX4 A; 3GBP A; 3O9Z A; 1XD2 A; 2MJL A; 2QZJ A; 5E9Q A; 4PO8 A; 2YXL A; 3LX6 A; 1NN1 A; 3RVN A; 2LXN A; 3K8O E; 3ZXY A; 4YAJ A; 2C7Q A; 1JZT A; 3F47 A; 3QXH A; 1A5A B; 1YD7 A; 4UXI A; 3I2K A; 2FVX A; 2VJD A; 4DBL C; 2ZSJ A; 1ZC4 A; 1KR2 A; 4C1N A; 1POW A; 2W2V A; 3ACD A; 2MSD B; 1OX4 B; 4ZU1 X; 2DQY A; 2IP2 A; 3EVE A; 2VEO A; 3BAF A; 3F3Y A; 3K16 A; 2DT5 A; 3N8K A; 4YJF B; 3AKZ A; 1CJC A; 4KWH A; 4F9T A; 2R97 A; 2WJV A; 1GZD A; 3A11 A; 4PO1 A; 5D5P A; 4HVL A; 2X22 A; 3E3A A; 3K30 A; 1XEX A; 1UOE A; 3DDO A; 4UVD A; 3SE0 A; 3W9Y A; 1MZV A; 2ISS D; 4QFM A; 2POQ X; 3RT9 A; 4NTZ A; 1GV0 A; 3DWG A; 4A6D A; 1TT5 A; 4ZOE A; 3TDM A; 2B6N A; 3DLP X; 5AYV A; 5PNT A; 4XGR A; 4JI3 B; 3I6I A; 4LNU A; 4MUA A; 1JH8 A; 1SFR A; 3IIK A; 1QH8 B; 2PX2 A; 4TT8 A; 4EO4 A; 5AW6 A; 5BSK A; 3OX2 A; 4BN5 A; 5AOA A; 3G8Y A; 3FB4 A; 1C9M A; 1FFH A; 1TM4 E; 2QJR A; 2CZC A; 2X8L A; 3B3F A; 3GY0 A; 4J7X A; 4YCJ A; 4B87 A; 4DMH A; 3T3I A; 4EFK A; 4F21 A; 1YO6 A; 1EP2 B; 1ZC3 A; 3N2G B; 3QVG B; 4JUA A; 1W49 A; 2C4V A; 3AQI A; 4JSR A; 3G0H A; 1Y8A A; 2AC2 A; 4TL7 A; 1FP6 A; 1WB4 A; 2WWX A; 3U2Q A; 1B41 A; 1EWQ A; 2GHT A; 5G0U A; 2HCF A; 1XTP A; 3BE4 A; 4D42 A; 3WYG A; 4K9S A; 3JQB A; 4HMP A; 4Z4I A; 1JVS A; 3CGG A; 5LBI A; 5BYA A; 2H4F A; 4YR9 A; 4PC6 A; 1P62 B; 3SQD A; 1G8J A; 2H2W A; 3EAQ A; 2JCX A; 2FW7 A; 2QTF A; 5A04 A; 3LVB A; 1DEO A; 2B82 A; 1G59 A; 3DUL A; 5AW0 A; 4B3F X; 3LUF A; 3QG5 A; 4BRS A; 3HZR A; 4HWK A; 2JD2 A; 4NBU A; 1T3T A; 4A0H A; 4BVF A; 2VYC A; 5IN4 A; 1LRK A; 4LK3 A; 2CEK A; 4TKM A; 5BYJ A; 1U7O A; 1JDS A; 4RMI A; 4R2H A; 2YUI A; 1Q74 A; 2HF9 A; 2O50 A; 4KO2 S; 4TXL A; 3PEW A; 2OGS A; 3GCZ A; 4QBH A; 3DXF A; 4TV8 F; 5DGC A; 2GAS A; 3WV4 A; 3G02 A; 3GEI A; 2Q46 A; 3H7I A; 2EFH B; 3H2H A; 3B6Z A; 2AR0 A; 1TQH A; 3X44 A; 2JBH A; 1DNP A; 4ZDW A; 1G0O A; 4PYL A; 1F8U A; 4ZTU B; 1N2C B; 1JE0 A; 2QN3 A; 4I3G A; 4L8L A; 4M1M A; 3RAB A; 3O73 A; 3MSX A; 1IJE A; 4H6X A; 5BW8 A; 3OO1 A; 2UZ1 A; 1MF7 A; 1DKU A; 1NUU A; 4HGR A; 4L2I A; 4RF3 A; 3IBG A; 1NI4 A; 3TIG A; 4WLU A; 4EP1 A; 1Y8R B; 3I1J A; 1YQE A; 4OSP A; 1UZN A; 5DTQ A; 5JXJ A; 2NSJ A; 1YCG A; 1TM2 A; 2QYH A; 5HOE A; 4RD4 A; 3ANX A; 4LYJ A; 4JI1 B; 4P0P B; 3V34 A; 5DCW A; 4CM7 A; 3CMM A; 3ESR A; 4JI5 B; 2EFD B; 3SR4 A; 1XG5 A; 1X86 B; 2OFW A; 2ZU6 A; 4LHS A; 5L51 A; 1I94 B; 1M2O A; 1NRX A; 4TMX A; 3HU2 A; 3X2F A; 5CDF E; 4GL2 A; 4X54 A; 2XCL A; 3AUT A; 3FZN A; 3MWE B; 4QGB A; 3HGT A; 3B2Q A; 1ZEJ A; 1UPA A; 2IP1 A; 1OXK B; 2PX4 A; 1GQ2 A; 3OE7 D; 4F72 A; 2V0C A; 1KJY A; 2QWX A; 5AVU A; 3O4F A; 4WBN A; 2VZ0 A; 4BLU A; 2HLP A; 2HM7 A; 2PCL A; 1PO7 A; 2VP9 A; 5BQ5 A; 3WOV A; 4AJH A; 5IBQ A; 1H2B A; 3SX0 A; 3PJG A; 3WV5 A; 1B6G A; 3VHX B; 3BF7 A; 4FFV A; 3BB1 A; 3DDS A; 1XVT A; 1EG2 A; 2YS6 A; 1DQN A; 4A7P A; 1LSO A; 5AKK A; 3U19 A; 1K5H A; 1TMG E; 2AOX A; 2VJN A; 1WBJ B; 3MFY A; 5AVK A; 1TUB B; 2P0A A; 3U7J A; 1CZO A; 1WG8 A; 3BD7 A; 4NOL A; 3TNF A; 1CVR A; 4XOU A; 5E1B A; 4AF8 A; 1Y9J A; 5KI6 A; 2JAS A; 4G8J A; 4CKC A; 5CJJ A; 2ZYI A; 2R60 A; 2Z8Y A; 4L4Z A; 1PSZ A; 2I91 A; 3PI6 A; 5H5J A; 1AH2 A; 2YHB A; 2IEY A; 4M83 A; 4NAH A; 3HUF A; 4RH1 A; 5CUR A; 2CV1 A; 1Q17 A; 1NF8 A; 2WIJ A; 2YYS A; 3QW2 A; 3A9C A; 2QZZ A; 4HEQ A; 2PKY X; 1F06 A; 1GRN A; 2QX7 A; 4AUL A; 3IFS A; 4KX3 A; 5BV1 A; 1NG1 A; 3BMQ A; 2FN7 A; 4PSD A; 4A6E A; 5BN3 A; 4BXO A; 1Q20 A; 1E1M A; 4JQS A; 2BJW A; 3KZX A; 3KTF A; 4YUX A; 5EQT A; 3NBU A; 3AV3 A; 5JPC A; 4G1U C; 1XCL A; 4CQL B; 4ND5 A; 3GJ4 A; 4MYH A; 1ZUN A; 5LYJ B; 4UAR A; 3L6B A; 5E7Q A; 1R6D A; 1TZM A; 2QT0 A; 3UAX A; 1JRL A; 1SUW A; 3OEC A; 1Y5I A; 4TM8 A; 4FMB B; 1ECJ A; 3PQD A; 4NV1 A; 2PZG A; 4JGI A; 2B1J A; 2WJZ B; 4NNJ A; 3SBD A; 2V7E A; 3DM9 B; 4UTN A; 2NU6 A; 4XI9 A; 3JS5 A; 3AH8 A; 4IDO A; 4JA2 A; 4ZVO A; 1X80 A; 4ZVM A; 4ZC8 A; 1QVZ A; 4A9A A; 5K0J A; 2YY6 A; 2DBY A; 1QG6 A; 3C1A A; 4LDJ A; 2YVK A; 5ALH A; 2O8E B; 1UQU A; 4F9E A; 2J33 A; 2Z2J A; 5GM2 A; 1M83 A; 4Y2Y A; 1I7S B; 2OG2 A; 3NKL A; 1FP7 A; 4KWT A; 3ZDE A; 5IH3 A; 3VZQ A; 3N75 A; 4O4H A; 3ZS8 A; 3TZE A; 3EIX A; 1RLI A; 2HB6 A; 2J9G A; 5LDD C; 4EYS A; 3EA0 A; 3DHV A; 3KZC A; 2NQ8 A; 4PC7 A; 3BRQ A; 4XTR A; 2X0I A; 5LP6 B; 4R2L A; 3P9S A; 1HA3 A; 3C15 C; 3D7S A; 4OQD A; 5SYM A; 4XGQ A; 3LHS A; 1UFR A; 3CC8 A; 2QU8 A; 3PPX A; 4O4I A; 4Q3L A; 2XD4 A; 5FHR A; 3CR7 A; 2XHZ A; 3DMH A; 3E27 A; 2WHR A; 3S2U A; 3EFB A; 3K0H A; 1I3O A; 2R1V B; 5CLF A; 2Q9B A; 4RY8 A; 3S2E A; 1PD1 A; 5C53 B; 1W73 A; 1UJ4 A; 3EWS A; 3R5F A; 3QXS A; 2IUS A; 4CG2 A; 1MW8 X; 2HQO A; 3OJE A; 2VPW A; 1FMF A; 5ALS A; 1BKY A; 3SMV A; 4RKW A; 3T7T A; 2P4H X; 3PVC A; 2CL0 X; 2P9Z A; 2W0M A; 2WJA A; 2D5I A; 3DOC A; 3H9J A; 5AM1 A; 5KYU A; 1LZL A; 5BUC A; 1IPH A; 2BHS A; 4BQI A; 2CLC X; 4GLF A; 1O95 C; 5THX A; 4DSA A; 5L3Q A; 2WQ6 A; 1YZU A; 4U9U A; 4YUW A; 4EA6 A; 3HEM A; 4EW6 A; 2ZJ8 A; 3QJ5 A; 2Q62 A; 3OFN D; 4XRG A; 3PTA A; 1GL7 A; 1JQK A; 2IYY A; 3TTS A; 1M7B A; 1T4G A; 5KTP A; 2E4Z A; 1JG6 A; 1PVD A; 1V0J A; 3EQ6 A; 4IF4 A; 3WC4 A; 1CUI A; 3JAM A; 1T36 B; 2K2Q B; 3PKI A; 1ZGG A; 4IJW A; 5ALL A; 5SZJ A; 1EFC A; 4QQK A; 3T5T A; 2GCO B; 3UT5 A; 3WST A; 1JDJ A; 1K6X A; 1XEW Y; 4B4D A; 3FFT A; 4CM9 A; 2IHY A; 2V3W A; 2G1U A; 5JSY A; 2JIZ D; 5BNW A; 3WLL A; 3ZGY A; 1IOW A; 1U7H A; 1V1R A; 3F74 A; 3T0F A; 3MT8 A; 5A4K A; 4IRU B; 3DFI A; 5LMC A; 1W0J A; 2CMG A; 2DFT A; 4B0O A; 1Z16 A; 1FDO A; 4C4A A; 4KI7 A; 3M6W A; 3NZE A; 5TV2 A; 2CXT A; 2NT4 A; 1RXE A; 2P3S A; 2VRC D; 2XLC A; 4N5I X; 4RV4 A; 4XOI A; 3RYF A; 4M55 A; 5KNC D; 1CUS A; 2IUF A; 4Y9T A; 4LZL A; 1BE6 A; 1D07 A; 3D64 A; 1FTW A; 2IYX A; 2QXY A; 1RMQ A; 1NBI A; 4OOE A; 5KYT A; 1EP1 B; 4HGO A; 1E79 A; 2CEA A; 3WFJ A; 4JZY A; 4CE6 A; 3CNE A; 3NOV A; 2AT1 A; 4WFI A; 1F48 A; 2FW9 A; 2QTW B; 2W7T A; 2WUV A; 1K2V N; 2VP3 A; 4YUV A; 5IBP A; 3VH4 A; 4CET A; 2LDX A; 5D3Q A; 1JFV A; 2FPO A; 3Q6M A; 5L3S A; 1QTN A; 1M72 A; 1E3M A; 4D3F A; 2QYS A; 5LJI A; 1N92 A; 2QM3 A; 2ZYW X; 3M5W A; 3PHL A; 4PPF A; 1FSS A; 3UCF A; 4DLS A; 2OG1 A; 3T3N A; 1BYI A; 4GMH A; 1ADG A; 1W59 A; 1YI1 A; 2YOG A; 3BXZ A; 2WD7 B; 2HT1 A; 1VJ5 A; 1AK9 A; 4ND6 A; 4C0X A; 3UMB A; 5SZI A; 2GLT A; 2Y8E A; 4ZO4 A; 3U4C A; 1WW3 A; 3ICO A; 4JXZ A; 2RGB A; 3MRS A; 4DPL A; 5C32 A; 3E5R A; 1YAJ A; 1CDD A; 2JH9 A; 3KYC A; 3KR9 A; 4ZVK A; 2WWG A; 4Z8Y A; 1WF3 A; 5AHO A; 5HTK A; 5T6M A; 5EJ4 A; 1AKU A; 3SY8 A; 1EGA A; 3WK5 A; 3ZY6 A; 1KS2 A; 2CWW A; 5KOJ B; 1E1R A; 3GAF A; 4G5G A; 3H5O A; 5UKD A; 4KQJ A; 1Z3I X; 1WRB A; 5FWA A; 4L08 A; 5IC5 A; 3V8R A; 5EZS A; 2PLA A; 1YQQ A; 13PK A; 2F8M A; 1C3O B; 2G7M C; 3OTG A; 1PS8 A; 3PHH A; 3RJ9 A; 1GO2 A; 1ORR A; 4F5Z A; 2JKV A; 1J25 A; 1SXI A; 2HO4 A; 3UT6 A; 4RIF A; 3O47 A; 1JHU A; 3FZQ A; 2HXY A; 1ECG A; 4TMY A; 4Y9U A; 3ONW A; 2IKS A; 3ZH9 B; 1NR5 A; 1WLY A; 1NN0 A; 4OAO A; 3TUJ C; 4R68 A; 5F1Y A; 1XXH E; 3RT1 A; 4NAR A; 5HWS A; 1GG5 A; 5CRI A; 4DAN A; 1X3S A; 1PJT A; 1SZC A; 5A9S A; 2B2C A; 1RXY A; 1HOX A; 3MWE A; 2EKZ A; 2HA0 A; 2SEC E; 4GNR A; 3L04 A; 3TAH A; 3ZWX A; 1W36 C; 3K9Q O; 5JNU A; 2DFK B; 4YT8 A; 3TZL A; 2VOE A; 2CMK A; 1K4Y A; 5DT2 A; 5AW4 A; 3DYB A; 2QR2 A; 3BGI A; 3AEK A; 1QQA A; 3VSE A; 2JIB A; 4MR8 A; 3W5U A; 1DMS A; 2FQW A; 1N6H A; 2V97 A; 4QYS A; 4Q9I A; 4WVF A; 5FJJ A; 1D0V A; 4WY2 A; 3SF8 A; 4BOY A; 2V9P A; 3IPC A; 5B6J A; 1EBU A; 1ST3 A; 2AHR A; 5CVD A; 5A3V A; 1TQ2 A; 4TT3 D; 3HWW A; 1HE5 A; 1S1C A; 1T8S A; 2WM8 A; 2Z30 A; 4LC0 A; 3QIA A; 2Q6K A; 3T4U A; 4MR7 A; 4GWL A; 3FUU A; 4GR5 A; 1UWV A; 1PT9 A; 4S0X A; 3FB1 A; 4MRO A; 4Q9N A; 2YBE A; 4GPA A; 2AK2 A; 3B85 A; 2CLF B; 4CBV A; 2R86 A; 3CLS D; 4I50 F; 3PNQ A; 2OBL A; 2WHP B; 3BXX A; 3CV3 A; 4RS4 A; 4XH9 B; 1ENY A; 2MIN B; 5E3J A; 1KAM A; 5FFM A; 4NXN B; 5CK4 A; 4EEK A; 3MTB A; 1XEF A; 1X1Q A; 3TLX A; 3ZJ5 A; 1W58 1; 2XE8 A; 5HIA A; 4PFK A; 2F8T A; 4PZC A; 3PZR A; 4NX2 A; 1T4D A; 2X0K A; 3D1V A; 4MCJ A; 4AMZ A; 3WL8 A; 5DNV A; 5J5C A; 4DEX A; 1IJK A; 4IJQ A; 3T7D A; 3CEH A; 1GAQ A; 3WHP A; 2XXA A; 3LBF A; 2NU9 B; 4OJN A; 5EP2 A; 3ZDA A; 1WHS A; 3T3H A; 1FBN A; 3FRI A; 1E2P A; 2M98 A; 4IUB S; 4EJS A; 1E6L A; 1KNQ A; 4KYT A; 1AUO A; 2AD1 A; 5E7I A; 5FQN A; 3WWD A; 3Q6N A; 1THG A; 4IH9 A; 4JP3 A; 2C3U A; 2QTV B; 1AQ6 A; 4UEK A; 3BWY A; 2QIO A; 3ABS B; 3ETE A; 4LBW A; 3OM9 A; 1X7Z A; 2ZE6 A; 5J59 A; 4P53 A; 3USZ A; 4DR1 B; 2QBY A; 3BK1 A; 3BUL A; 3DVA A; 4WA8 A; 2IS4 A; 3WK4 A; 1IN6 A; 4GXQ A; 1I13 A; 5TME A; 1PJB A; 4IG4 A; 4I66 A; 1KHT A; 4JN6 B; 3ZE6 A; 2CJW B; 4PAG A; 4WW0 A; 2PSD A; 3IVX A; 2YZ7 A; 3BWX A; 3MII A; 4CTN A; 2ZSU A; 1FQW A; 2RKB A; 4ZOL F; 3EA5 A; 1U0E A; 5ECD A; 4UEW A; 4XD2 A; 1L8Q A; 2VFA A; 5HJ2 A; 4BKP A; 3T95 A; 3DAG A; 3EEB A; 1MPT A; 1RII A; 1SVI A; 1N4A A; 1CS0 A; 1FTN A; 1G98 A; 5KYN A; 1Q1G A; 1DXH A; 1HLG A; 1Y4Z A; 4B47 A; 1X39 A; 4H59 A; 1E2K A; 4DIK A; 5EJ7 A; 1UAN A; 4IIG A; 1T8U A; 2H2G A; 4RCY A; 2M2I A; 1L0B A; 3Q7R A; 1RXC A; 3LBI A; 4QFD A; 1JEQ A; 2D5A A; 1ODK A; 2FDX A; 4IDA A; 2ZYT X; 3MOG A; 4GHW A; 3OCV A; 3KB1 A; 4IZ6 A; 4ZVL A; 4Q4L A; 3UCE A; 5KTS A; 4TLC A; 5KND H; 3WDL A; 1X42 A; 4MZD A; 4YP9 A; 1FP4 B; 4KK3 A; 3ROK A; 4AG3 A; 4EQ6 B; 3R75 A; 1RWV A; 2ZRI A; 1PD0 A; 1SZJ G; 4M0D A; 2BHT A; 2J5B A; 2ONK A; 5B37 A; 2NPF A; 1A9Q A; 4NIM A; 3PUJ A; 1GTR A; 2BGG A; 1OAK A; 3ASY A; 1DEA A; 5FV7 A; 5KCM A; 3HK2 A; 1GAX A; 5DLE A; 3MG9 A; 2C5E A; 1ZRN A; 1OPX A; 1HDI A; 3GEB A; 3M4Y A; 1KDR A; 4AP5 A; 4D8U A; 5KNB H; 1U1F A; 1PS0 A; 4F0J A; 3ZZ1 A; 5JH7 A; 3AFN A; 3GLV A; 2CK3 A; 3QX8 A; 1G8S A; 4MRR A; 4CEJ B; 3ZDF A; 5GMO A; 4IWN A; 4EAT A; 3VTK A; 3CZ5 A; 1M34 B; 5JQG A; 2QU7 A; 3PUV A; 3O4H A; 4CMA A; 4YRR A; 3DXX A; 5G1N A; 7ABP A; 2ODB A; 4RKF A; 1ZR2 A; 4QYM A; 3EZG A; 4PG8 A; 1FUU A; 4PYM A; 2QSZ A; 1Q0U A; 4XAQ A; 4G63 A; 3NAG A; 4TZ6 A; 2OYR A; 4LYG A; 5DU2 A; 3I9V 6; 4NAB A; 3UK2 A; 1N6L A; 3AR9 A; 2GHI A; 4JEI A; 3GXB A; 3VR6 D; 4V1C B; 1G8P A; 1EUY A; 2YJZ A; 2ZQY A; 4OX9 B; 2OEP A; 3HU5 A; 4GOK A; 5EI5 A; 2RHJ A; 4ES6 A; 1ZAK A; 3EPK A; 2D7J A; 1OPR A; 4PLC A; 2H17 A; 4J7C A; 5ALK A; 4LV6 A; 1YF3 A; 2FB9 A; 2IE0 A; 3KEM A; 5GPB A; 2R7N A; 4EEN A; 4UTO A; 1AZS C; 2RMK A; 3EB9 A; 3Q3V A; 1L5V A; 3GLI B; 5DCY A; 2GSD A; 3DWI A; 1ZP6 A; 4RD6 A; 1W96 A; 5G0V A; 4K9M A; 1F3L A; 1KZ9 A; 1E66 A; 3EXH B; 2E4U A; 5F9M A; 2LW7 A; 2R79 A; 4UVG A; 4RDX A; 4EFM A; 2IUT A; 3KJN A; 1BE1 A; 2EKQ A; 4FON A; 4H0F A; 5F76 A; 3H1F A; 2PUG A; 2YFK A; 4N54 A; 3LY9 A; 1EEH A; 1A9P A; 5CHY A; 5AI8 A; 3V4O A; 4QTY A; 2ARK A; 5JS7 A; 3LWB A; 1K0F A; 2CHE A; 2GJA A; 2VYN D; 2JEO A; 2J45 A; 4MLP A; 5ESX A; 3DYI A; 1GKU B; 3ZW7 A; 3GAZ A; 5DHQ A; 2ZAY A; 4I1P A; 4EIW A; 4JKO A; 3LI2 A; 1XCO A; 4PYJ A; 1TCC A; 1VG9 B; 1PKE A; 3ZV6 A; 3EOD A; 3KP1 A; 4DS3 A; 2RJO A; 5DTE A; 1EK2 A; 4J94 A; 1P9L A; 4EI9 A; 4CL2 A; 1NLZ A; 2C5S A; 5E6U A; 3EN0 A; 1XDZ A; 4HLN A; 4QG7 A; 2O5V A; 3UB4 A; 1NAT A; 2CND A; 3ZCS A; 3EVG A; 1S2N A; 4EKE A; 4BPB A; 3PRK E; 1CWU A; 3ROE A; 3TIF A; 2W70 A; 2P9B A; 5I7A A; 5CEJ A; 2I1V B; 3R4C A; 1N2S A; 1NI3 A; 1MCZ A; 3C97 A; 5EVJ A; 4YNB A; 5C37 A; 3OZI A; 2GBF A; 1ZO8 A; 1UT5 A; 2CJP A; 1EAY A; 2FWA A; 3K15 A; 1E2Q A; 3DDW A; 4WXR A; 2HRQ A; 4JCO A; 4WPG A; 1BCR B; 4E46 A; 5BWC A; 2HR1 A; 3ZTT A; 1K3T A; 1GT6 A; 2V7T A; 2WVA A; 3TM5 A; 3QWB A; 4ZZ2 A; 1W1I A; 3KVV A; 1W35 A; 2ZDG A; 3GMS A; 3HUT A; 3S1L A; 4MW5 A; 1XDW A; 2O8E A; 3V00 A; 1KBQ A; 2QR3 A; 4RD3 A; 4IBO A; 4AD8 A; 2G76 A; 1FDS A; 1A7J A; 2UAG A; 4LKR A; 3C43 A; 4NYI Q; 4G5O A; 3RLI A; 5GOZ A; 1T9C A; 1XR3 A; 2AUN A; 3ABQ B; 3D2L A; 3L0Z A; 4MTL A; 4OHM A; 4CMJ A; 2D00 A; 3BTS A; 5CX1 B; 1LU9 A; 3ENK A; 3QIV A; 1PN3 A; 1TZC A; 1PLJ A; 2ID8 A; 1LJU A; 1U2R A; 4C4Z A; 3QHP A; 3PUH A; 2HCT A; 5K45 A; 3BJM A; 3GFQ A; 3U1T A; 2BTE A; 1TC1 A; 3HFS A; 1L5Y A; 1SG0 A; 1RAE A; 4YI3 A; 1Z6G A; 1BMT A; 2XQI A; 2YV2 A; 3CZQ A; 4RVC A; 3U4D A; 4F45 A; 4KDC A; 5K7S A; 2G4C A; 2G9R A; 4P98 A; 1X8X A; 4URY R; 1YAS A; 3WGM A; 2VAT A; 3QEB Z; 4BHT A; 2QHX A; 2ONZ A; 3V2U A; 5HZM A; 4IYJ A; 4U02 A; 1MZP A; 5FTL A; 1MAW A; 4OY3 A; 2Z3V A; 4DAO A; 1U1I A; 2HAD A; 5JR3 A; 3U1B A; 1KI9 A; 2VC2 B; 1HO3 A; 1LTQ A; 1W29 A; 4GMF A; 4PFQ A; 5A5D A; 3K70 D; 2WFL B; 1NWC A; 4O2B F; 2QLF A; 3I0Z A; 3W1V A; 5AUN B; 3KQK A; 4PX5 A; 4B76 A; 1Z83 A; 4AZT A; 1XCG B; 2Q6I A; 1EXV A; 1M1Y B; 3V4L A; 1AC5 A; 3ZYZ A; 2BFF A; 1SXQ A; 4YT2 A; 4NP6 A; 3N2L A; 2PKQ P; 4ARU A; 5KPD A; 1P17 A; 4EDH A; 4HH3 C; 4M1T A; 4XM2 A; 5EJE A; 1PR9 A; 2B1Q A; 1LLD A; 2CN1 A; 1IK6 A; 8AT1 A; 3O31 A; 1FFG A; 5FWD A; 4B5L A; 3UBT A; 2C95 A; 4CKB A; 1ZJ4 A; 2FUN B; 1RQN A; 4OLA A; 4B82 A; 5J58 A; 5EWM A; 2RHR A; 5JS8 A; 5KQL A; 5K8I A; 4Y30 A; 5CYG A; 1E2D A; 3OQN A; 1DC8 A; 1H4Q A; 5L3V A; 4UU1 A; 1TF5 A; 5JCO A; 3OCJ A; 2I3A A; 4DGS A; 3ML1 A; 2IV3 A; 1W4Z A; 4KZP A; 1TL7 C; 5K0F A; 1Y9Z A; 3H2Z A; 3O3M B; 2CJW A; 4PTL A; 3P26 A; 2ONC A; 3G2N A; 4GPI B; 3LW7 A; 4DID A; 1XTJ A; 4LCB A; 3OU2 A; 2G17 A; 3K5P A; 3MB5 A; 4Q7J B; 5EIW A; 2XB9 A; 2P4E A; 3FM9 A; 3QWI A; 4HQ6 A; 1EIN A; 5C8Y F; 2H52 A; 2I6X A; 4FFL A; 1H3F A; 4J9U E; 1YS1 X; 3IOB A; 2HIG A; 2ID4 A; 1ZLY A; 2BGK A; 1JAX A; 3QKR A; 3BWD D; 4EG8 A; 2OCG A; 2EER A; 1HRD A; 3MJT A; 4Q02 A; 3STU A; 3I23 A; 7ADH A; 5H4G A; 1QIM A; 4FCC A; 1LSU A; 2HO5 A; 1NKT A; 2P4S A; 1WMF A; 1XA0 A; 4S0Z A; 2O07 A; 5DTR A; 2FZK A; 4MVX A; 1P4H A; 3PIC A; 4E22 A; 2G70 A; 1SPB S; 2JGR A; 4M7O A; 5JCX A; 1ZWL A; 3X2E A; 4B71 A; 2PJU A; 2B4K A; 4XM1 A; 1F4P A; 1Z8H A; 3ABO B; 2HQ1 A; 3T4P A; 3PC4 A; 2WSS A; 1GNS A; 1VBF A; 1FP4 A; 1EDE A; 2V6J A; 5IEB A; 2ZGV A; 3NNS A; 3FPG A; 2B1I A; 1OV4 A; 3MBI A; 3FCC A; 2P2Y A; 3FPS A; 1CHN A; 4P5D A; 5TII A; 3CR3 C; 5IAA A; 4TV8 A; 4EQ6 A; 3ZDB A; 2OUK A; 3E2I A; 4EB6 A; 3EQ7 A; 3TM4 A; 1DXO A; 2IXG A; 4M0E A; 5COA A; 5JZ9 A; 4AKG A; 1IDN 1; 1AGY A; 3JD1 A; 1K7F B; 1SOU A; 3OOH A; 5CM8 B; 4BBP A; 7REQ A; 2FK8 A; 1AQJ A; 2F67 A; 3PZB A; 1VRW A; 2F9M A; 2ZZK A; 3GLT A; 4OXN A; 5EML A; 4AS1 A; 4PR3 A; 4FUQ A; 4L8V A; 1ESE A; 3LT5 A; 4O25 A; 4ARO A; 2OLK A; 4R9R A; 5KPJ A; 3ZDZ B; 1M67 A; 1EG7 A; 3Q85 A; 2QPO A; 1KBZ A; 2TSY B; 2XEX A; 3GJ6 A; 4OLH A; 1C8L A; 4LCE A; 3A14 A; 2GBG A; 2A7X A; 4Q7K A; 1E6W A; 1ICE A; 3GQB B; 3LYA A; 2XKB A; 1K6M A; 1ZCA A; 1KI8 A; 3PK0 A; 4M22 A; 1OFU A; 3MMS A; 4D9N A; 1O54 A; 3NIW A; 4WT7 A; 3WAD A; 2NAC A; 3OCC A; 3A6E A; 1RYM A; 5DGH A; 4JBI A; 4IDC A; 3AEU B; 1U6I A; 1ZBD A; 3CSS A; 1O4V A; 4PLV A; 3V6L A; 3Q8X B; 3TFW A; 3ZCT A; 2VSN A; 4RL6 A; 4BNM A; 4D9B A; 3QBD A; 2W6C X; 5DRT A; 5FA6 A; 5K8J B; 2GCG A; 2ZVP X; 1ZMX A; 1E1Q D; 3ICW A; 2FPP B; 4XCQ A; 2V6I A; 3SM3 A; 2Y0E A; 3I04 M; 3NTV A; 3CY6 A; 3GGC A; 3EXH A; 4OFX A; 4WOJ A; 5T57 A; 1NE7 A; 5LD2 C; 1HFE L; 3RT7 A; 1ACM A; 1A2Q A; 4I7E A; 3LX4 A; 2IYA A; 5F2O A; 4ZOC A; 1YA6 A; 1P7Z A; 1SCJ A; 3BRW D; 2B7C A; 4KMS A; 2J3J A; 1UQT A; 3KQP A; 3N0T A; 3UKO A; 4CL3 A; 4ENU A; 5LQC A; 2R0Q C; 3DS8 A; 4KGV A; 4KRY A; 2CW5 A; 3AWD A; 4YAU A; 1CQX A; 1G7S A; 1YJC A; 3CSU A; 1V1M A; 4QAR A; 4X1Y A; 3OQP A; 4O1M A; 5IK2 A; 4AB1 A; 4WJL A; 2J3H A; 5FF9 A; 4D9E A; 3HN7 A; 3VR5 D; 4HH4 A; 2CLE B; 3H1B A; 1A77 A; 1UBH S; 2CE2 X; 3QBT A; 1G6K A; 4KLN A; 1NW4 A; 3G5U A; 3HBN A; 1YVD A; 4Q3E A; 3IIL A; 1PWY E; 3ROF A; 3X1Z A; 5D3Z A; 3P71 T; 5DP2 A; 2BRU A; 1TZJ A; 3LXX A; 3C3X A; 2DJI A; 2IXF A; 5ALY A; 4M38 A; 2B35 A; 3S1A B; 4DLT A; 1G2O A; 1GZ3 A; 3GUC A; 4TMW A; 4E30 A; 4UAS A; 4EJM A; 2DC1 A; 3OD1 A; 5FT9 A; 3RTC A; 1OLU A; 3E48 A; 3BIF A; 1D04 A; 1GN9 A; 1I6M A; 3DMT A; 4MYD A; 1Z18 A; 1QAN A; 5CPF A; 5G40 A; 5IT0 A; 1WSA A; 2JGM A; 4IQF A; 2W2L A; 4B4U A; 3GWZ A; 4BEJ A; 3VBE A; 1R5N A; 4DR5 B; 2CXQ A; 3PGL A; 1SXG A; 2F2T A; 2GUG A; 1V8P A; 3VSC A; 3STT A; 5CFR A; 5CFO A; 3FMP B; 1O6B A; 2VJO A; 4YMW A; 1M6H A; 2ASV A; 4V0K A; 3ZXI A; 2PR2 A; 1YC5 A; 4O1I A; 1DCT A; 2P8S A; 4KU4 A; 3QVS A; 1R10 A; 5JCZ A; 3C3M A; 3GV0 A; 2QTT A; 3W8R A; 3ZHQ A; 2C8V A; 3OKC A; 3GHY A; 3JAR A; 3MUO A; 4PN7 A; 1RS0 A; 4BE7 B; 4AJ2 A; 3G9U A; 2IPN A; 2AQI A; 5AT1 A; 4JST A; 4PYQ A; 2YWB A; 4NYM Q; 3A3P A; 2ZB2 A; 1QMG A; 4LXX A; 4X63 A; 3LX8 A; 2C2K A; 1ATI A; 5EMH A; 3DTV D; 2ZS9 A; 3T9B A; 4ROR A; 4PN3 A; 4GXR A; 4ZXP A; 1JYE A; 1PJA A; 1RZV A; 4KBG A; 3SEQ A; 5JWO A; 1Q0Q A; 2H2H A; 1X9I A; 3O9M A; 3PE6 A; 3PNM A; 4G5Y A; 5E6R B; 4EPR A; 4K9P A; 4OUD B; 3KYD B; 1E8D A; 1N34 B; 1JHZ A; 3U26 A; 1X7D A; 2FEW B; 3HRI A; 1H65 A; 3HY3 A; 2DT8 A; 3R0I A; 1QAQ A; 1N9Z A; 3OQV A; 3R0Z A; 1YDM A; 4XYE A; 2H1C A; 2CF5 A; 1PHR A; 4L0D A; 2ZB7 A; 3PP8 A; 4CF7 A; 2W6N A; 2ZKI A; 2UYH A; 3U4Q A; 1ONB A; 2ZRE A; 3AVQ A; 4UTX A; 2PVS A; 1M66 A; 1U80 A; 4FCT A; 5B6I A; 4QEC A; 2YZG A; 3LK7 A; 2OPH A; 1QR6 A; 4NZY A; 3REF A; 4G1V A; 1ZX1 A; 3MMN A; 3U3D A; 2DWO A; 4GKJ B; 3JAS A; 1QHN A; 3C1T A; 1LF0 A; 5LIP A; 4X90 A; 3IOD A; 3RRJ A; 1PR3 A; 2I62 A; 1XA4 A; 3JTM A; 1YMV A; 5SIC E; 4C06 A; 1JE1 A; 4KVZ A; 2QTR A; 2ZBU A; 1Z2P X; 2ILT A; 5KMG B; 3TH5 A; 1ORD A; 4RLH A; 4RAB A; 3U4A A; 2WT9 A; 1UZU A; 4MQF B; 3C5H A; 2GEV A; 2Z8X A; 2R6F A; 4J6F A; 2BR4 A; 3AII A; 5ES3 A; 1CUJ A; 1RQL A; 1JEE A; 2BEL A; 2AVN A; 1ADJ A; 2CY0 A; 2VJA A; 4RM3 A; 1KQO A; 3UMG A; 5K6R A; 1TCA A; 3I1F A; 5I48 A; 6Q21 A; 3DOU A; 4HGN A; 1BQ4 A; 5D1H A; 1JDU A; 2QW0 X; 4IHJ F; 4UNN A; 2VK1 A; 3ZCU A; 3ZD6 A; 5HR5 A; 4HGZ A; 2BL4 A; 5CU7 A; 2GM3 A; 3H2Y A; 3HHP A; 3NHY A; 1X77 A; 3SHT A; 2GPN A; 3TVT A; 1E2G A; 2X1L A; 2CNN A; 5P8W A; 1V8F A; 1BMF D; 3BFJ A; 3L1K A; 1UKZ A; 3MLA A; 4M1K A; 4YP1 A; 2EZ4 A; 1QCY A; 1ORE A; 1LPO A; 3I37 X; 3RUP A; 3BWV A; 3PTZ A; 4ATN A; 1SHZ A; 5JD5 A; 1XTQ A; 4HUJ A; 5LBU A; 4Y8V A; 4BUZ A; 1AHI A; 4MCI A; 4TLE A; 1PK9 A; 1MAU A; 4X7R A; 4CV0 A; 1IDO A; 1RHM A; 3Q8W A; 2A30 A; 1J09 A; 2XAA A; 3OP4 A; 4YMS A; 1XTR A; 1ZN8 A; 2JHF A; 5FO1 A; 2W2Q A; 1R00 A; 3TIV A; 4IIL A; 1IHO A; 3NAM A; 2D0I A; 2HSZ A; 3CX3 A; 3KVY A; 4MR9 B; 3TB6 A; 3TMK A; 3ZI4 A; 4CQJ A; 3JYO A; 1ML3 A; 2FN9 A; 4GG1 A; 3AXS A; 1QK3 A; 4J1S A; 3TZH A; 1A1S A; 1MHP A; 3TB4 A; 3PXX A; 4WOP A; 1PQP A; 3T1Y B; 1C7J A; 2B9E A; 1OF1 A; 3ND7 A; 3PFK A; 3QA3 E; 2EJW A; 3H42 B; 4Q7M B; 4HPX B; 4BP9 A; 3SJC A; 5TB5 A; 2AI3 A; 3JAW A; 3LUJ A; 3EHE A; 1BU8 A; 3G68 A; 3VNV A; 4MVJ P; 1RAF A; 1ZVJ A; 4U7D A; 5TMB A; 1X2B A; 2D1S A; 3FDZ A; 4FKZ A; 5H1Y A; 3RFV A; 3TNY A; 1JR3 A; 4M49 A; 1MG5 A; 2O5R A; 2BFD B; 2QUZ A; 3QWF A; 4NEQ A; 5JSZ B; 1Z0A A; 1KQ3 A; 1QBG A; 5CDU A; 2BT4 A; 4CMI B; 4P34 A; 5KYW B; 1OB5 A; 3O83 A; 4BMV A; 3P5B A; 5FPK A; 1DX6 A; 3FFX A; 3OEW A; 4U39 A; 3U7F B; 2B92 A; 4I0W B; 1FLA A; 1GCA A; 3V3K A; 5K0G A; 1TVK B; 2OE2 A; 3A6G A; 4K87 A; 2PAF A; 3KU1 A; 4UCQ A; 4ZZ0 A; 2X0R A; 1NNE A; 1CF2 O; 2LIV A; 1OIX A; 3NUG A; 2JGD A; 5MFP A; 5CCE A; 2XZO A; 5CNJ A; 2HW8 A; 4C88 A; 2L18 A; 4PEV A; 2PUE A; 1WL8 A; 1KJI A; 3NM6 B; 4KYU A; 2WIK A; 3OHP A; 1FDW A; 4QOG A; 2VF2 A; 4RE6 A; 4R84 A; 1XAL A; 4G01 B; 1H9C A; 3K0A B; 4CM8 B; 1CQP A; 1M8K A; 4ZEO A; 2C0P A; 3I4F A; 4IDE A; 1SM4 A; 2HY7 A; 1IN7 A; 1HQC A; 1LRL A; 1ZJ5 A; 2WPD D; 2RGX A; 4N7B A; 3AR4 A; 1K92 A; 2D2V A; 1F14 A; 2AWO A; 1DSS G; 2IHT A; 3BK2 A; 3ESB A; 3K6S B; 4RHY A; 1LQU A; 3UA3 A; 4JL6 A; 1UWC A; 3L3B A; 4N18 A; 1Y81 A; 4JQZ A; 1X7W B; 3VPB A; 2NZJ A; 3WLJ A; 3LHK A; 4TKV A; 2VR1 A; 4ZCI A; 5BP7 A; 3PPI A; 4KDR A; 1FFW A; 3ENV A; 4C2B A; 2AW5 A; 3RTA A; 1CBU A; 4E5N A; 4F86 A; 1OR8 A; 3Q8K A; 2HLH A; 2XMB A; 2ZS7 A; 5IG2 A; 2WW8 A; 1P5R A; 4PZL A; 4YB8 A; 2OP1 A; 1D2N A; 1N2E A; 1TLF A; 1FSZ A; 2HK7 A; 5EVC A; 2PME A; 1QOQ B; 3RRY A; 2REO A; 4I50 A; 3K32 A; 4ZVU A; 4J0H A; 5CDS A; 4NBS A; 1K0U A; 1XMV A; 4MVJ M; 3CKY A; 5TY0 A; 3U0V A; 4MWB A; 4BNW A; 4N5L A; 3KJF A; 3M4N A; 4M1Y A; 4N7J A; 5T64 A; 3RHF A; 3EPJ A; 1G21 B; 2A5J A; 4HGY A; 4BTE A; 1DBW A; 4K81 B; 4M76 B; 1TDJ A; 1NW6 A; 2Q27 A; 2XOK A; 3LNT A; 4B9E A; 4XWK A; 4KR3 A; 4R01 A; 3PYL A; 3CWC A; 4BV8 A; 4YP7 A; 1P5F A; 4IF6 A; 1JLL B; 2ZSH A; 2E1P A; 4Q3B A; 3ICC A; 1FF9 A; 2BMF A; 4HFJ A; 2GAI A; 1PFV A; 2DZD A; 5LR6 A; 4AJN A; 1JXA A; 1B8U A; 2VPX A; 3GLU A; 1P3D A; 4B3M B; 1Y8Q A; 3WQB A; 5B1Y A; 3UEN A; 2C29 D; 3H97 A; 5B1I A; 1TTT A; 2XT9 A; 2A0Y A; 2GAO A; 2PRK A; 2YW9 A; 3GCW A; 4UM9 D; 5HHT A; 3UEO A; 3UB2 A; 4U0J A; 1A9Z A; 3PDU A; 5JLJ A; 4MWT A; 5C31 A; 1G66 A; 2UU9 B; 4Z54 A; 3OIW A; 2V65 A; 4PX2 A; 1HZZ A; 3CPJ B; 2VLB A; 4BGU A; 4PZA A; 5E38 A; 5LZ4 A; 2SBT A; 1UBJ S; 2XUQ A; 3EVF A; 3MWG A; 3HCC A; 3BHN A; 3FNB A; 3MJH A; 4LW8 A; 4MWZ A; 4TTA A; 4WJZ A; 3V1Y A; 5JMW A; 2X0E A; 3WXM A; 4RZ2 A; 4LMR A; 1X1R A; 4JLV A; 3HA3 A; 4ZOL A; 4LGY A; 2FWI A; 4I4T B; 5DVZ A; 2QD1 A; 2IS1 A; 3G2K A; 3UG6 A; 3IT0 A; 2C5G A; 2QG9 A; 4GBG A; 2VO1 A; 3DMF A; 1E2U A; 4YXW D; 3ZD7 A; 3KO8 A; 4N65 A; 3KCN A; 3AY0 A; 3GCG A; 1OH6 A; 1U0F A; 2XR1 A; 1WW8 A; 4JOC A; 1QDL B; 2O5E A; 1BFK A; 1Y7T A; 3QTT A; 4GGM X; 5AVJ A; 3WNQ A; 1LDB A; 3NAT A; 1TRK A; 3N70 A; 3UC5 A; 1Z2E A; 2OEN G; 4CDG A; 2I54 A; 3WQR A; 4LWO A; 4QJ5 A; 3STV A; 1BEE A; 1HDG O; 1T5T A; 2HDO A; 4DHW A; 2YC2 C; 2O54 A; 1KAH A; 1ZOL A; 2Y68 A; 2ZWI A; 3OAA A; 3MW8 A; 2XA2 A; 1KVQ A; 1ZIC A; 4QO7 A; 2AE2 A; 1IEV A; 3I32 A; 4EJF A; 3TQQ A; 1IC6 A; 2IH2 A; 2J7P D; 2CVO A; 2FCA A; 3KUD A; 4CN9 A; 221P A; 2P1Z A; 2C9M A; 4Q12 A; 2IV2 X; 3TRD A; 2COU A; 2ZPA A; 1AT1 A; 3HJC A; 3MER A; 4RS3 A; 4BX2 A; 4DST A; 1DTW A; 3F1L A; 4G24 A; 3WBF A; 3WRY A; 1JVB A; 4KTV E; 3IJP A; 1MC5 A; 1Z25 A; 3A10 A; 1RKU A; 4FEA A; 4RES A; 3AIO A; 1HEY A; 5FWX A; 1FPQ A; 2H48 A; 3AR3 A; 1ADB A; 4PLG A; 4RUK A; 5KC9 A; 2WIC A; 2C9D A; 3E1G A; 1SKA A; 2HIH A; 1B1A A; 2MDT A; 3A32 A; 1R9J A; 1NAS A; 3HCN A; 2YFH A; 3L7C A; 4L15 A; 4Q4H B; 4RHT A; 4B09 A; 1XRO A; 2JFG A; 3FN4 A; 4MDY A; 1OAO A; 2IF2 A; 3WXB A; 4ZIC A; 2ZUC A; 4LIS A; 4TMF A; 5ILO A; 1L5W A; 1ZGA A; 3ZBQ A; 5B3J C; 2J5Q A; 5AHX A; 4FYM A; 1LBQ A; 3R7S A; 1EFU A; 4Z53 A; 3TLZ A; 4BND B; 4DML A; 2XRY A; 3EAF A; 2WE7 A; 3QX9 A; 1NP3 A; 3M1I A; 1GKK A; 1N8K A; 3HN2 A; 1VP9 A; 1PGP A; 2I4I A; 2P9Y A; 3PDI B; 4MRM A; 3A58 B; 2C4H A; 3OHS X; 3F49 S; 4MGY R; 2P2N A; 4LRY A; 4RMG A; 3RYT C; 3GLF E; 2XYP A; 2O2Y A; 4QXQ A; 2UDP A; 3JSX A; 1RI4 A; 1U28 C; 3H65 A; 5DV9 A; 4CUJ A; 4TLB A; 3FLA A; 3GLH B; 4PRK A; 4LNA A; 2DY1 A; 3P27 A; 4B4W A; 2B21 A; 3A31 A; 4WNI A; 5DRY A; 4D3S A; 5JYB A; 4O2A F; 3UJH A; 3IMG A; 3L4S O; 2AZD A; 1GKL A; 1GA6 A; 1R4M A; 4IIV A; 5EWJ A; 1XL1 A; 4V24 A; 3I5A A; 1XDB A; 3JVE A; 1IMO A; 3PEE A; 1DIH A; 1K3F A; 2CL5 A; 1MC4 A; 1U0G A; 4J76 A; 4Z8X A; 3VR4 A; 3WWO A; 1NVK A; 1I6L A; 3SWW A; 4P7B A; 5DB4 A; 1KZG B; 2EZT A; 5HKB A; 1NQW A; 3MD0 A; 3ZLB A; 1ML4 A; 2AN1 A; 2BBS A; 2C2X A; 2K2W A; 2OOR C; 3B8E A; 2MYP A; 1I8T A; 1KJ9 A; 3DFU A; 2BGU A; 2VYV A; 1TCV A; 3C7A A; 1XDS A; 1V45 E; 5IT4 A; 2GJ4 A; 1SXJ D; 4QT4 A; 3L0I B; 2OWD A; 3BIX A; 3TLM A; 3HCA A; 5CDH A; 4I1V A; 3B3A A; 2JLA A; 2ZRO A; 4Q1C A; 4Y18 A; 1QGO A; 4C3O B; 1ZSN A; 1SUE A; 4NJM A; 4B6G A; 1X7Y A; 1V9C A; 4UMF A; 1PDV A; 1IN1 A; 1GNT A; 4C4R A; 1P3J A; 3MOS A; 4WML A; 4MVJ I; 4FN4 A; 2ZYR A; 1QS0 A; 3FNH A; 4URW R; 2H7Y A; 3OCW A; 3W05 A; 3WQH A; 4POA A; 5A3Q A; 3K5H A; 2BSX A; 2GK6 A; 1GXS B; 3P9N A; 2NZU G; 3FH0 A; 4XB6 B; 4BP1 A; 1R0Z A; 1Q14 A; 3TR0 A; 5JSZ A; 1QHE A; 1YMU A; 3IBF A; 3ILY A; 2P9C A; 4KSC A; 5DLY A; 5FNZ A; 4UDX X; 5H59 A; 4EW1 A; 1UM9 A; 3HCW A; 1X9G A; 4MIE A; 1FYV A; 1E94 E; 1SK9 A; 1YI8 A; 4UUN A; 3LUA A; 3WGV A; 3UH1 A; 1OLO A; 1G3U A; 2X2P A; 4ZQB A; 1AKT A; 4NCX A; 1IPE A; 1TVK A; 2QRU A; 1YRL A; 2HJR A; 3K5W A; 2Z98 A; 1WXX A; 2QJF A; 4D8Y A; 3UIE A; 3UTD A; 4OYY A; 1F12 A; 5AKL A; 2AOW A; 3I9N A; 3OPQ A; 4WHM A; 4N47 A; 2P2D A; 3ABI A; 4QFB A; 4MMX B; 5DX0 A; 1G1M A; 2NAP A; 3ESQ A; 4ZAV A; 4JBY A; 2AIR A; 3QFR A; 5FX2 A; 5TPZ A; 2QUJ A; 2IY9 A; 1PJ2 A; 4ETN A; 1U11 A; 2EJC A; 2R6A A; 3FVQ A; 4ZI7 F; 2ZRK A; 4RND B; 3V4Z A; 1EFK A; 1P9R A; 1JFL A; 2H1F A; 2PLW A; 4TM7 A; 1EP9 A; 1EMD A; 2GMW A; 2HD4 A; 4IID A; 5EP3 A; 2FZG A; 1BJR E; 1G9X A; 3UQZ A; 3O3O B; 5M5B A; 5AI6 A; 5A03 A; 2N0S A; 3ZCP A; 1GEG A; 1NLF A; 1XHE A; 1E4E A; 1Z56 C; 2X2F A; 1U0S Y; 3H2J A; 4YQE A; 3TTT A; 9ABP A; 4C87 A; 2Z5F A; 4GX5 A; 1CY4 A; 2J3K A; 1RXW A; 4HWG A; 3N87 A; 3HDT A; 1W30 A; 1Z0F A; 5TR9 A; 3AUS A; 4MUH A; 4A16 A; 4NNP A; 2PVC A; 1YZN A; 3AP2 A; 1YB7 A; 3AM5 A; 2PIA A; 5CLG A; 4RF5 A; 1PDW A; 3EWI A; 1HV8 A; 3DE6 X; 4O4J B; 1I4E B; 3HNP A; 1OIV A; 4O2B A; 3SOZ A; 4F4H A; 1DT5 A; 1Z63 A; 1PEK E; 4RA9 B; 1T1S A; 1T5L A; 2EKP A; 4O0M A; 1WQ1 R; 3PHB E; 1LQT A; 4WNA A; 3RVS A; 3ZBT A; 1WDW B; 1DKR A; 2BBO A; 3L1I A; 3RYH A; 1N5R A; 4JT2 A; 2QM0 A; 3OLX A; 2OV2 A; 1NNL A; 2VCH A; 4LOK A; 4O0L A; 4BO0 A; 4EO9 A; 1KMC A; 2FSS A; 3JAT B; 4D25 A; 2VT7 A; 2Q4O A; 3UBM A; 3EX7 C; 1V95 A; 3IV9 A; 3RS0 A; 3T3M B; 4ZPX A; 1OZG A; 3STX A; 1AF4 A; 1QGU B; 4UTV A; 2NV6 A; 2RAU A; 2OGW A; 3E3O A; 1QJC A; 4EKI A; 1NBO A; 3N1U A; 4IG9 A; 1A4I A; 2CLS A; 1M8G A; 4FN5 A; 2XGG A; 3JD0 A; 3R44 A; 3IT3 A; 4EPX A; 1RQ7 A; 5JNT A; 4K6G A; 1WPG A; 2O3R A; 3IP1 A; 2PUD A; 3LX5 A; 4MQU A; 3WP0 A; 5CBK A; 4DA4 A; 1E60 A; 2ZI4 A; 3SHW A; 3N5K A; 3ZDY B; 5JPD A; 2I6G A; 2JC3 A; 1NUS A; 4O5A A; 4E31 A; 1MOQ A; 5KJ1 A; 4YUY A; 4URZ R; 3EZN A; 3L8Z A; 2GDJ A; 2JAT A; 4YRI A; 4ZRE A; 5IEJ A; 5JAD A; 5KNC H; 1XAG A; 1Q3H A; 1XZN A; 3W2G A; 4C6T B; 4RHX A; 1CYE A; 3EZL A; 1ECF A; 3C5C A; 3I6Z A; 1SA0 B; 4IZO A; 4RH3 A; 3K71 B; 4NS1 A; 1YJ5 A; 4B4K A; 4WCF A; 5C8Y A; 3S4Y A; 3U09 A; 2I33 A; 4G0M A; 2OQV A; 2AOT B; 5KDL A; 2DET A; 5SZH B; 1L7Q A; 2YWG A; 2G0A A; 3MAG A; 5DHU A; 5LP6 C; 4OK5 A; 4ZHQ F; 3EZ2 A; 3R7F A; 2Z8Z A; 5JT1 A; 1U7W A; 1DE0 A; 2AQH A; 3DKR A; 3O7M A; 4A2P A; 4RY2 A; 2J59 A; 3L8M A; 1VFP A; 2W6I D; 3I09 A; 4U2C A; 5B58 T; 4JH0 A; 3LOU A; 3GUZ A; 4AKI A; 4G5H A; 2CJF A; 3BCH A; 5DND A; 1I36 A; 5IUK C; 2J9F B; 3M34 A; 3LJK A; 2VLI A; 4PC1 A; 1X38 A; 4RIH A; 3QMP A; 2E89 A; 4L4Y A; 3W8E A; 3CNL A; 4MDH A; 2O8N A; 3QTL A; 4LA4 A; 2NO5 A; 3FE2 A; 4NV0 A; 4OA8 A; 1Y9D A; 4D8T A; 4DLP A; 2FN4 A; 4ZMS A; 4JY7 A; 1HSO A; 3EGX B; 1QZZ A; 3NOR A; 3Q9S A; 3LFU A; 4RFS B; 1RC8 A; 1U2D A; 3RVP A; 2Q41 A; 3PFI A; 1A9Y A; 1BTO A; 2C2H A; 3CE6 A; 2I1W A; 4FSX A; 4EM6 A; 3IQE A; 3ZIA D; 4JAS B; 3I9V 1; 3TRH A; 4FB5 A; 3R74 A; 3VAA A; 1NXQ A; 1SCA A; 3VOA A; 5M04 A; 1ZNQ O; 2V4Z A; 2V5A A; 3OVM A; 3P2Y A; 1K7C A; 1LK0 A; 4NMH A; 4TV9 B; 1L7M A; 3QJY A; 1QHF A; 1OI7 A; 4GGO A; 3TKY A; 1M1U A; 2CXO A; 1NN5 A; 2PBZ A; 3I72 A; 3L6E A; 4G65 A; 4O2R A; 2O19 A; 2GTP A; 3KZO A; 4FOL A; 5ITW A; 5JJK A; 3D3W B; 2PT6 A; 2BGT A; 2BE7 A; 2JFY A; 1KS9 A; 5DNA A; 3PXG A; 1ZGC A; 2GN1 A; 2J3I A; 1Q92 A; 2QA5 A; 3CH4 B; 4HAT A; 3RH0 A; 4OMD A; 1DHT A; 1CB0 A; 2YQ5 A; 2C31 A; 4LF8 B; 4HOY A; 3HJF A; 2OZL B; 4U4A A; 5I4K A; 2C2G A; 4KWI A; 1BW9 B; 3HCO A; 2HMA A; 3ZHT A; 1WZE A; 2P1D A; 3EQZ A; 4KG7 A; 3B82 A; 1BX3 A; 5AKH A; 1TUI A; 1JPV A; 2J6B A; 3PGK A; 3TMY A; 3ECC A; 4X20 A; 3WID A; 3ZHO A; 5BUQ A; 4LX0 A; 1VJG A; 3WLQ A; 1S4M A; 1FFE A; 1SCD A; 3ZJ4 A; 4YAG A; 2Q3J A; 3N0W A; 1W6R A; 1T34 A; 2P6N A; 1A69 A; 5T4B A; 4FYJ A; 1SOW A; 3THS A; 3TXY A; 1HDE A; 1YUL A; 4DCT A; 1DEX A; 4M9U A; 4RQA A; 4KLC A; 4KVF A; 1RYZ A; 2PMC A; 4A8J B; 3NC1 C; 2QM7 A; 2SCU B; 4OI1 A; 3DB2 A; 4UQP A; 4ZJU A; 1NC1 A; 3GJS A; 5HKL A; 5KOY A; 2QG4 A; 4A6P A; 1GFI A; 2QX6 A; 2QZ4 A; 4I14 A; 1C1X B; 3OUU A; 4QIH A; 3I58 A; 1HE2 A; 1UAG A; 3SOP A; 1I2A A; 3T06 B; 3TBH A; 1JQA A; 4V38 A; 4QNN A; 3FUV A; 3C45 A; 1CV2 A; 3P0E A; 4RE5 A; 4UHC A; 3SYL A; 2G25 A; 4RTM A; 3ZWM A; 1XNH A; 1XMP A; 2RAV A; 3E3M A; 2DPL A; 3O8B A; 5E0O A; 1PE0 A; 3HCE A; 5JXH A; 4ER3 A; 2G36 A; 4BMS A; 1MX9 A; 4HNU A; 3UV3 A; 5AVT A; 3QVT A; 3NX4 A; 5EMM A; 1ZL0 A; 4RGQ A; 4EG3 A; 4HBG A; 1GAD O; 3FSJ X; 1QPZ A; 3H6H A; 1RJE A; 4Z1O A; 4G0V A; 4MNT A; 2X4L A; 3QVJ A; 4AY5 A; 3K4Q A; 1T1I A; 3WYA A; 3U6K A; 3ND5 A; 1EGH A; 2EU8 A; 2YIC A; 4F1J A; 2J0U B; 3D9Q X; 4AXB A; 4KMR A; 1GQS A; 1SUC A; 4PPG A; 5JAS A; 1HNX B; 3RRN A; 3DV0 B; 1QII A; 2WJH A; 2OHO A; 4GPB A; 4G0Y A; 2FG6 C; 1K06 A; 2PU7 A; 3HAD A; 4WZ6 A; 1WET A; 1KP2 A; 1WB5 A; 3CUV A; 2IL1 A; 1CDZ A; 3KVO A; 1Q19 A; 3G85 A; 3M6M D; 3L84 A; 3VYV A; 4BQR A; 4BMP B; 2N47 A; 3KHU A; 4H1U A; 4HNR A; 4MS1 A; 3SS9 X; 3OQ6 A; 4BNV A; 4EL0 A; 3GL9 A; 4DWO A; 5JVD F; 5K0L A; 3P9P A; 4GR9 A; 5K3C A; 1LC3 A; 1J0A A; 2G9Q A; 1NVT A; 1WDM A; 2I78 A; 2JFF A; 2YBD A; 3AEQ B; 1PZE A; 3PT5 A; 1NXO A; 5B6P A; 3OU6 A; 2I2F A; 2CF6 A; 2GRJ A; 2R68 A; 3E7P A; 3EZF A; 4JUB A; 5J1S B; 2XMQ A; 1RTT A; 4Z1M A; 4DOM A; 1RRP A; 5BOS B; 4G0W A; 3WIE A; 4GZL A; 1RM3 A; 1VKO A; 3RBX A; 4B35 A; 6ADH A; 1G7T A; 4WLE A; 4EEZ A; 4PQR A; 3JWP A; 3C8H A; 4AJJ A; 1TCU A; 5LZ7 A; 3H8J A; 1RC1 A; 3LOQ A; 5AW8 A; 1J9Z A; 2PAA A; 4B73 A; 1E79 D; 3OSU A; 16PK A; 4C0K A; 1QF7 A; 2NZV G; 5CGQ B; 2OGU A; 3VUF A; 2EYQ A; 3FGN A; 2XCI A; 3BS4 A; 3E35 A; 2JLU A; 2BKL A; 4HDM A; 2BMV A; 3S5J A; 3I4C A; 4XCV A; 3ZHV A; 2JZC A; 3T3D A; 1T2C A; 2YAS A; 4LDR A; 1JIK A; 4B3J A; 2ON6 A; 2QI9 C; 3G8B A; 1NG9 A; 4JWK A; 1EGQ A; 5TRT A; 4FFP A; 1EX6 A; 4LK2 A; 3D7L A; 1UIN A; 3I2Y X; 4EB8 A; 3LS2 A; 4NEC A; 4W99 A; 1YKG A; 3ONN A; 1X94 A; 4XKJ A; 4K03 A; 5BSU A; 4OGK A; 2IGB A; 3ZK8 A; 2G67 A; 4U3J B; 3R0V A; 2FP4 A; 2VUU A; 4JJG A; 3KQM A; 5FTW A; 5GV7 A; 1L1R A; 3BMN A; 3KLB A; 3GJT A; 1NTR A; 2FSP A; 4PS1 A; 3ANS A; 4DDK A; 1MJG A; 2G9Z A; 1E1R D; 1LK5 A; 2B2N A; 2PL1 A; 3SR0 A; 4Z4H A; 4FFW A; 3AVX A; 4RNC A; 1TOA A; 5MN5 A; 5KBY A; 4FGK A; 3TFO A; 2OO5 A; 4G36 A; 4RXT A; 4OHP A; 2WNI A; 3EG9 B; 1WB6 A; 3P96 A; 4BVE A; 4C0A C; 4R54 A; 1OXM A; 2B2X A; 5BUR A; 1IXS B; 4HGQ A; 4RHF A; 1GAR A; 4JYM A; 1WM1 A; 2VCE A; 2QOR A; 3LC2 O; 1E5D A; 2PV0 A; 2IOF A; 3EGV A; 3PRH A; 4JFA A; 4INA A; 4YCB A; 1NVW Q; 3PFW O; 4ZAN A; 2IEZ A; 4IHJ A; 4WKP A; 1GQY A; 1E32 A; 5MFZ A; 1YR8 A; 4MVY A; 1USU A; 4XVH A; 2TMD A; 3FDI A; 1B43 A; 1WQ4 A; 3L01 A; 4BNI A; 2GXU A; 3OPN A; 1RP7 A; 4G86 A; 4IWY A; 4O2A B; 1G16 A; 2AOT A; 4JLP A; 3OF5 A; 3OTW A; 4PVD A; 3TQR A; 3N39 C; 4KQW A; 3GUH A; 4B33 A; 2HA7 A; 3CRV A; 2CLD X; 4XM0 A; 1Q21 A; 1RRK A; 2ZPU A; 3AM4 A; 3NV7 A; 1SXJ E; 3V8M A; 4D4J A; 1EWT A; 3ZW9 A; 5JNS A; 2ZE5 A; 2PKX A; 4ALJ A; 1FFS A; 3TSA A; 2VBI A; 2PNF A; 5KSP A; 2YDY A; 2GWR A; 2GXA A; 3DL4 A; 4CW7 A; 3IAS 6; 2CYA A; 4B2L A; 5AUP B; 2ZWP A; 3I61 A; 1EX1 A; 4NK4 A; 2MHC A; 3IKM B; 4PIN A; 4TQG A; 3AUX A; 4O1H A; 5HKK A; 2WRZ A; 5K0B A; 1H1L A; 4R3W A; 2QM8 A; 3D8B A; 3PC7 A; 2ZI5 A; 2IYR A; 5E2A A; 2VPQ A; 4RFS A; 3RSL A; 4DJD C; 1U0B B; 1F20 A; 1XO1 A; 4D2I A; 2P0E A; 4DA9 A; 1K8R A; 1SG6 A; 3IE1 A; 4J90 A; 1HGX A; 4K6H A; 1HE4 A; 1I5E A; 1VHT A; 3O8D A; 5D8D A; 4G5X A; 2ZP1 A; 3F9I A; 2PPV A; 4TN5 A; 4BGV A; 1OXU A; 3NWJ A; 4I9F A; 2PQM A; 1T2E A; 4FJ2 A; 2YHI B; 3LDG A; 4HDQ B; 5LCX A; 5ICE A; 4OHF A; 3K05 A; 4JT9 A; 2ZZM A; 1Z8N A; 3CON A; 4XJV A; 3ICE A; 2AC7 A; 1HLF A; 2QLB A; 1SHL A; 3HPR A; 5JNL A; 3VDN B; 5CML A; 4U7G A; 3ZVR A; 2R00 A; 1AIP A; 3GJ0 A; 4QDU A; 2JI4 A; 4Q3C A; 2V8O A; 2TRS B; 3CN7 A; 4Q24 A; 3RUA A; 4V0M A; 3AKC A; 4PR0 A; 5IDX A; 4CIA A; 1ULA A; 1ZOO A; 3ADP A; 3DMY A; 4RVO A; 4B7C A; 5K8T A; 5KQR A; 5LXT F; 1X8L A; 2FK7 A; 1XA3 A; 3ZYC A; 1V8D A; 1OWM A; 2OZL A; 4F3C A; 4WAS A; 5ELX A; 1JMK C; 1MRZ A; 1H3N A; 1N5K A; 1TO1 E; 1ESC A; 1M0W A; 2ZTU A; 1GL6 A; 2PSJ A; 4JLN A; 2WVJ A; 3OPM A; 1LDM A; 2IXB A; 5JCJ A; 3KPU A; 4ZMB A; 1XU4 A; 2A4M A; 1A96 A; 4FD3 A; 2YZK A; 3VRH A; 4KW7 A; 4LTD A; 3O7W A; 1C0P A; 2BFP A; 3PT9 A; 4AX8 A; 4BYM A; 1WME A; 4FMD F; 4MUG A; 2XUZ A; 2ZFU A; 3ABZ A; 3C3E A; 1JAH A; 2KI5 A; 2IS6 A; 1QNF A; 4F3N A; 3KZ1 E; 5IFK A; 1C3E A; 1QID A; 4Q4D A; 4BJ5 A; 4C6S A; 1HYQ A; 4I5E A; 1NX6 A; 4M0P A; 1ZS9 A; 1BN7 A; 2ID9 A; 3DZC A; 3M3P A; 1XI2 A; 1VGV A; 3E22 A; 4LFA B; 2DVM A; 1OHH A; 2B25 A; 2H0A A; 5J9F A; 3Q11 A; 1GQQ A; 3ZU5 A; 4CNB A; 2FMF A; 4U2B A; 5CK5 A; 1D0S A; 4BX0 A; 5IAJ A; 1RJD A; 2MYU A; 4OYL A; 1IRX A; 4NE2 A; 4EMB A; 1RPZ A; 1H6C A; 5EG5 A; 1T8P A; 5AWF C; 2HSJ A; 2HSE A; 1ZSY A; 5I9B A; 2Q4D A; 2JCG A; 4NUL A; 1A5S B; 1K88 A; 1PL0 A; 3DD5 A; 3DOJ A; 4N01 A; 3X1K A; 1IB0 A; 4YJH B; 2O7R A; 4TMK A; 1MO2 A; 4BDT A; 4GFD A; 1E9R A; 3EF3 A; 4MV3 A; 5LQN A; 4ND8 B; 4E7O A; 2HCJ B; 5IDY A; 4DLZ A; 1PV1 A; 4Y1R A; 1XZF A; 3S8Y A; 3VTZ A; 2DPG A; 3K53 A; 2XB8 A; 2PA3 A; 5AES A; 5BVG A; 3IDS A; 1QK5 A; 2H4Z A; 2C54 A; 2YC4 C; 3GFR A; 3PQ3 A; 4KRX A; 4OGW A; 4QDK A; 4ZT5 A; 5FOQ A; 5K4G A; 3AVV A; 2HMT A; 1CB7 A; 2HGZ A; 2ABW A; 4P38 A; 4HQL A; 3ILS A; 4N0E A; 3WL5 A; 1XZM A; 1GTV A; 1UBO S; 3A4L A; 4IHK A; 4UBN A; 4JEN A; 3BD6 A; 4UZS A; 5M29 A; 5KS2 A; 2CVH A; 2MWM A; 3AUY A; 1T0H B; 3BDI A; 3FW1 A; 3IIE A; 4IGE A; 2Y3I A; 4N4A A; 3TE6 A; 4DBV O; 3JQA B; 1TIK A; 4CL0 A; 1TAH A; 1MAH A; 1BS9 A; 2YCB A; 2I8A A; 2K7Z A; 3KKM A; 4P1Z A; 4FOS A; 4WXY B; 1NJ8 A; 2A14 A; 4UAT A; 1A2K C; 2DUW A; 3DE1 X; 2GA8 A; 4QGH A; 3P9R A; 1N36 B; 2FPG A; 4WI1 A; 4FZR A; 4EJ2 A; 5J4Y A; 4UUA A; 2R2G A; 2PKW A; 2A9J A; 5KOH A; 3VAY A; 1NBM A; 1RJ9 B; 3W6U A; 3BMC B; 5SZK B; 5JJC A; 3DOF A; 3TG8 A; 3THY A; 3UXE A; 1N2O A; 2RK3 A; 4RXU A; 4ZZI A; 4TMT A; 2QSY A; 3WHI A; 4HNV A; 3F8S A; 4B7U A; 5KND A; 2EIX A; 2CLO B; 3EMK A; 4DMM A; 1CFJ A; 3EXG 1; 1AX9 A; 1PJL A; 1D2M A; 5D87 A; 1Q7R A; 2R66 A; 4DEY A; 3HXO A; 1SKY E; 5HKA A; 4EHK A; 3ZJV A; 2C7G A; 1K66 A; 4FGJ A; 2OXT A; 1PLK A; 1GCI A; 2ODT X; 2PGO A; 5B3A A; 1IY2 A; 3RHT A; 4Z5X A; 3ZGX A; 1NW5 A; 1YS9 A; 3RNO A; 3HKE B; 3UXH A; 4WN9 B; 4NEH B; 3KXI A; 1G5I A; 4ZDI A; 5KNB A; 3RAG A; 2AD5 A; 1LJ8 A; 1S96 A; 5JWR A; 1EQA A; 3SJZ A; 1DCF A; 2JLX A; 5EUQ B; 5TVK A; 5JZM A; 1BEU B; 4W1R A; 1QVW A; 4HMQ A; 3PDW A; 3BIC A; 1JU9 A; 1T1J A; 4D82 A; 1BXG B; 3CUK A; 4LRS B; 2I7T A; 3U16 A; 4MEA A; 4DDV A; 1Z7B A; 2VZ8 A; 1UWK A; 4C2T A; 3T7R A; 1WV0 A; 2J5K A; 3QEM B; 4EFN A; 3V1T C; 2X19 A; 1H3E A; 1TTH A; 3AW9 A; 3R0X A; 4GVX A; 3KOM A; 5ECW A; 1QOR A; 3IST A; 3Q40 A; 3O5A A; 2RH8 A; 2WUW E; 2NNW B; 1N2I A; 3L7L A; 2XQV A; 3LY5 A; 3M89 A; 4PAO A; 4R7G A; 3SXU B; 1MXF A; 1WUI S; 1NPT O; 1V4V A; 3RWB A; 1UMD B; 1E3S A; 3FYD A; 1G20 A; 4J3J A; 3W2T A; 4Z4F A; 3A1S A; 1YCH A; 2D1Y A; 5HE9 A; 4X65 B; 3KDA A; 5L7W A; 5I9T A; 2JCV A; 4O2A A; 4UE3 S; 1GOT A; 4PPQ A; 4KQX A; 4YDQ A; 3FAK A; 5TGT A; 4C0S A; 4E03 A; 2ZIV B; 1BOO A; 3LLS A; 2O8C B; 1O97 C; 4CMH A; 2V4M A; 1FX0 A; 3QSG A; 4W5H A; 2I6Q A; 3A2L A; 2YJN A; 1LB1 B; 4MP8 A; 1JSZ A; 1CJK C; 3IKV A; 4E13 A; 3UAO A; 2VTB B; 3MTE A; 2IDJ A; 4YPO A; 1WCW A; 1ATZ A; 1RI5 A; 1ZD1 A; 4BO9 A; 3K09 A; 2YJG A; 3EUL A; 3SVL A; 1VQ1 A; 1X1E A; 2HBQ A; 3CLV A; 4XGI A; 3VR5 H; 4EG5 A; 4DR6 B; 2V98 A; 1YI0 A; 4QQF A; 2REB A; 4LDZ A; 4QUL A; 4BB6 A; 2YJ5 A; 1II8 B; 2WU0 A; 3Q0I A; 4BQE A; 2I03 A; 4MKZ A; 4HRX A; 4LRT B; 3BG5 A; 3HXQ A; 3VR1 A; 1MVN A; 2I2D A; 3NBK A; 1SMK A; 5LIR A; 1QYR A; 3HLK A; 4N9Q A; 4TXN A; 3KKQ A; 2QTN A; 3R9I A; 4W5N A; 1R0C A; 2NYU A; 4V2U A; 3QF4 B; 1B00 A; 2HOQ A; 4G07 A; 3OQO A; 2ZIA A; 1XJ5 A; 3EH2 A; 1C80 A; 4E51 A; 2OBB A; 3P20 A; 3A28 A; 3GRO A; 5JPI A; 4QL3 A; 1T10 A; 4NU6 A; 5C34 B; 1JFS A; 2Q4B A; 3RBV A; 4CI0 B; 2JHA A; 3RG8 A; 5I3Z A; 4C76 A; 1ID8 A; 4Q1A A; 1XMJ A; 1YAC A; 2H08 A; 3CNB A; 5HSG A; 1U2Q A; 3FBW A; 3GJX C; 4W7H A; 3A4V A; 3EYW A; 1RPT A; 5BV2 P; 3C7C B; 1ZGB A; 2RHC A; 4WXM A; 4Z9Y A; 4NG5 A; 4ZKE A; 5EKT A; 5KSY A; 2J0V A; 1ZN9 A; 4EAA A; 4GKB A; 2J3M A; 1SEV A; 2J3E A; 1VLQ A; 1EIZ A; 4XXH A; 3U3M A; 3UNX A; 5AQ1 A; 1P9O A; 2VRW A; 1NV8 A; 3PNO A; 3V1K A; 3OML A; 1VL8 A; 2Y1G A; 4S1V A; 4FP9 A; 2JFZ A; 4BNZ A; 4CV2 A; 2IIQ A; 2XF8 A; 4JJ4 A; 4AMG A; 2JJ1 D; 3ROD A; 1RIF A; 4DTC A; 4CU5 A; 2EQA A; 4DDU A; 1BCS A; 1JHO A; 1T35 A; 3HYC A; 4XA8 A; 2DWP A; 7MDH A; 4ZAZ A; 4MWE A; 3VU9 B; 3T99 A; 5GON B; 1YQW A; 4RBN A; 1NJ6 A; 1RAI A; 1Y4S A; 4LEC A; 3MIN A; 2W4Q A; 3VKF A; 1S2G A; 1BHQ 1; 3OIU A; 3EIW A; 4ZVN A; 1G8F A; 1RZ3 A; 1WHT B; 4QTZ A; 1D9X A; 2QRJ A; 3V80 A; 3U7G A; 1R8Q A; 2BGJ A; 1PZM A; 1Z0I A; 3EOB I; 4P36 A; 1KJW A; 2HC9 A; 2I3D A; 2DXI A; 3NKF A; 4RKR A; 1Y63 A; 3K8Q A; 3D6F A; 4KIU A; 2DR2 A; 1OFI A; 4QLB A; 3K96 A; 2A9F A; 3WGN A; 4ALK A; 3K1Y A; 4B3G A; 1C7R A; 4D4G A; 2O8D B; 2B8W A; 2R5F A; 3GJZ A; 4UHE A; 2BP7 A; 2L2Q A; 4QRD A; 5JRD S; 1SXH A; 1SBN E; 4DLA A; 3N28 A; 4CEH B; 3PLR A; 2A94 A; 3LFH A; 2FR8 C; 4IV0 A; 1W48 A; 1BIF A; 3GUV A; 4UHJ A; 1VBM A; 1DV1 A; 3UAY A; 1SBH A; 2IQI A; 2X5X A; 1JHQ A; 2RD2 A; 2RHB A; 3UJ8 A; 3BTU A; 1IH8 A; 1H4V B; 1PQY A; 2JFP A; 1O6C A; 1J8Y F; 2PZA A; 5H80 A; 1KFJ B; 3W3Z B; 5CR9 A; 5JJI A; 1LDY A; 3BEO A; 1Y80 A; 2FZ5 A; 3OES A; 4IZH A; 1BHO 1; 3DKV A; 4C6H A; 3IOM A; 3GLR A; 2FEA A; 1Q1B A; 4GX2 A; 3INT A; 4CID A; 4ED9 A; 1YON A; 3V1L A; 4ID3 A; 4PFJ A; 4A0R A; 4V0S A; 4D9F A; 4X91 A; 3WJA A; 4QBT A; 2DUN A; 3IT2 A; 3NOX A; 4PLZ A; 4DR3 B; 1C9K A; 5EHI A; 3CR8 A; 1GRR A; 4H7J A; 5JDC A; 3N8G A; 5IYZ B; 4FC7 A; 3JQA A; 4S2E A; 1DLI A; 3N3B C; 4L16 A; 4DNQ A; 2PRJ A; 2ZWO A; 3TZ6 A; 3X1D A; 1JFF B; 2IPM A; 4UEO A; 2FEX A; 1R0B A; 2H3H A; 1FS6 A; 1C5K A; 1U0N A; 5KIN B; 2ZPT X; 4ZHQ A; 3CVY A; 4J49 A; 3U13 A; 2PGJ A; 1ION A; 2HKI A; 1DAF A; 3WRX A; 2TMK A; 2BME A; 2ETV A; 3CG4 A; 5AM5 A; 2CBZ A; 3SZR A; 3BMC A; 5DCU A; 2QV0 A; 1SVS A; 2WDP A; 3DAP A; 4TYW A; 5D5Q A; 2IIV A; 1KI3 A; 4M9C A; 3I76 A; 4ZEV A; 2VZL A; 4N49 A; 2CDN A; 2ZRL A; 2W8B A; 4IDH A; 5JRZ A; 2XUP A; 1ZK4 A; 1E9E A; 1SKU A; 2XKA A; 3GRC A; 4TZT A; 2QJT A; 1JG3 A; 1FQK A; 5H83 A; 5IK2 D; 3ESC A; 2GB4 A; 2EX1 A; 3ASV A; 5CXS A; 4GC5 A; 1JX6 A; 1L4N A; 1FSP A; 1E1C B; 3JZI A; 3AJR A; 4IVP A; 4DC9 A; 1NQT A; 4TVO A; 3KQT A; 5FL7 D; 2D5C A; 2VAW A; 3PHJ A; 2OVF A; 4U2K A; 2IKQ A; 1JXO A; 3EFH A; 2YZN A; 2JAO A; 4TYQ A; 1ZEM A; 3L0C A; 4Z2Y A; 2HWK A; 3L77 A; 3TCK A; 4YPF A; 4DM3 A; 4NPA A; 3CW9 A; 3TG2 A; 3AF6 A; 1K3U B; 2E4X A; 2YXE A; 2VDW A; 3AES A; 2CDA A; 1E5L A; 5BNT A; 1EUC A; 2C83 A; 4REQ A; 1T2U A; 3GLG B; 4M54 A; 3FYU B; 1L7D A; 2CUT A; 2HQS A; 2XRM A; 2GF2 A; 1NQA O; 5I7E A; 2A0W A; 1IOZ A; 2Q3F A; 6PFK A; 5JXW A; 3QE3 A; 3BTO A; 5F12 A; 4G2N A; 2BCG Y; 1MC8 A; 4WEC A; 1DIG A; 3R7W A; 2O23 A; 4F4I A; 1E4V A; 4Q5I A; 5FOV A; 3SXU A; 5KX5 F; 4JVM A; 5CDF A; 2IEA A; 2XB2 A; 4F3R A; 5BR7 A; 1ZJZ A; 1VSV A; 1U0R A; 1X74 A; 1Q11 A; 1BJT A; 4OMU A; 4NNO A; 1U8F O; 5L7Y A; 2QXU A; 3SNK A; 2AX3 A; 5GZ6 A; 1SK8 A; 4Y2H A; 1JF8 A; 4WES A; 3NTU A; 1Z4J A; 2A59 A; 4PM4 A; 2YHS A; 5IKE A; 3W1E A; 3ADC A; 3HI6 A; 1L5M A; 3OID A; 3RRM A; 5G3Z A; 3HYN A; 3MS7 A; 1EA7 A; 5DHT A; 1LGY A; 2QRP A; 3R9X B; 1IU9 A; 4R1N A; 1IIB A; 4B3X A; 3V4R A; 4WLF A; 3TQC A; 2FQQ A; 3EXI B; 4W5J A; 1Q0R A; 5BR1 A; 4OB6 A; 4LY3 A; 1CQZ A; 1P2F A; 4I1S A; 4QXP A; 1P44 A; 4AS3 A; 2A5L A; 4K7J A; 3RRO A; 1NGS A; 4DA8 A; 5ARB A; 2WJ3 A; 4JK3 A; 4HS4 A; 5EJ9 A; 3D0R A; 4C7J A; 1Q7B A; 2GPY A; 3RBZ A; 2VWP A; 3I6M A; 4IGK A; 4ZAY A; 4LV5 B; 4IZI A; 1U8A A; 4LJ5 A; 4W5T A; 5TQ2 A; 1KZ4 A; 5K8C A; 3W5S A; 2LP4 Y; 3GVI A; 4M88 A; 4H7E A; 3IQX A; 1GPJ A; 5K03 A; 5BN4 B; 3FF4 A; 3DMG A; 3H04 A; 2LE0 A; 1XJC A; 1Z73 A; 4NMX B; 4AU1 A; 4GH5 A; 1MB2 A; 3UJ7 A; 2ZYH A; 1Q2X A; 2C5F A; 4QED A; 1DZI A; 2AS0 A; 3KTO A; 4PXY A; 5CLL A; 2AKY A; 3DDZ X; 1K75 A; 4I1O A; 4KPG A; 5TVG A; 1EFA A; 3IRU A; 1L3I A; 1MPX A; 3H5V A; 4MIT A; 1LEH A; 1XQ6 A; 4FK8 A; 2UVP B; 4LZN A; 3CKK A; 3NGL A; 2O8F A; 1LK7 A; 1QCC A; 2FZ4 A; 4KE7 A; 4LDP A; 3G7S A; 2RHW A; 3Q3J B; 1P2T A; 1TX4 B; 4PVC A; 4MB0 A; 4MZX A; 5B4U A; 5DH9 A; 2A57 A; 3RYI B; 4XAS A; 5JVD A; 1P0P A; 3QE0 A; 1DAI A; 1PJS A; 3AKD A; 1XDU A; 2VP2 A; 3U9S A; 1P1I A; 3AFI A; 1MT0 A; 1GU0 A; 1HUQ A; 1OE0 A; 3FA2 A; 4MRN A; 4G4S P; 3LF1 A; 3RSQ A; 4MB2 A; 1NY5 A; 2I7C A; 4YAE A; 2XGM A; 2OHG A; 3G0I A; 3UGT A; 4FEG A; 3WGH A; 4GZ5 A; 5CLI A; 4QUE A; 2KM1 A; 2KS6 A; 2O8B B; 2NQT A; 1NW9 B; 4MQE A; 4HYE A; 3RKS A; 2O6L A; 4AY3 A; 1XX6 A; 2XAU A; 4XRR A; 3C6M A; 3YAS A; 4L9R A; 1II9 A; 2P9Q A; 4KXM A; 3UIC A; 1SKM A; 3GNT A; 1LWN A; 1AMO A; 2RHD A; 2HDN B; 4TKU B; 5ES4 B; 1N2Z A; 3BN3 A; 4V0L A; 3ONF A; 2QLN A; 4BLP A; 1OYV A; 3HDQ A; 3F67 A; 3O8Q B; 5JTT A; 1EFD N; 3HKB B; 4N5H X; 4IYR A; 4YP5 A; 1AKY A; 5SYN A; 2X9N A; 5AI5 A; 3ZI7 A; 1JHA A; 2OHF A; 4R2M A; 3Q7T A; 1JHV A; 2CWD A; 3V2H A; 4B3C A; 2XTJ A; 4RQU A; 1ABE A; 4IIJ B; 2XWQ A; 4MI9 A; 4ZVS A; 4AN1 A; 1ZIY A; 5LL6 P; 2V7G A; 1YUN A; 5AVX A; 1L8L A; 5HRC A; 4CEH A; 2PWB A; 3NIB A; 3OQJ A; 2XDA A; 1IRI A; 1CQW A; 4PCU A; 1BDB A; 4QJB A; 3KJX A; 1KK3 A; 3HYL A; 1NN3 A; 4HLU A; 5I4B A; 3FUT A; 4LY6 A; 3HKC A; 2C8K A; 3G8C A; 2J4F A; 4LWZ A; 5BOV A; 2J32 A; 1Y6Z A; 5IH7 A; 1RHQ A; 3E6A A; 2XMG A; 1II0 A; 3DMR A; 3H2K A; 3QFC A; 4PHT A; 5CVE A; 4Z9A A; 4FYX A; 4Q01 A; 1J2T A; 5A7G A; 2IY8 A; 4MV6 A; 4P31 A; 4RHE A; 4HVT A; 4TTJ A; 4D9H A; 4ZJP A; 5HIK A; 5JAU A; 2X1M A; 1JQD A; 5KHA A; 2AZ4 A; 4NQZ A; 3MK3 1; 4EYK A; 1J70 A; 5ERG B; 5BMV B; 1W8I A; 2BZG A; 1Z26 A; 3AF2 A; 4QSM A; 1G2Q A; 2X0F A; 4EYO A; 1HG0 A; 2W43 A; 1GA1 A; 1BRM A; 4IJV A; 2P65 A; 5CB4 B; 2QP9 X; 4HNT A; 3RHS A; 1D9Z A; 4IQ4 A; 4C7O B; 2GAA A; 1RRG A; 5F1K A; 4JEL A; 3DJY A; 2BEV A; 2MSN A; 5EMK A; 1NBH A; 1NTO A; 2ZQ5 A; 2GZ2 A; 1QWS A; 1TMY A; 3GEP A; 3WZM A; 2WZD A; 2DSN A; 5CT6 A; 3WIU A; 3ZDC A; 3MGB A; 1GPL A; 4OBX A; 4G4J A; 4PX3 A; 3RC1 A; 3TQS A; 1RLT A; 4ZTB A; 1R12 A; 2PMK A; 3KTA A; 5HUH A; 4J2O A; 2GD1 O; 2HIM A; 4IP0 A; 1NIJ A; 1U0H C; 1QKJ A; 4DMG A; 1OI2 A; 4GP9 A; 3QPD A; 4FGS A; 3RU7 A; 2PLR A; 2QTA A; 4IVQ A; 4QXR A; 5A1T A; 1BVU A; 2O2G A; 4AQD A; 3OEE A; 1HZJ A; 2EYU A; 2JFW A; 2LBP A; 1BRO A; 2M6R A; 2QQF A; 1OXT A; 1R8J A; 1JBE A; 1CY2 A; 1G7R A; 2Q0Q A; 4Z49 A; 1Y4U A; 4JIT A; 2ZJF A; 3N53 A; 5KNL A; 4EZI A; 5AUO B; 3OPV A; 2WIN I; 1T3H A; 5CFZ A; 3A2K A; 2PWY A; 3BSW A; 4OHR A; 3RSO A; 3U3R A; 2YVU A; 4P4S B; 4X8H A; 3VHQ A; 2A78 A; 3G49 A; 4MGI R; 2VHE A; 5DBG A; 4BC0 A; 2H1S A; 1JLS A; 1R8X A; 3JQ7 A; 2P0Y A; 3PJF A; 4TVK A; 4WWH A; 3SC2 B; 3P9Y A; 1I7N A; 4NHO A; 1E1C A; 2VF7 A; 3H99 A; 4P5P A; 5EJI A; 5B53 A; 4HFM A; 4BC1 A; 2X6R A; 2OQI A; 1DIN A; 1NYR A; 1QHH A; 5I7O A; 2VJY A; 1HEI A; 1J1U A; 4EGJ A; 3ONV A; 5FI1 B; 3TOS A; 2FFB A; 1QP4 A; 1B8V A; 1QC5 B; 2XOK D; 1SXJ C; 2H00 A; 4Y6G B; 3TLK A; 3X0D A; 1K27 A; 2Q1U A; 3D31 A; 4UOZ A; 4LOJ A; 4EKD A; 3UAN A; 1H69 A; 5FV4 A; 4IS2 A; 2D06 A; 4HFR A; 7GPB A; 3C6X A; 1Y89 A; 4PAW A; 3SYK A; 4CM5 A; 3CT6 A; 3NBM A; 4P4T A; 4LF7 B; 2BI4 A; 4L22 A; 4RPH A; 1T2D A; 2WP8 J; 1P61 B; 5DM1 A; 1Z07 A; 1VKR A; 1REQ A; 4BNG A; 4KR2 A; 4HUT A; 1JJ7 A; 5J3J A; 3HDY A; 2DX7 A; 2K5U A; 4P9N A; 4ALI A; 4B48 A; 5KTO A; 3ZWC A; 2B5W A; 2PZ8 A; 1A8S A; 1E3L A; 3FYH A; 1B0P A; 1W85 A; 2YW2 A; 3IUM A; 1U7T A; 4JQO A; 4WEO A; 5T4E A; 1RAB A; 1KE1 A; 3JTE A; 5AKE A; 3W5I A; 1U04 A; 5I7F A; 1Y98 A; 4BX9 A; 1BVR A; 4MAR A; 5A2Q A; 3T41 A; 2ZKL A; 2ECO A; 3L4K A; 5AHR A; 2YZM A; 5HS6 A; 4E7P A; 4EHJ A; 3LXR A; 4LDQ A; 3LP5 A; 3C5V A; 3E0Q A; 1IQP A; 2XDY A; 2V7X A; 5AOB A; 1V3T A; 5TE9 A; 5HVN A; 3RWO A; 4WGG A; 2CVZ A; 1XHF A; 5HJ7 A; 3WYB A; 4LJA A; 3EMV A; 3VPC A; 4FM9 A; 1XRP A; 1C9N A; 1FYE A; 1TJP B; 4NOM A; 2AJD A; 2YWC A; 2CHG A; 1SCN E; 2VQS A; 5HM8 A; 3BJE A; 4EHH A; 1U9I A; 3DEF A; 5FH9 A; 1IXR C; 1DL5 A; 3NNN A; 4OAT A; 4AO6 A; 3U9L A; 3R7L A; 3BL6 A; 4BKQ A; 4U2G A; 3OAA D; 2EZ9 A; 3SKV A; 2IEB A; 1TK9 A; 1YMQ A; 2BJV A; 3TL3 A; 3UMS A; 5KRE A; 5KHL B; 1WKC A; 2KW5 A; 3TIN A; 2XRZ A; 3S6J A; 1ZNX A; 1UJN A; 4F4U A; 3JWI A; 3P0F A; 2IHK A; 3OLC X; 5EKX A; 2A1I A; 5FEU A; 3G6K A; 4Z9X A; 3ND9 A; 3DT6 A; 2CUN A; 1MQ9 A; 1QH1 B; 3Q5D A; 5HKO A; 4QIC A; 2AK3 A; 1Y1S A; 3D8U A; 5BN5 B; 2HRR A; 2Q9U A; 4EYQ A; 4EKG A; 4RZU A; 5LLT A; 1JDP A; 1KO5 A; 1AUX A; 1RKQ A; 1P75 A; 2DQK A; 3KPY A; 3O4R A; 1XYV A; 1OXV A; 1VB5 A; 3QKW A; 4Y1G A; 4C7B A; 3T3P B; 1KY5 A; 3AUF A; 4K6K A; 3NH9 A; 3I01 M; 4OXI A; 1QLH A; 3Q41 A; 1GKI A; 3RVL A; 1PNV A; 3Q98 A; 2NG1 A; 5CXQ A; 5BQO A; 5HK9 A; 2BI8 A; 4D98 A; 1JG1 A; 2B6H A; 4EOG A; 1R64 A; 3E7F A; 1MJF A; 1QRD A; 4ZQG A; 3S7T A; 4D02 A; 4DIM A; 3ZE9 A; 3VSB A; 3WYD A; 1YV9 A; 3SK0 A; 2XCR B; 3W77 A; 5DKF A; 2CHU A; 4BLV A; 3BXE A; 3GRV A; 2QTZ A; 3KC2 A; 1KP3 A; 2HMS A; 1SC4 A; 4NBK A; 2GSW A; 3TYX A; 2XT6 A; 3KAP A; 1UEX C; 2EP8 A; 3E18 A; 1XZD A; 4DCS A; 2CCG A; 4IKP A; 3GA7 A; 2WTA A; 4U7P B; 3CCF A; 2P2S A; 4JLO A; 1G9S A; 4ZAG A; 2NT3 A; 1HSZ A; 1KMH A; 2RGG A; 2ZIW B; 1S3I A; 4HDN A; 1KVT A; 1LW7 A; 3C24 A; 3TDK A; 1FFX A; 1LPM A; 1TFR A; 4E94 A; 2QEN A; 3L46 A; 3HR7 A; 1J9E A; 4NBL A; 4Z4C A; 2BF4 A; 3O8O B; 4NG4 A; 1HFK A; 4REN A; 2H9I A; 1CHD A; 3QE2 A; 5G0T A; 3VR4 D; 1ORT A; 3ZE2 B; 1RDF A; 3G9T A; 1W47 A; 4DA6 A; 2W2T A; 1VLZ A; 3TOV A; 4UEJ A; 3RIX A; 4MKX A; 1KJN A; 5H3L A; 1AKM A; 5LFA A; 2YNM A; 1I4L D; 3SQM A; 1UKE A; 4YAO A; 2Q2V A; 4O2L A; 3WB1 A; 2A9K A; 2VYV D; 1ZVV A; 1JWB B; 1SCB A; 1Z74 A; 3GFB A; 3IJ5 A; 1E3W B; 4UUB A; 5FD8 A; 3TKL A; 4DN6 A; 4RA6 B; 4TUY F; 3K0C C; 2WUF A; 3ZHX A; 5ADW A; 4YON B; 1B3R A; 4HTF A; 1OZH A; 1Q50 A; 2JF0 A; 4GM0 A; 3MA0 A; 3P9Z A; 1UOO A; 2MSL A; 4BPR A; 1TA9 A; 3M70 A; 2J5R A; 3CRZ A; 3DC2 A; 2WB2 A; 8OHM A; 1YKF A; 2I3Z A; 2R1T B; 1JLK A; 1COW D; 2D1R A; 5KYX B; 1XEW X; 5CP8 A; 1R66 A; 5BSM A; 5C8X A; 2V1U A; 1NJS A; 9GPB A; 4XW2 A; 2KWI A; 3PNV A; 3GMD A; 3NO4 A; 2BFB A; 2WIL A; 2OCP A; 2V5V A; 2XSZ A; 3EBP A; 4CR6 A; 2X5O A; 2CCK A; 4UPD A; 2P11 A; 5CT4 A; 3B1F A; 1GR0 A; 1MOS A; 4XTC S; 1N9G B; 1PTK A; 1XLT A; 3LP6 A; 4R8R A; 5ALE A; 4L1K A; 2YWJ A; 4C14 A; 2C77 A; 2OXR A; 3OE7 A; 3DHW C; 5EGS A; 2VBC A; 1MJ5 A; 1RRM A; 4Y2U A; 2W6P A; 2ECQ A; 4IFB A; 4ZFN A; 1HTT A; 2O7Q A; 1I33 A; 1JV2 B; 2CL6 X; 4YDS A; 1JN0 A; 3ODP A; 5CB4 A; 3AJD A; 2D2E A; 3M5P A; 1JXM A; 1VDM A; 4HAY A; 1TYD E; 3RVO A; 1SB8 A; 3UH0 A; 4AY4 A; 2UYC A; 3JD2 A; 1RAG A; 3MWL A; 3OY2 A; 3C48 A; 3HA2 A; 5BV8 A; 4I55 F; 2HF8 A; 1QSG A; 1WSD A; 4RU1 A; 3IAS 1; 1VLL A; 1RYH A; 5FPS A; 4QXA A; 5ETJ A; 5TPZ D; 4P22 A; 1DGQ A; 3QYJ A; 3PYX A; 3FPB A; 3ANT A; 2A1U A; 3HJH A; 2AOU B; 3SKY A; 3OIC A; 1YAH A; 2FRX A; 2PZK A; 3KNZ A; 5FA8 A; 2GBL A; 3W4J A; 4Z51 A; 1FVO A; 4JEU A; 4P3K A; 4EHF A; 3CF4 G; 3ID6 C; 3VJL A; 4N5D A; 3BRC A; 4PXA A; 4UBO A; 3ZJY A; 2R11 A; 3ZV4 A; 4A10 A; 4JI8 B; 5EWL A; 3ZWQ A; 2VQD A; 2O5C A; 4ZO7 A; 1U6J A; 2DF8 A; 1EFR D; 4MPJ A; 1Q6Y A; 2YMP A; 3E58 A; 3E2M A; 1GUZ A; 1N2J A; 1YR9 A; 3L8D A; 1C4O A; 1FJG B; 3A6H A; 4FZ5 A; 4P4S A; 3COP A; 1H2H A; 3QE9 Y; 1FIU A; 3U3Z A; 2JI6 A; 3WE7 A; 4NCA A; 4FDU A; 4CG0 A; 2C1E A; 5FLK A; 4OFB A; 1T2V A; 1SU8 A; 2WG1 A; 4X1I A; 1GNR A; 4U2F A; 1US8 B; 1G7C A; 2XDW A; 3CVX A; 3D2B A; 1KMN A; 3WGQ A; 2WYH A; 4ZYZ A; 3ZR4 B; 1PL7 A; 1PBT A; 1NAI A; 2Y1K A; 5JNR A; 5KIA A; 1HT2 E; 3CN9 A; 4BN4 A; 2ZAI A; 4Y2G A; 3RAE C; 1L4M A; 3C5W P; 2ZLB A; 1YRA A; 1B6R A; 1RJF A; 2NSY A; 1VPD A; 3HSK A; 1U2T A; 1U5A A; 1RQG A; 2ECP A; 1FYF A; 3FX2 A; 3KAJ A; 2ZAM A; 4R41 A; 2PXH A; 4YED A; 4YRE A; 2O3J A; 1CZU A; 3U1V A; 4DLV A; 4J0C A; 3NCT A; 1X7G A; 5K05 A; 4ICI A; 1JAI A; 3RYC B; 1A97 A; 4Q47 A; 1JAY A; 3AJ9 A; 5IRC D; 3W2H A; 2NXW A; 4RSR A; 1S8F A; 2QRV A; 4M2L A; 2JLC A; 4Q3D A; 4IXM A; 4LVC A; 3W8F A; 1X1T A; 5HQ3 A; 3EZY A; 3QEK A; 6REQ A; 1HWY A; 3RGW S; 2BGL A; 5D7B A; 4L8W B; 1VE5 A; 5BSV A; 3G88 A; 1JB1 A; 3RFF A; 3ROP A; 1KVU A; 1E0J A; 3K72 B; 3RU2 A; 2CVQ A; 4QOF A; 3ZID A; 4C0H A; 3RSF A; 3FTE A; 4GYJ A; 2V74 B; 3M4Z A; 4F8J A; 5BQ3 A; 5DEO A; 3ENZ A; 5A7F A; 4MS4 A; 3PV0 A; 3L1U A; 4DIB A; 4RHU A; 5C9Z A; 1JEO A; 1IY8 A; 3I4L A; 3PCX A; 3S2F A; 1N8S A; 3RV3 A; 3ACC A; 1VJC A; 4HDR B; 3CX6 A; 4RCV A; 2FPM A; 4PPN A; 1HFW A; 4Z0T A; 5CA0 F; 4UUM A; 2JLY A; 2ZRN A; 4CLR B; 4BZW A; 1KYT A; 4QUI A; 1LBH A; 1S3F A; 3HP7 A; 3FBT A; 3FOC A; 4FE4 A; 1O8C A; 3CFY A; 4NON A; 2AFR A; 1KJJ A; 4FYP A; 3D43 A; 3AB3 A; 5A0F A; 3OFS A; 5C1D A; 4YIH A; 3D6H A; 4JUD X; 3AI2 A; 2DFN A; 4DLY A; 2CFT A; 3UMM A; 1Z2C A; 1TAU A; 4TOI A; 1ISP A; 4JUC A; 3OC0 A; 3L0D A; 2I2B A; 1ZH2 A; 1M7G A; 3K6R A; 3IJR A; 5HBG A; 3MS4 A; 3V1U A; 1GFS A; 3R6L A; 3HL0 A; 4JQY A; 2JL1 A; 4B83 A; 3CLR C; 1KI4 A; 4E4Y A; 1YR7 A; 5JAT A; 4B6J A; 1GUA A; 4OJY A; 3GEM A; 1FLN A; 4EHI A; 1BXR B; 2WD9 A; 5DMX A; 4JUI A; 1D03 A; 3JQD A; 4N39 A; 1WVG A; 1DCM A; 3GD7 A; 3A4Y A; 4INJ A; 4R1P A; 4Q9L A; 4EG0 A; 5CFP A; 4WZA B; 1A72 A; 3R3W A; 3A1C A; 2QRO A; 4KGZ A; 2XQJ A; 4Y1F A; 1NM5 A; 2VRC A; 1AS0 A; 1F3O A; 2HT6 A; 3OM1 A; 4YML A; 3QLE A; 1J21 A; 3BKW A; 4LVN A; 5AIC A; 3JPY A; 2BFE A; 2IYE A; 1NRI A; 3IJE B; 1DWP A; 4PQY A; 2ZJ1 A; 2FOX A; 1FCJ A; 1PFY A; 1MW9 X; 1E5T A; 2AZT A; 4WLO A; 2JCB A; 2OXC A; 4PON A; 3JQ6 A; 4GP7 A; 4U9I S; 4XDG A; 1R0S A; 4GMX A; 4KOA A; 4N0P A; 1EB9 A; 2H7X A; 4IM7 A; 4QAT A; 3TGP A; 5CFN A; 3DUF B; 4DCU A; 1USI A; 2Z43 A; 3PQF A; 1EJ2 A; 3PGJ A; 3QPC A; 4FNG A; 4D4H A; 2C0C A; 3LA8 A; 1K5P A; 1P7W A; 3AW8 A; 4COO A; 3ZLJ A; 4LSG A; 4B31 A; 1N77 A; 4OHY A; 1RRV A; 3JYH A; 1U3V A; 3EVZ A; 3M82 A; 5EHQ A; 1NUT A; 5C5C A; 5DM7 0; 1U3T A; 4KNW A; 4JJF A; 4RDC A; 2P68 A; 2OV1 A; 3O8O A; 4DSN A; 1FDR A; 1YK0 A; 1BJK A; 2CZG A; 2YRW A; 3NJ8 A; 4EHB A; 1CQI B; 5DUL A; 1DT3 A; 1XF1 A; 2YWH A; 3R9X A; 4OMF G; 2Z5V A; 2PQ2 A; 2NVO A; 1IBS A; 1J56 A; 4U5Q A; 5K51 A; 3KQS A; 4CI9 A; 1YTU A; 2FX5 A; 3L7B A; 5L4S A; 1EXD A; 5EE3 A; 2ERC A; 3EVC A; 2NV2 B; 5AJV B; 3ZZN A; 1E3W A; 2WMN B; 3SZU A; 4NDN E; 5JQW A; 1LUA A; 2WUD A; 1NNU A; 3K4H A; 2Y1L A; 2OCL A; 3NBA A; 4ZIR A; 3LY7 A; 4H6V A; 2I34 A; 3P19 A; 1H0K A; 2Z9B A; 4DPM A; 3EJ6 A; 3ER6 A; 4C04 A; 4Z7O B; 2OEC A; 2JGL A; 3K5I A; 1E7R A; 1SG9 A; 2Y4O A; 3DDU A; 3I2H A; 3KXP A; 2NU8 A; 1PFG A; 2C8J A; 4YQZ A; 1XGK A; 3DKP A; 1RZU A; 3MGG A; 4HKT A; 5EKU A; 3GEG A; 4JT4 A; 4MIO A; 4ZK6 A; 2O59 A; 5ALM A; 2IEI A; 2PH1 A; 1CX9 B; 2A67 A; 3IUY A; 1DAM A; 3N76 A; 4FLE A; 4L0C A; 4MX8 A; 3M9L A; 5E9W A; 3W1W A; 4B75 A; 4BFV A; 5IJW A; 4R69 A; 3D8H A; 1ZJW A; 4B7A A; 1VTK A; 2VSY A; 1Y7P A; 3PD0 A; 4BVB A; 2CVF A; 2WT4 A; 1TK2 A; 2VPI A; 2A1T R; 4DA7 A; 3DZG A; 1N7G A; 4ESO A; 2BJH A; 3T5C A; 4G8B A; 2WM9 B; 3UFX A; 4BCB A; 5KX5 A; 3E53 A; 3W5J A; 1T4C B; 1QIE A; 4OYH A; 2B30 A; 2PWZ A; 3WO0 A; 3A6P C; 2EZ8 A; 3D1J A; 3VI4 B; 3S1S A; 4ZC1 A; 3KVN A; 3HJG A; 1EDD A; 4EJS C; 3WGT A; 2FSH A; 5IUJ C; 2IBM A; 4AZW A; 1L4B A; 2ZRC A; 1UC4 B; 2O2E A; 3ND8 A; 3SPX A; 2REQ A; 4KYV A; 4MV8 A; 4DMK A; 3E8S A; 1GVN B; 1V37 A; 2FWB A; 2ONY A; 1F1B A; 3DEK A; 3NCH A; 2C78 A; 2WWT A; 5LQJ A; 2EJU A; 1UBS B; 4RVU A; 2M9M A; 1QO7 A; 4YVF A; 3X1W A; 5IPF A; 3ZGZ A; 2F3R A; 1JPN A; 1GUB A; 2Y92 A; 3BVP A; 1SUP A; 3FRH A; 2F3S A; 3LK6 A; 3LLV A; 2J3Q A; 4LOL A; 5CB6 A; 1T9B A; 1K8Q A; 2AHO A; 3HWX 1; 3H2I A; 4UJ5 A; 4X3O A; 1VHW A; 4JI6 B; 1Y7I A; 4AK9 A; 3OIY A; 3AL2 A; 3KET A; 5JSF A; 3WTB A; 4F9F A; 3FNF A; 1KI7 A; 1A8P A; 3GJ8 A; 2KBF A; 1DRK A; 2QAG A; 2V6G A; 5CCD A; 4YFY A; 3UJD A; 1QMP A; 3HDG A; 5JE8 A; 1GPY A; 2IU3 A; 3RVQ A; 3DF7 A; 1ISH A; 3K9H A; 3QPA A; 3ADB A; 1VJD A; 4PYN A; 3WJ1 A; 1CQJ A; 3FGZ A; 3HHE A; 2EAR A; 3LVF O; 4E2Z A; 1E3Q A; 3RM3 A; 1I14 A; 1P2U A; 1PJZ A; 1YU6 A; 4IUY A; 4KPU A; 4MPP A; 2CLL B; 1F60 A; 2W5U A; 3QI2 A; 4NPC A; 8LDH A; 5J2U F; 1QGD A; 1C2Y A; 2E0I A; 2BEU B; 1ST2 A; 1T3S A; 1N6I A; 2W48 A; 5EE5 B; 2HA2 A; 3KLO A; 1EVJ A; 4QBI A; 1WUT A; 2WVG A; 1NXV A; 1XMS A; 4S2U A; 1Q84 A; 2BTQ B; 2RFL A; 3EGD A; 5A5E A; 2JKZ A; 3PUW A; 1RPN A; 4C89 A; 2VDL B; 4QPN A; 1GT9 1; 1P73 A; 2QE6 A; 3AI1 A; 4DFD A; 1N6P A; 1EM8 B; 1VYH A; 3OP1 A; 1M0S A; 1LI4 A; 6NUL A; 2ECF A; 4ZU6 X; 1W36 B; 3QIT A; 1A27 A; 3LW8 A; 5ALP A; 2ACW A; 2RHO A; 2TMG A; 3G0G A; 2QD2 A; 4MW6 A; 4J0I A; 5GVS A; 4XJX A; 3C3Y A; 4O29 A; 2OBY A; 3EIF A; 4DNP A; 1WVM A; 1TLL A; 3C6Y A; 3N8D A; 4IKE A; 1L5Q A; 1OXB B; 2YWR A; 4CQM B; 4ADS G; 5K8U A; 3O2J A; 1OXS C; 4BY9 E; 4CKY A; 1P1H A; 4IIN A; 4LI0 A; 4JIM A; 4OPM A; 3STH A; 1R4N B; 3VI3 B; 1DKO A; 1I80 A; 2CDB A; 1TZ2 A; 3GJQ A; 3JQG B; 1MJN A; 5HCU A; 5AW2 A; 1KL7 A; 2H1H A; 5TLS A; 1Q22 A; 3ZPX A; 4J6E A; 1NXT A; 1WAB A; 2ODP A; 2A9Q A; 4QVG A; 1BEZ A; 1B2R A; 4HDR A; 4FB4 A; 3EEF A; 3F9R A; 3NIG B; 2GAR A; 5HEK A; 1ENP A; 1DUI A; 5IKP A; 2A9O A; 1U8E A; 3UWM A; 4ARZ A; 3IAF A; 5L40 A; 1KEK A; 4CM4 B; 4MO2 A; 3SFT A; 2WQ7 A; 4L6I A; 1HDR A; 1QTI A; 1M1Z A; 2UZ0 A; 3H3F A; 4XB1 A; 5DBF A; 3V1N A; 1O1Y A; 2YWX A; 2J87 A; 4KIJ A; 4WN3 A; 1QLW A; 4L3W A; 3VU8 A; 5HBR A; 1IUO A; 1BE8 A; 5J2T F; 2MT9 A; 3EZ7 A; 3H3J A; 4JB8 A; 4EI8 A; 3U5Z B; 2OME A; 3CIF A; 1T3Z A; 3KX2 A; 4XPZ A; 4DRY A; 3IPX A; 2XA9 A; 1WO8 A; 5K2M A; 4W5W A; 3X1X A; 1NJ5 A; 5THQ A; 3O2N A; 2BAG A; 2ZT6 A; 2EGG A; 5CLQ A; 1GTZ A; 2M1Z A; 1XRJ A; 1YS2 X; 2G5T A; 2OFP A; 3W6N A; 1MGP A; 3DE5 X; 3WL6 A; 3QPV A; 5EPU A; 3L7N A; 4F97 A; 2XHE A; 3EF1 A; 3OIV A; 3LPL A; 5CBP A; 4OH7 A; 3VYU B; 4BWV A; 5CEF A; 3L0B A; 4Y1E A; 5SYA A; 2HNB A; 3V9A A; 4AJE A; 3LQF A; 1CVI A; 3KV3 O; 5EFO A; 3UJB A; 1X13 A; 1XVA A; 2BN4 A; 1NZJ A; 1QQ7 A; 3ENN A; 1OWO A; 2J09 A; 1AU9 A; 4FMC F; 3SGL A; 3CMC O; 3OSZ A; 3WS7 A; 2DR0 A; 1GZU A; 1GIL A; 1YJ8 A; 1W25 A; 4OXX A; 5E3I A; 2VQ3 A; 4R5M A; 2AQ8 A; 3PJD A; 1LBT A; 1XYG A; 2J31 A; 3AIK A; 4MSU A; 2G9U A; 3KZN A; 2N1M A; 1KAR A; 1QYD A; 3LG2 A; 4MUX A; 2AF3 C; 4F4D A; 2W42 A; 2N3Z A; 5FUY A; 2Z5G A; 4ND3 A; 5G1O A; 1R74 A; 2HMY B; 1RSZ A; 4M87 A; 2FSF A; 1CDO A; 4UOE A; 3K31 A; 3IEL A; 1W5F A; 3DUF A; 2FLW A; 3KL2 A; 4LIP D; 1FRZ A; 5TT0 A; 3RSC A; 3U40 A; 4CEI B; 1XQ1 A; 3H5T A; 3WL3 A; 1JQJ C; 4TZK A; 2XH8 A; 2X0D A; 2ESR A; 3E5N A; 3LBN A; 3NDJ A; 3M1A A; 2E85 A; 5IF3 A; 3UHJ A; 2I7X A; 3NYI A; 4DYH A; 4U2D A; 5JCJ D; 3FI9 A; 3U4H A; 5REQ B; 1QRT A; 2W2N A; 3IEX A; 2FEO A; 2Q5F A; 2QQ0 A; 3K0E A; 1VG8 A; 2ZIF A; 4DWW A; 4XDZ A; 3Q15 C; 3AFG A; 1P45 A; 2EDA A; 4US1 R; 4TTI A; 4YRS A; 2Q50 A; 2CV2 A; 2FHP A; 3LQY A; 1JTF A; 3VXK A; 2X9L A; 1L4L A; 2QEQ A; 4TWI A; 3EPH A; 1OWN A; 1ODT C; 1A5T A; 5JAQ A; 1OB2 A; 3RC3 A; 2O8V A; 3OZF A; 1V8B A; 5AK5 A; 4BNY A; 1EK1 A; 3GRZ A; 2PBL A; 1G21 E; 5FOT A; 1RRC A; 4DE5 A; 4YH1 A; 1GLV A; 4DTI A; 1NJG A; 5DWV A; 2X7J A; 2VHT A; 1EB8 A; 4BWY A; 4MWC A; 3VYS B; 4Q7Q A; 5CG2 A; 4GT1 A; 2EP7 A; 3BIL A; 2AKA B; 2YIV X; 5KJE A; 5I9L A; 3UV7 A; 5JV5 A; 1OHH D; 4O02 B; 2RBK A; 4HZU B; 2WU4 A; 3JVD A; 1KGS A; 3I73 A; 4UUO A; 4DCG A; 3K0S A; 2IUU A; 2XKJ E; 4QUJ A; 4B6P A; 4Z7N B; 1MB9 A; 2FFA A; 3OJ0 A; 1L7V C; 2PGA A; 3IT1 A; 3P0J A; 3Q72 A; 1U5C A; 1DOH A; 2FJP A; 2DE0 X; 2FLV A; 4FHZ A; 3KWM A; 4NMW A; 4NSN A; 4XE5 A; 1PR5 A; 2XYR A; 2ZIT A; 4HP8 A; 3REQ B; 4FI3 C; 4KQH A; 1YTL A; 2EDC A; 1PNO A; 4XFD A; 4GYY A; 4L4R A; 3OTO B; 4WXE A; 3ZHB A; 3AER B; 2W90 A; 3ZC3 A; 3BIO A; 4B1F A; 4HK4 A; 4WFQ A; 3HB4 X; 2NO9 A; 3A6J A; 4IFI A; 2UZ4 A; 3SSM A; 1ZH4 A; 4DSZ A; 2WV7 A; 3SDZ A; 1JTV A; 2JAP A; 2ZJZ A; 4TWJ A; 3TRI A; 2CD9 A; 1BNC A; 3JWH A; 4LXG A; 3U4I A; 5EQD A; 3N2Y A; 5K3O A; 3DDN A; 5GGY A; 3LBH A; 2A88 A; 4X8M A; 5CNI A; 2R8X A; 4M1O A; 5BW5 A; 5DNW A; 4EDF A; 2IIT A; 1AGR A; 1Q1E A; 3K2B A; 1H0H A; 3KS9 A; 4RAQ A; 1YOV B; 4B6Q A; 1Q77 A; 1J24 A; 1KYX A; 1R7R A; 3EFO B; 2B0J A; 2E82 A; 3ON5 A; 3SS7 X; 3PEV A; 3PII A; 4BLW A; 3H05 A; 2ATI A; 2E4V A; 5K3F B; 1TEV A; 3KLM X; 1EE9 A; 4G5S A; 4ER5 A; 3N2K A; 4MRA A; 2FA9 A; 4XQE A; 2O0K A; 4J0F A; 4ZJY A; 5IHX A; 3HY6 A; 4BFC A; 4NJE A; 4TRR A; 3UT0 A; 2YQ4 A; 2H2D A; 5DBO A; 1M6V A; 4MW7 A; 5KWV A; 5LOV B; 1E5V A; 4FHG A; 5C5I A; 3VDX A; 4G81 A; 1YDE A; 1J4A A; 2WP0 A; 3AF0 A; 4ARV A; 3RGG A; 3U62 A; 2O0M A; 2ZIX A; 1RVV 1; 1U8T A; 2R6G A; 4URX R; 3TOZ A; 1B16 A; 3M6V A; 3G0D A; 4R98 A; 1TW3 A; 5KVS A; 3NWB A; 3IR6 A; 2AMY A; 4L5J A; 1UKA A; 5ALQ A; 2LRH A; 2A0K A; 3P2J A; 5JO9 A; 1M33 A; 4O3F A; 1VPL A; 4COK A; 5LLD A; 3IEI A; 3QUT A; 3DNP A; 1GDT A; 3S68 A; 1IWP B; 1CE8 A; 4KXV A; 2IOY A; 1N2G A; 3C1O A; 4BEB A; 4D99 A; 2PT5 A; 1UBG A; 4P3P A; 3I42 A; 2PBF A; 3K9W A; 3VM6 A; 2IYV A; 3E9M A; 4DJE C; 2BEU A; 3G7U A; 3WNB A; 4DY6 A; 2HJV A; 2VQE B; 1GJR A; 2AI2 A; 4B3B A; 1HPL A; 3W5C A; 2BTQ A; 3H0C A; 5E4F A; 2PZB A; 3NEY A; 2DHQ A; 2J07 A; 3FIJ A; 1YB6 A; 1CCW A; 1BWQ A; 1ZNZ A; 2A5F A; 4PLP A; 4BNT A; 4J4B A; 1IHX A; 1JIQ A; 3AL3 A; 3PHG A; 1P18 A; 1WY7 A; 3GLH E; 4R3N A; 1V6A A; 3FZG A; 3K73 O; 2WF9 A; 4LY9 A; 3ZTY A; 4HWP A; 4X1K A; 4B2I A; 4WBN F; 1E5Q A; 3PSH A; 4UXJ A; 4B6O A; 3HMQ A; 2EIH A; 3LWD A; 4Z99 A; 4YLE A; 2EQ5 A; 4KB5 A; 3NRJ A; 3ING A; 3P0I A; 2ERY A; 1W4C A; 2Q2O A; 2ZRG A; 3QFS A; 4E5V A; 2NXJ A; 1PLL A; 2WNH A; 3DDC A; 2PS0 A; 5K5E A; 4OHK A; 1C3J A; 1Y7H A; 4OLF A; 3NT4 A; 4I9G A; 2Y1D A; 3CQ9 A; 4UAJ A; 4XIJ A; 1DAO A; 1E5X A; 3TZC A; 3UKD A; 2QP3 A; 2NO7 A; 1GPK A; 3VNB A; 3EVD A; 3HR4 A; 3K7E A; 3R1I A; 3PFH A; 1CIP A; 2I4T A; 4G4I A; 2YHU A; 2ZM5 A; 4MI6 A; 1KEW A; 1TIB A; 3BE8 A; 3WK8 A; 5L6M B; 2VF5 X; 4IDP A; 1KPG A; 4LZA A; 4FX5 A; 4F91 B; 4UBK A; 3HVJ A; 2VT2 A; 3GRU A; 3H8V A; 1VHL A; 2D8M A; 5EZY F; 3RFC A; 4M8N E; 1F5N A; 1WUY A; 4KFR A; 3PWZ A; 2ZJ4 A; 4L74 A; 4XWY A; 4NY1 A; 3GID A; 5IAB A; 1KQP A; 1NST A; 1DKN A; 3LQR A; 4NS4 A; 1APB A; 3MBO A; 3RUI A; 2M6K A; 4TUY A; 1BWS A; 3AMV A; 1Y5M A; 4FOO A; 2GCE A; 2XA1 A; 4H7P A; 4I81 A; 5FOW A; 2DWQ A; 3PVZ A; 3VPX A; 1M6W A; 4LGV A; 2D0D A; 2LV8 A; 1UC5 B; 2ZK7 A; 3SIX A; 4JL8 A; 4ZCQ A; 4BMY A; 1XC7 A; 3HBJ A; 1S8A A; 2IYW A; 3CIO A; 2WOJ A; 2K87 A; 2JZN A; 4LXT A; 5CNM A; 5EE9 A; 2V0G A; 3DFF A; 3S29 A; 4QRH A; 4UFN A; 4ER7 A; 3R3J A; 3Q3C A; 2BM1 A; 1UK7 A; 3VC2 A; 4UD2 A; 4PSC A; 2LCQ A; 4YRK A; 2I55 A; 1ZIN A; 3OMW A; 2AXE A; 2JKY A; 2A86 A; 2R4Q A; 1ZRS A; 4IHA A; 1X6V A; 2OV4 A; 2E9S A; 1BKS B; 5BUS A; 2YV3 A; 3SFZ A; 6GPB A; 1W4R A; 1V16 A; 2BRU C; 2ZTH A; 5DHR A; 4IL5 A; 6CHY A; 5ICG A; 4RGV A; 3CF2 A; 3QUB A; 1LQ2 A; 2XQF A; 3KUX A; 3SEF A; 2FDR A; 3P9X A; 5E1D A; 3WY9 A; 2L69 A; 3R5T A; 1PT8 A; 1JKJ B; 4BMC A; 3GLF A; 4MUM A; 5DGD A; 2MSK A; 1O9H A; 1QFX A; 4I55 A; 2PY6 A; 5E9A A; 2BR5 A; 3IDO A; 2ASE A; 3TR6 A; 1M74 A; 4KFS A; 4H3S A; 1DQS A; 2BUC A; 3HO6 A; 1ZH8 A; 2X8J A; 1WMS A; 3K0F B; 4LE0 A; 3EZ9 A; 3BRW A; 1M73 E; 1VP3 A; 4GEC A; 2P91 A; 4O4I F; 4F60 A; 1SJ9 A; 3M6L A; 5CVU A; 3H11 A; 1P72 A; 1M1N B; 3OPY A; 3WMF A; 4MQ6 A; 3DCY A; 3LPD A; 4WAU A; 1FNB A; 4C5B A; 3LV8 A; 1FDI A; 3VR5 A; 1MB0 A; 4HVA A; 4M6W B; 1L7E A; 1P1F A; 1TUG A; 1CEQ A; 2YXD A; 1VEC A; 5AKI A; 1G6Q 1; 1GWN A; 2BEW A; 5EPO A; 1DXQ A; 1OVM A; 2XZP A; 1LX6 A; 4WK0 B; 1VPW A; 4DUZ B; 3EC1 A; 3LKD A; 4KFU A; 3PUK A; 1M10 A; 2WSM A; 1FNN A; 1PQ4 A; 2VNI A; 3A0R B; 3UKJ A; 1AJE A; 1GAE O; 1B0A A; 1V3Q E; 1FL9 A; 4MIK A; 3H8S A; 3DBR A; 1NVD A; 5DOV A; 4KCG A; 1ESM A; 4OQY A; 4QX6 A; 4E5M A; 1PP4 A; 1YB4 A; 4RAO A; 2FWM X; 4GPT A; 4D57 A; 1VJT A; 1DIO B; 5LB4 A; 2WG2 A; 5CH3 A; 3ZWD A; 1HBJ A; 1W6U A; 1RM0 A; 1YR6 A; 3JQP A; 2CKD A; 4PAQ A; 1XJQ A; 2GCN A; 3BJI C; 4MUB A; 1Z13 A; 2QME A; 3R23 A; 5CVC A; 2YDX A; 5LNK 6; 2IOD A; 3LVR E; 1DX9 A; 3IV7 A; 4E5P A; 5AK0 A; 5ALU A; 6YAS A; 3ZU6 A; 2JJY A; 5DI3 B; 2O52 A; 1PGQ A; 1WCX A; 3NET A; 1ZUD 1; 4ISB A; 3ZU3 A; 4B46 A; 5DF1 A; 1A95 A; 5JXI A; 4YYZ A; 3GFS A; 1XWI A; 3G2P A; 3SE7 A; 2Q8N A; 1KDY A; 4BNK A; 5B7G A; 3RVR A; 1ODI A; 1BE0 A; 1R15 A; 3ZWW A; 5L45 A; 1LV7 A; 5I4C A; 1CJM A; 2Q3H A; 1KSJ A; 1NT2 A; 2GYW A; 3EY4 A; 5JIB A; 3R6W A; 1PR4 A; 4UAG A; 3OET A; 1J1I A; 1I3L A; 1V5G A; 1Z82 A; 1ZJY A; 5JRJ A; 1RM5 A; 1WZN A; 3LT1 A; 2NO4 A; 1V5W A; 2WKR A; 2CNB A; 3IME A; 1ZVM A; 5E4J A; 4ETI A; 1LFD B; 5B58 C; 2UXB B; 1T6B Y; 5IJ0 B; 3HJA A; 1N8T A; 1QGY A; 3A6D A; 5FNV B; 5DN6 A; 3SZ3 A; 2OOK A; 4DYV A; 5CD9 A; 1R88 A; 1YRQ A; 5LD2 B; 1NZF A; 1T6N A; 3ENQ A; 2AOV A; 4OAP A; 4I5I A; 3WLN A; 3JQA D; 2LCI A; 3GGG A; 4HUQ A; 3DE4 X; 4OQQ A; 4ON9 A; 3IMF A; 2AGV A; 2JAQ A; 4HNQ A; 3SQL A; 4I4I A; 3WZ2 A; 3LHI A; 5CA0 A; 2EBH X; 4BZQ A; 2FG5 A; 3QWH A; 1V1J A; 2G28 A; 3ESY A; 2PD4 A; 7YAS A; 1T9G S; 2IYL D; 3L8Y A; 3A9W A; 1IXK A; 3GZJ A; 3H5W A; 3I2J A; 5TPW B; 2GPT A; 1IBK B; 3FLE A; 1ZI9 A; 4XSV A; 1FVH A; 2OO3 A; 3H83 A; 1GYR A; 4DV0 B; 2Q7D A; 4GF5 A; 3KE9 A; 3SQN A; 1S9H A; 3H87 A; 3LDH A; 2AXN A; 1Z0J A; 4FFB B; 4HI0 E; 3RFX A; 4M0M A; 4EEX A; 1F0P A; 3KKP A; 3B7W A; 3Q87 B; 4RPI A; 1G60 A; 1E7Q A; 2ODQ A; 3PNL A; 1X7X A; 2M1X A; 3NBX X; 2AUT A; 2X3U A; 4HAZ A; 5C5D A; 4BE9 A; 2XMS A; 3GDO A; 3MTQ A; 4YCN A; 4UTP A; 2HTF A; 4BX3 A; 4LYH A; 4JDP A; 3WDM A; 4RVF A; 2ATE A; 3G7N A; 3OLY A; 3B3Q A; 4IW3 B; 3U48 A; 3T1V A; 2QJO A; 4RTJ A; 1DAK A; 5AUQ A; 1I51 A; 1L5R A; 5K0Z A; 2J4C A; 4HBZ A; 4V0M E; 2H0R A; 4Q9H A; 3OZE A; 3EQ9 A; 4H1T A; 4KQM A; 4YMV A; 3AOE A; 4CKW A; 3GJ3 A; 4WY8 A; 5E6S B; 2P9G A; 4R0N A; 1Y3B E; 2O1E A; 3KYJ B; 5DTI A; 4PIV A; 3KZM A; 1XNJ A; 1MAB A; 3DQZ A; 3IQW A; 1ZCB A; 4INO A; 5ER6 A; 2BGI A; 1CER A; 2ISQ A; 5LMN B; 4BRU A; 1DG9 A; 5IP5 A; 4KWC A; 1LSS A; 1MQA A; 1GHT A; 4BN0 A; 1JHD A; 5FUW A; 1ICI A; 2FW1 A; 4XPI A; 4WOK A; 2JS7 A; 3PQE A; 2IHN A; 1Y8C A; 4OI4 A; 1E20 A; 1Z37 A; 4Q4J A; 4ARI A; 3NAN A; 1R0R E; 4KJZ A; 2Q0S A; 5EIA A; 2QLL A; 2GD6 A; 4E15 A; 3T34 A; 1K5G A; 4MQL A; 4ISC A; 4DV5 B; 1A8Q A; 1KPI A; 4CM3 A; 1I7L A; 2ZTS A; 4MS3 B; 3T6K A; 4UZE A; 1BC5 A; 1P7Y A; 3MAJ A; 4QO8 A; 2XNC A; 5KVQ A; 1RI3 A; 4P7F A; 3S99 A; 4AG5 A; 3QV2 A; 4XD7 D; 4FNO A; 1ISJ A; 4HDK B; 1JLH A; 3FMV A; 2R6D A; 4PHG A; 1YS7 A; 1AHP A; 1C1D B; 4B43 A; 5KJC A; 4UNI A; 1DBI A; 2JFQ A; 1NSF A; 2H54 A; 5LWH A; 2BIS A; 3CO0 S; 1PZG A; 1DO8 A; 1Z4L A; 1M9N A; 1IK4 A; 3KSR A; 2C2Z A; 2BD0 A; 5EYP A; 4LMA A; 2GD2 A; 4EG7 A; 1QX4 A; 4RWZ A; 1U7X A; 4E2W A; 3VGJ A; 4HX2 A; 4QFC A; 5E2I A; 4GLB A; 5FUM A; 1RQP A; 2RHG B; 3A06 A; 4F9G A; 5D3M A; 4BBZ A; 3WAJ A; 1G8M A; 3B1V A; 1NU6 A; 4QQN A; 5LYJ F; 1CR1 A; 1WVI A; 3HFJ A; 1U2G A; 2FP3 A; 1FXW F; 3L92 A; 4TS1 A; 4ZO8 A; 3H9Q A; 4C9B A; 4KN9 S; 5DLD A; 2D3T A; 1CNF A; 1QP8 A; 1ZW6 A; 3IH5 A; 3RRE A; 4XGN A; 3FNR A; 3VIS A; 3THX B; 4YRA A; 2OAQ 1; 3URM A; 1NVG A; 2ZI9 A; 4LC1 A; 4J8P A; 1PWE A; 2YY5 A; 2JK1 A; 3W2E A; 4CR7 A; 2QVB A; 1Z2O X; 3A6K A; 3G5M A; 3QI7 A; 3FI0 A; 4CM6 A; 4TRO A; 3I9L A; 3LP8 A; 3WB2 A; 2WK1 A; 4FC6 A; 4GGP A; 4QUB A; 2ORI A; 4HVC A; 4BVJ A; 1UMC B; 1STO A; 3OX1 A; 4LAF A; 3EOZ A; 2R1U A; 1P7V A; 3GP5 A; 4NDA A; 1Z6Y A; 4YT4 A; 4YV0 A; 1M2W A; 2XUK A; 2WPD A; 4H4C A; 2TYS B; 4DMR A; 4XVU A; 4EMT A; 1YQ9 A; 3I30 X; 1XRM A; 4G2T A; 3CW8 X; 1P7C A; 2AMV A; 2R62 A; 5AI4 A; 2NVU B; 2OCA A; 4TSK A; 2CD7 A; 4GBJ A; 2EX4 A; 2EV9 A; 2C94 A; 4UFO A; 4KSY A; 3B89 A; 4W7I A; 3UYL A; 2WM2 A; 5M34 A; 3I34 X; 4RAN A; 2WJI A; 3MWF A; 2YN2 A; 1CFZ A; 1G8G A; 5F21 A; 4LEI A; 3WI7 A; 4YMH A; 1O6F A; 1P4S A; 5JD4 A; 1JWA B; 1U4O A; 1QRR A; 1QQT A; 2QRM A; 3TDN A; 2YIJ A; 1IB6 A; 4M0U A; 4Z4Q A; 1E8N A; 2V9Z A; 3D5Q A; 3OA2 A; 4NAT A; 3M49 A; 3LE8 A; 1NE2 A; 2R8P A; 1L6I A; 3ZUP A; 1L7X A; 3K3P A; 4KC0 A; 4Y2S A; 4REL A; 4Z7Q B; 3ZX5 A; 1EE2 A; 2GZS A; 4WA7 A; 4WCD A; 4QEK A; 4OB7 A; 3BIJ A; 3ROZ A; 1J6U A; 3O3N B; 5HK8 A; 4MKF A; 3ADO A; 3KQO A; 4FEE A; 5CVT A; 4ZVT A; 1WUJ S; 4BYG A; 1XPR A; 1T25 A; 2F62 A; 3PQ5 A; 4HCJ A; 4MGE A; 5B3K A; 1ZI6 A; 3EDO A; 3DBL A; 4XDY A; 3H75 A; 4HRY A; 1PYG A; 4YAI A; 3U39 A; 4ROS A; 1ZFN A; 3E8X A; 2VNA A; 1H5Q A; 2VJC A; 5AQE A; 4AKT A; 2MU1 A; 4KQ1 A; 1J0B A; 3MTG A; 5H9W A; 1R3D A; 1KYV A; 2BH2 A; 2XBL A; 3D6A A; 2XF2 A; 2X4I A; 1TD9 A; 3G2Q A; 4LX8 A; 1E6K A; 3UK1 A; 2QRK A; 4C2A A; 4EA7 A; 3QX7 A; 5J2U A; 4M3N A; 3ROQ A; 4ZYX A; 2AN9 A; 3QTB A; 1EWY A; 1T15 A; 5LWQ A; 1XEA A; 5G02 A; 1FML A; 1JJB A; 2GUW A; 2X86 A; 2WFZ A; 1SRQ A; 3NON A; 4QVI A; 1GSH A; 1GFZ A; 4G8D A; 1J07 A; 4OCZ A; 2QV7 A; 3T9F A; 1PT6 A; 4RV5 A; 1G42 A; 1O2D A; 4C30 A; 3BBP A; 4FFM A; 5B2Z A; 4YXW A; 2DJ5 A; 2ZUW A; 3ODG A; 3GVP A; 2X42 A; 1O9B A; 1DC7 A; 4PZU A; 1AKV A; 5H81 A; 4BJH B; 4C0B A; 2H57 A; 1PJC A; 2F17 A; 4IIX A; 4FXO A; 1T0I A; 1YR3 A; 4A5O A; 3NVK I; 1OAO C; 2RC6 A; 2CLM B; 2R85 A; 2Y0D A; 2DBR A; 1FQO A; 3UF0 A; 1R6U A; 2ZE8 A; 1QVV A; 3H84 A; 3AY3 A; 3MWD B; 2I4M A; 2DUJ A; 2HBZ A; 3HEB A; 4BFU A; 1N33 B; 1ID1 A; 4U7F A; 1PF7 E; 1U81 A; 1XXH B; 4LMP A; 5IW8 A; 1KDO A; 1PR1 A; 3D0K A; 3IAA A; 1FSG A; 1YID A; 1GL9 B; 1DDI A; 2O48 X; 3PI7 A; 3KJI A; 4GBM A; 4O4L B; 4PAN A; 5DDI P; 1Q2U A; 3T4X A; 4F92 B; 1Q12 A; 3U44 B; 1G17 A; 3FPF A; 5DMH A; 3UXY A; 2WAY A; 1I9G A; 4LQC A; 2XYG A; 2ILV A; 1MO3 A; 1AO3 A; 2A8Y A; 8ABP A; 3AYX B; 3HCP A; 2GK3 A; 2HQB A; 1ODF A; 3CO5 A; 4RLS A; 3J7A C; 1O5O A; 5J2T A; 3FHT A; 2C84 A; 2C1Z A; 1BGW A; 1YL7 A; 1M0E A; 3H0K A; 1Y4A E; 1XTK A; 2CB0 A; 1V71 A; 1XMI A; 3W3A H; 1Y8Z A; 2B8E A; 3GFV A; 4K2H A; 3JWG A; 3HNO A; 2N1B A; 3P4X A; 2I76 A; 2QMU A; 2XDQ B; 3BX1 A; 2HRD A; 3FLM A; 3TPZ A; 1HE3 A; 4PG6 A; 3EEO A; 3A05 A; 3GDW A; 4KXQ A; 1W9H A; 3IQD B; 1H1T A; 1R9N A; 2IXT A; 5E4T A; 3H4V A; 1N4D A; 3WIV A; 5A6S A; 4PTY A; 2UUB B; 4HL6 A; 1VSU A; 2CNO A; 4JVO A; 4XUC A; 1UK6 A; 4CTO A; 3L8E A; 3MTA A; 4H7F A; 5HNK A; 3PNB A; 2ZT7 A; 2B3D A; 3LLU A; 5TF4 A; 1TKA A; 4DDW A; 1S4Q A; 3JPW A; 4I9U A; 3UPD A; 1O94 D; 1Z4K A; 3C6B A; 1JYS A; 4C2U A; 1F8F A; 5AFS A; 5ALZ A; 2D59 A; 2ZIU B; 5BWN A; 5B57 C; 2F8S A; 1LDE A; 1LDN A; 2P3O A; 3KAQ A; 1GNP A; 6ABP A; 1NP7 A; 3ZKW A; 3DLA A; 5L53 A; 4DJ5 X; 4GDF A; 2AI1 A; 1A9T A; 3IE2 A; 2HPV A; 2WE8 A; 1XZH A; 4QXO A; 1EFP B; 2OT3 B; 3RF7 A; 1NUP A; 2FI1 A; 4GI1 A; 1XEL A; 3IPL A; 5CCX A; 1PK8 A; 2Q1S A; 1YQT A; 1QRU A; 2IUY A; 4YCA A; 4HDS A; 5EJM A; 3GLS A; 3GRA A; 3B1W A; 2Q2N A; 4JKI A; 1IVN A; 2VJL A; 1XPJ A; 1FDT A; 1MH1 A; 4EBB A; 3UMY A; 1OIW A; 1JLR A; 3ZV5 A; 4NEN B; 4GOX A; 4BL5 A; 1X19 A; 1RBZ A; 5HTO A; 3R6T A; 1SH7 A; 4I6G A; 3G0C A; 4NFY A; 4K6O A; 3NAS A; 1VKN A; 3QVM A; 3Q3G E; 4LTM A; 3F8T A; 4J91 A; 1NQ5 A; 3HSY A; 1CY7 A; 2WHY A; 1QHG A; 4IRM A; 1MMF B; 1O58 A; 1W4B A; 2GOY A; 2HEI A; 2P6O A; 2QI9 F; 3E82 A; 3GF4 A; 3G2I A; 3BMO B; 4H35 A; 2JJM A; 1NOI A; 3QEN A; 3L9X A; 3L79 A; 1ZES A; 2F43 A; 3GPC A; 1BDM A; 2JID A; 4G26 A; 3D4L A; 4B81 A; 2IGT A; 5CAE B; 1GSO A; 1ZM7 A; 2L82 A; 1K7Y A; 2ZCJ A; 2VXC A; 1G19 A; 5IZC C; 5FWY B; 1YSC A; 5H02 A; 4X6X A; 4RUL A; 1IQR A; 3DLC A; 4MJX A; 3V97 A; 3O95 A; 3I64 A; 1FMT A; 1M2G A; 1O7J A; 4PG7 A; 4KP7 A; 1PX0 A; 1UJ2 A; 3IVA A; 2ZJ6 A; 4G1F A; 2SKC A; 4A6X A; 2CNL A; 1HDO A; 4RK1 A; 2LR0 A; 4OHC A; 4TN1 A; 4DCM A; 1ELO A; 3MB8 A; 4NH0 A; 4AF6 A; 1WPQ A; 4MIC A; 2HSD A; 5EJ2 A; 3SD7 A; 2HK9 A; 5I7U A; 2JAR A; 1QV7 A; 5A06 A; 2NN3 C; 4Y0V A; 4DNF A; 3WGK A; 4UNR A; 1TE2 A; 3HLT A; 4W8G A; 3CZM A; 1P0F A; 1RHR A; 2PG3 A; 3BGD A; 5DHA A; 1JKX A; 4BVK A; 3ZDD A; 3FWN A; 4YPV A; 1KZH A; 1EVE A; 1BD4 A; 4M1Q A; 4M0L A; 1BHS A; 1UK9 A; 1WR8 A; 2JJQ A; 4Q6W A; 4ECA A; 4EGQ A; 4DB2 A; 4I6J A; 1VCO A; 3GJ5 A; 1IL0 A; 1JEJ A; 2I3G A; 2YZ2 A; 1IVY A; 3ZLV A; 1WSB A; 4N48 A; 2VJM A; 1U2E A; 2QRH A; 5KWE A; 4M8S A; 1FTQ A; 5B1D A; 3KLP X; 1VYV A; 1JWL A; 3DD1 A; 3UAW A; 2HHH B; 4KO1 S; 4L75 A; 3FYS A; 4YJG A; 1WBB A; 3I0N A; 1DQR A; 3F3K A; 4BUC A; 1FC0 A; 3COY A; 5HRU A; 1ZXX A; 1DMR A; 4DNX A; 4ZYW A; 1X7H A; 1GZ6 A; 1YBA A; 2PZL A; 4R9S A; 5TX7 A; 2N46 A; 1XLU A; 4PO9 A; 3H9B A; 1N5L A; 2VDR B; 3VPD A; 5K01 A; 1C7Q A; 1M2N A; 2OD9 A; 3FVR A; 1S2D A; 2W71 A; 2EGU A; 5K6A C; 3AHJ A; 1JEY A; 3GVC A; 4TTP A; 2XUF A; 2N9U A; 5G53 C; 1C9D B; 1GAJ A; 2GI4 A; 2J67 A; 5GAD l; 4LJ9 A; 4V3C A; 1NHG A; 1U5B B; 2RGU A; 1FFC A; 3GI1 A; 2MR6 A; 3OI7 A; 3CWQ A; 3ICV A; 1OLX B; 1FDU A; 4F2P A; 3ESD A; 1H85 A; 5AW3 A; 5FL3 A; 1KI6 A; 2IIP A; 4ZA2 A; 3RRB A; 3WNV A; 1OZF A; 3ZLU A; 3L49 A; 5ADU S; 2P67 A; 4MV7 A; 2ZAG A; 4F93 B; 2DH6 A; 3VJM A; 2PX6 A; 3JQ8 B; 4PZD A; 4LZW A; 1F9E A; 5G4L A; 2YNM D; 5HUJ A; 5CT8 A; 4MDE A; 1IBC A; 2R8E A; 3VKW A; 2IHJ A; 3CX8 A; 1O96 A; 8MHT A; 3IBT A; 4NLL A; 2QY9 A; 5FA5 A; 2X7X A; 1N6K A; 3OD5 A; 2AEM A; 4DLW A; 4HQR A; 1PRY A; 3GFP A; 4UDN A; 5BSG A; 3EGI A; 2V96 A; 5IVK A; 3GZN A; 4RPK A; 2BBW A; 3I1I A; 5HXA A; 5GAJ A; 3NW9 A; 5CB0 A; 3U4G A; 1XPU A; 5A05 A; 4JQC A; 2I2C A; 2FR1 A; 1CU1 A; 4HXF B; 3RAR A; 3EXG 2; 1TM3 E; 1E59 A; 3M81 A; 1EDO A; 1H66 A; 2IEL A; 2X5N A; 3DOE A; 4IHQ A; 4UBL A; 2XSZ D; 5TQ0 A; 3W7V A; 1PS9 A; 1E9C A; 2IU4 A; 3KTD A; 4DT1 B; 1K6I A; 1H2E A; 4XWW A; 2NTV A; 1TMO A; 1MIH A; 3N0V A; 2OD7 A; 1Z5G A; 5AKY A; 4S05 A; 2EW1 A; 3ILV A; 3O74 A; 4DCP A; 3OZB A; 1OIL A; 2ZWV A; 3ZDX B; 1OCE A; 3RS4 A; 4H0C A; 4KBF A; 3FRO A; 1WCI B; 1YZT A; 5C18 A; 3E9Z A; 2Y4N A; 1UKB A; 3JQE A; 2HV8 A; 3IKJ A; 5KTK A; 2IZ5 A; 1NYL A; 3V3T A; 3DM5 A; 2R3S A; 5IVD A; 4XFJ A; 3KR2 A; 3WGL A; 5GJW C; 1EX9 A; 1M2J A; 1Q5P A; 2W6I A; 1JBK A; 3F5F A; 4OBE A; 4BDS A; 5JY6 A; 4ER0 A; 2IU6 A; 3FWK A; 1MNA A; 1XK6 A; 3MIL A; 3RIE A; 2IHV A; 4O59 O; 3D4I A; 5EQJ B; 3T15 A; 4BMO B; 2XUI A; 1FTS A; 2J41 A; 1T5H X; 4EQZ A; 1JKI A; 2OV3 A; 5ISM A; 1XCB A; 1TEX A; 3CJT A; 3F7T A; 2DG2 A; 1PW5 A; 1SCU A; 4O4L A; 3TLC A; 1DDG A; 5K0S A; 4U63 A; 2WAO A; 4X71 A; 2VH2 A; 3RJ5 A; 1XTT A; 5BVH A; 1RT9 A; 3DCN A; #chains in the Genus database with same CATH homology 4Q39 A; 3S2Z A; 3H1E A; 4F3T A; 4R83 A; 3GBV A; 3S1S A; 3L6R A; 3E4C A; 3ANY B; 3SZO A; 3AHI A; 2ES4 A; 2XMR A; 2XMC A; 1R27 A; 4KXW A; 2XZD A; 4PGF A; 4Q3O A; 2DLN A; 4P82 A; 4GM4 A; 3TW8 A; 2QVY X; 3A9S A; 1T8W A; 4S1T A; 4JEM A; 2I80 A; 4L28 A; 4RLQ A; 2LZ0 A; 4XIF A; 4J0K A; 1VKZ A; 2G63 A; 4LDA A; 1MDF A; 1RXU A; 2FN8 A; 2J9Y B; 3OPY I; 1EPU A; 2V5U A; 4CCS A; 1L5F A; 3FCX A; 1WD7 A; 2XRF A; 1C0E A; 4B5K A; 2PAN A; 1OLS B; 2C58 A; 4QYI B; 1Q83 A; 5LKA A; 1H2Z A; 4BXO B; 1ZMR A; 2HU7 A; 5EMN A; 1R3U A; 3VNA A; 4D3X A; 1DBQ A; 4Q6B A; 1OX4 A; 3TQI A; 2VZE A; 1FVF A; 3SE7 A; 2J3D A; 1NXP A; 2DST A; 5K2O A; 1QIK A; 4G97 A; 3FSG D; 5B3J A; 2H51 A; 4D4I A; 4EVR A; 3T8Y A; 2Q3C A; 3LQ4 A; 1A9R A; 2XCO A; 5EXE A; 3IE0 A; 2CGE A; 4L4U A; 2AEE A; 2QS9 A; 3OO0 A; 5IC4 A; 3OQH A; 3ILH A; 1CG6 A; 4RK5 A; 5DFA A; 4QEL A; 4X92 A; 4JU8 A; 2D6F A; 4FGL A; 1ODS A; 1PR6 A; 5DJ5 A; 3O1I C; 5DNC A; 1RPJ A; 1K86 A; 1S8O A; 1QOZ A; 3I39 X; 1V11 B; 4RM2 A; 3HDV A; 2Q0X A; 1OU4 A; 4BTV A; 2HRE A; 3JZF A; 3IXQ A; 1CPY A; 3C74 A; 4KRG A; 3CCC A; 3TOC A; 4ENP A; 1VE6 A; 3G9Z A; 4YEC A; 4BAU A; 1WKV A; 2VSQ A; 1A9O A; 3IVR A; 1L4E A; 3GWG A; 3C8D A; 3SIR A; 2WLS A; 1USW A; 1GPU A; 3AHE A; 3BL8 A; 4LMB A; 5CCE A; 3GFF A; 3HZO A; 3TSY A; 3ISV A; 3CYF A; 1LX7 A; 4N0Q A; 2GHR A; 2BZS A; 3HLY A; 1QIF A; 2GEF A; 3BJR A; 3LLC A; 1CEY A; 1C30 B; 2HXG A; 2GBC A; 1IHD A; 4KE6 A; 3ETC A; 1K7E B; 1GGK A; 4JU9 A; 3T52 A; 3VMM A; 2JGE A; 1Y5N A; 2Q5Q A; 5HF8 A; 2VPY A; 2D5L A; 4QPJ C; 4EA8 A; 5EIE A; 4EB3 A; 3K9B A; 2JLB A; 4F5E A; 5ALO A; 4R8K A; 1AB6 A; 4EB0 A; 1DCK A; 3WA6 A; 1QB8 A; 1VPE A; 3C3A A; 2QB5 A; 3EXE B; 4HO1 X; 2C42 A; 2IY7 A; 3B52 X; 5DBH X; 1VFN A; 3F71 A; 3BFP A; 4L51 A; 4F4F A; 3GOQ A; 3STV A; 1YIO A; 1EI9 A; 4OFW A; 3IA2 A; 4R2J A; 1TCB A; 2Y9P B; 2KHZ A; 3PX7 A; 2WTN A; 1VXR A; 1Q0Z A; 3T9D A; 1TKR A; 1A4X A; 5JS1 A; 3F5M A; 3R41 A; 2FW0 A; 2D1F A; 3RR2 A; 2R1V A; 2GBP A; 3HO8 A; 3C3B A; 1AMU A; 1XZB A; 3H1P A; 5IBR A; 4H7K A; 1YA4 A; 2WUG A; 1BVH A; 5LNK 1; 3IBC A; 5IC6 A; 1OLS A; 1TJN A; 4Z4G A; 1P6Q A; 2PUB A; 3IDA A; 3WKD A; 2VVT A; 5AKJ A; 3NOQ A; 5EHZ A; 2D2X A; 3V42 A; 3C70 A; 5AXB A; 2JVJ A; 3F6S A; 3V7F A; 1T36 A; 5HFA A; 4CCY B; 3CEP B; 3QQ5 A; 5AKZ A; 1YA8 A; 2QL7 A; 5ALI A; 1K8Z B; 1EHY A; 1IUP A; 2NPO A; 3DJF A; 5T3Y A; 4DMF A; 3G8D A; 5LZ5 A; 1OX6 A; 2RIP A; 4FYI A; 2FEK A; 2X2O A; 1BD3 A; 2YCL A; 5KWS A; 2HZB A; 4KN5 A; 3MGA A; 4REO A; 1J33 A; 4BCD A; 3HXM A; 1AKQ A; 4X24 A; 4C01 A; 3JWE A; 4OBB A; 1EYZ A; 4QUG A; 4C5C A; 2WIG A; 1XYY A; 2ACK A; 2RJ3 A; 3S8E A; 1HQD A; 1QX3 A; 1G2P A; 1YBH A; 3CZA A; 4Q34 A; 2ZSK A; 1H22 A; 1RP1 A; 2J9X B; 1IOV A; 1HKH A; 2CLI B; 2O2I A; 4LXY A; 1FW8 A; 2Z1D A; 1C3O A; 4UHH A; 4KQ9 A; 5IVH A; 4TS9 A; 4KPY A; 3P45 A; 4MV1 A; 3DHY A; 5CXX A; 1JMY A; 2JFX A; 1DV2 A; 1AKR A; 4QU5 A; 3MYY A; 5H9V A; 5IU6 A; 2LPM A; 4RKQ A; 3RQI A; 1KQF A; 4F11 A; 4DCJ A; 3EQ2 A; 4DJF C; 2FCR A; 2MYT A; 3EBL A; 4LYY A; 4ZWN A; 1AG9 A; 2Q6V A; 1FFD A; 1FVX A; 2VRN A; 3K20 A; 1Z7E A; 4HAI A; 5JAO A; 2Z8Y M; 1JHP A; 2EHJ A; 3VSD A; 3DLT A; 4QAS A; 1V2H E; 3FZY A; 4AO7 A; 2X15 A; 1JU4 A; 2WHP A; 3GAA A; 2GM5 A; 4WBX C; 5AM4 A; 4AUE A; 3PF8 A; 4AZ0 B; 3HXK A; 4KE9 A; 3URK A; 4FX2 A; 3ZOZ A; 1CY8 A; 2ALJ A; 1UDR A; 4D56 A; 1L5Z A; 3OOY A; 1O57 A; 2WSY B; 4GR4 A; 5JD6 A; 4GXN A; 4LXH A; 1VA4 A; 3NXK A; 3Q3E A; 1AQL A; 3X43 A; 3KHT A; 4WUT A; 1ZIX A; 3KYI B; 1TYZ A; 2QD5 A; 2BKV A; 1QGE D; 3O82 A; 1QO9 A; 2V7B A; 4AIR A; 2XEC A; 4F3K A; 4FEY A; 4RY9 A; 2IVF A; 1CUG A; 3D02 A; 4I4T F; 1Z36 A; 3FGP A; 4CGY A; 2ZDQ A; 3FSE A; 3GGJ A; 1NNI 1; 2MOK A; 2ZJ7 A; 1RNL A; 4N7M A; 2R7L A; 4KZK A; 2G8L A; 4JAV C; 1D6M A; 4KXY A; 1DBR A; 4E4R A; 3ZGN A; 1JP7 A; 5F7X A; 2XTA A; 4KV7 A; 1X3L A; 4YGX B; 1Z34 A; 5DBE X; 4H27 A; 4NEG A; 2WY2 D; 3OQI B; 2QOE A; 3UUE A; 3WWP A; 4RKY A; 1JJI A; 4GW3 A; 4EY8 A; 3EDC A; 1F4V A; 4LZO A; 1XT6 A; 3VR0 A; 4DM4 A; 3W04 A; 2RON A; 2VTV A; 4UBM A; 1OX5 A; 4JYP A; 2A9V A; 3B6J A; 3CZ9 A; 5FXM A; 4Z0N A; 3BXP A; 3CH7 A; 5IAO A; 3WFZ A; 1A3C A; 1ZWK A; 4NQR A; 5ADV A; 3ZV7 A; 2LND A; 1G2I A; 2X76 A; 5CBP B; 1ZR4 A; 3IEP A; 5HGX A; 2QR5 A; 1L5O A; 4F06 A; 1CUU A; 4AZ3 B; 3IUQ A; 5JAH A; 3DTV B; 1J0C A; 2HPH A; 3FUC A; 4EVS A; 2QSJ A; 1JHX A; 4GUD A; 4V2I A; 4K9N A; 2EFY A; 1J8Q A; 2FUK A; 2C3M A; 3NFR A; 1DZ3 A; 4RSM A; 2PGN A; 3WKA A; 5ALR A; 4AEC A; 4EY7 A; 4EHL A; 2M6S A; 1P0M A; 3QPB A; 2AJB A; 1TVM A; 1BCR A; 2ZUU A; 5BQP A; 2AJ8 A; 4EF4 A; 4K9R A; 3GA5 A; 1ULB A; 3N6X A; 3CXU A; 4KQK A; 4AXX A; 1EDB A; 2A0X A; 4RPC A; 2WUE A; 1PAU A; 1GPN A; 1GPH 1; 2PL5 A; 3IDZ A; 2PL9 A; 2Y9M B; 2P4U A; 3NHW A; 3HVR A; 2JBA B; 2E4W A; 2RKW A; 4MA0 A; 1YDG A; 4KAC A; 4L0M A; 4O4J F; 2WFL A; 2HQR A; 3BXG A; 3LGX A; 4WDQ A; 1PNR A; 3IP9 A; 4AZ0 A; 4MLC A; 1DQY A; 3KV1 A; 5KZM B; 4F2D A; 1BAP A; 4ME6 A; 3BRE A; 1H2W A; 5GQS A; 1SOM A; 1YHZ A; 4I4N A; 1CUA A; 4EY3 A; 3HJU A; 2JFU A; 4JLS A; 4Y2P A; 3PWT A; 2JIO A; 4AWB A; 4ZMR A; 2QLJ A; 1LNS A; 2MR5 A; 3C39 A; 5SY9 A; 2NV0 A; 4GM1 A; 2XOD A; 1GNL A; 3VU1 A; 1JQ5 A; 2C00 A; 5AKX A; 1IUN A; 1KEE B; 5HUQ A; 2CDR A; 1D5W A; 5BW6 B; 5CH5 A; 1Y6R A; 1VOT A; 4MPT A; 5B7Q A; 1A7U A; 5K20 A; 2HBR A; 2ZUV A; 3IQH X; 5KQG A; 3E7W A; 4BAZ A; 4QYX A; 3Q0T A; 4E4T A; 4RWE A; 2GPS A; 3IAM 6; 4M7W A; 4KAJ A; 3N2Z B; 2OV8 A; 1MTO A; 3AHD A; 3F6R A; 1ZD2 P; 4TV9 F; 3DGF C; 1WTC A; 4A5S A; 3M9W A; 4UDY X; 4E5D A; 2NZY A; 3QVL A; 1YK1 A; 1KBO A; 3LZC A; 1FS5 A; 1CUZ A; 1QK4 A; 1LPS A; 1R50 A; 5SY6 A; 3V55 A; 3ZGL A; 3CLK A; 1YFZ A; 4R31 A; 3UHF A; 1JPU A; 4NV3 A; 5K3S A; 3AHG A; 2FUG 6; 3QLV C; 2A33 A; 3S5U A; 1YCD A; 3E0X A; 3O21 A; 2X9Q A; 4LYD A; 5F4B A; 5FP0 A; 4KK4 A; 1R9G A; 5HJ5 A; 4BB9 A; 2Z9D A; 2YID A; 2HA4 A; 3OZG A; 3BSF A; 3DPS A; 2RI6 A; 4J43 A; 5EJ8 A; 4L8Y A; 3WA7 A; 3EUE A; 4EEC A; 2JGF A; 4ARB A; 2WKW A; 4FQ7 A; 3L8C A; 1UPF A; 3BF8 A; 3SLK A; 4LXU A; 3L7O A; 5D6N A; 3LRT A; 3SG0 A; 4M9R A; 3RPE A; 5C81 A; 3AZP A; 1RXI A; 3KEL A; 3TE7 A; 3QEL B; 1GGF A; 1G4H A; 1VCH A; 2Q5J A; 2HLN A; 3X3H A; 3DEJ A; 2PMB A; 1NVF A; 4FTW A; 4OIF A; 3GGS A; 1L5K A; 3HBM A; 2I87 A; 4KPF C; 4G0X A; 4WIN A; 3W9S A; 2PFK A; 2BFC B; 4JEH A; 3OWX A; 4GO1 A; 4OHO A; 2IX9 A; 1U1G A; 2H5I A; 3HB1 A; 5ALJ A; 2H9Y A; 3IUL A; 5T4F A; 2Q7X A; 1AKW A; 4N76 A; 4J0G A; 3C87 A; 2QKY A; 5AM3 A; 1RHK A; 3FZE A; 3UOW A; 4X93 A; 2W4I A; 2XVY A; 2JEY A; 1NC3 A; 4G4G A; 2WTE A; 3ZY5 A; 5IAS A; 2CHF A; 2PM8 A; 1ZL2 A; 5B55 A; 2OCK A; 3EA4 A; 2E6K A; 5C7Y A; 3ZHS A; 3OC6 A; 3A9R A; 2B61 A; 2P90 A; 1FLD A; 3KJQ A; 4MQF A; 3H18 A; 1WCY A; 3WDK A; 2QXT A; 1EWV A; 3QFF A; 4ENS A; 2GKG A; 3T94 A; 1YQU A; 3F97 A; 4QDT A; 1U4N A; 2XBU A; 5SY4 A; 4I46 A; 3AF5 A; 1VXO A; 2MYN A; 1AB5 A; 3N8I A; 3PR2 B; 3NHU A; 5E4Y A; 3HEA A; 4BWF B; 4WV3 A; 4MR9 A; 5BPH A; 4F2H A; 2B94 A; 4ZQI A; 5JSQ A; 4ECE A; 4AUM A; 1YR1 A; 3O4V A; 2H85 A; 4YV7 A; 3WK6 A; 5EJ5 A; 2OCI A; 4QAQ A; 2BFD A; 1RHJ A; 2WJW A; 4BX8 A; 3INY A; 4R0M A; 3UJ3 X; 4PYR A; 3U4M A; 2QVG A; 3VVL A; 1A50 B; 3IPA A; 1Q16 A; 2OC4 A; 1R9M A; 1ZD4 A; 2UZ1 B; 3IES A; 1NI4 B; 2BHN A; 4YS6 A; 3ENW A; 3TTW A; 3VJK A; 1X7W A; 1MAA A; 2J85 A; 4AP6 A; 3OF3 A; 3W79 A; 3LQ2 A; 2QW1 A; 1A2O A; 3B2N A; 1Z2D A; 2P9H A; 2VXO A; 4W1Q A; 2D1Q A; 3JU3 A; 2FJ0 A; 4NIC A; 2RI0 A; 1KRW A; 3HBL A; 4KXL A; 3ZEI A; 4CKC B; 5IUN C; 3UWS A; 1MD9 A; 1AHN A; 3MQ4 A; 2Y2V A; 5BN3 B; 2AYY A; 3E9R A; 3F6B X; 2QDK A; 3F6P A; 5AM0 A; 4AY6 A; 5DYY A; 4H7D A; 3OY7 A; 2CKM A; 1VLJ A; 1X80 B; 4KQG A; 2LIP A; 4U7H A; 2YWD A; 3O6V A; 1B74 A; 3NHJ A; 1E18 A; 3IP5 A; 1GGV A; 1HK7 A; 2GN2 A; 1KF0 A; 4L7Q A; 3A0U A; 4QUH A; 5AJT A; 2QVC A; 4FR2 A; 3QMW A; 2ISC A; 2Q29 A; 4DAE A; 4Q05 A; 4RFL A; 2QD3 A; 3O4G A; 5JIS A; 3C98 A; 4E1V A; 2HFJ A; 4IH4 A; 2IIB A; 1MX5 A; 2BUB A; 3N7T A; 2PUC A; 1VB3 A; 3CCB A; 3IER A; 2KPO A; 2PO7 A; 1FSF A; 1RWW A; 4L3V A; 1C4W A; 4H77 A; 1S1M A; 5C8Z A; 4QOH A; 1WEH A; 5DWD A; 5FPQ A; 2ZWM A; 2FEP A; 1QDU A; 3PDI A; 1BDH A; 1JSC A; 1ZAY A; 1U9Z A; 1CZH A; 4CFS A; 1WLS A; 2QVZ X; 2JNW A; 4OLB A; 3SX4 A; 3MM4 A; 4S24 A; 1V1R B; 5TGZ A; 4RVN A; 4R2W A; 2W93 A; 1NXS A; 2R48 A; 5C4I A; 4KFC A; 2MTB A; 3RK4 A; 4OP2 A; 2DGD A; 1GPW B; 5D2C A; 3RIM A; 3VR3 D; 2GZM A; 3D2C A; 2XUJ A; 4PTZ A; 4LRQ A; 4FLM A; 1VCM A; 1KJ8 A; 4ZRD A; 1KQG A; 5AK3 A; 1GXS A; 1HOR A; 2H5L A; 1ECC A; 4L5Y A; 3T5B A; 4M98 A; 1VE7 A; 3B12 A; 2HA3 A; 1ZN2 A; 2AF4 C; 4Y9R A; 1GQR A; 1CUY A; 4MEB A; 2NW6 A; 4OU4 A; 2FXV A; 2HN9 A; 2C2O A; 5CFQ A; 3DAH A; 4BCC A; 4R1Q A; 5AXC A; 1AMN A; 4IH1 A; 5ALF A; 5GON F; 4D9C A; 1PR0 A; 1DWQ A; 2I6F A; 4IRX A; 2BGW A; 3STY A; 5BQX A; 3O1J C; 3WRW A; 3ETJ A; 4BMZ A; 1NUA A; 2DPY A; 2OHH A; 2YYA A; 4RK6 A; 1ODC A; 5DK6 A; 4MR8 B; 1ZD5 A; 2PVP A; 3UK7 A; 3SF8 B; 2V0N A; 4P5E A; 4N0M A; 1XFD A; 3TII A; 3G6W A; 5AIB A; 3K9C A; 4MR7 B; 1YRH A; 3UJN A; 1QE5 A; 3AX6 A; 2XCT B; 4BTL A; 4BB0 A; 1F1J A; 5ALX A; 1BA2 A; 1MB3 A; 2WNS A; 1UPC A; 1NU8 A; 4TS3 A; 1SU7 A; 2OHV A; 2Z3Z A; 2RK6 A; 1BMQ A; 2KQU A; 3EQ8 A; 1MQS A; 5BRN A; 3VVM A; 1WHS B; 5IYZ F; 5EIX A; 4Q4C A; 3TD9 A; 5DW0 A; 2BZ0 A; 3PRY A; 1XE3 A; 2P9T A; 1X7Z B; 5I5E A; 3DVA B; 4R1L A; 4DG5 A; 3PQ4 A; 4HT3 B; 3TIX B; 2J9F A; 3ORQ A; 2OGZ A; 4IMP A; 1CS0 B; 2OLE A; 1RFG E; 2IP4 A; 3VGP A; 4P86 A; 2ZJ0 A; 2KX7 A; 4HR7 A; 2Q7O E; 2QX8 A; 1FJ2 A; 3O84 A; 3BGS A; 2DFP A; 4WKO A; 3L7J A; 1FUI A; 1YBF A; 1QQB A; 4OHB A; 2BPO A; 2QUA A; 3B6M A; 3DMP A; 3HB9 A; 1LKZ A; 3TZB A; 1QKK A; 1T0U A; 2PE5 A; 5FEM A; 3LHF A; 3PQ6 A; 4Y2R A; 1YR2 A; 4PRZ A; 5ABP A; 1SUF A; 2N75 A; 5DGT A; 3R3U A; 3FVT A; 5DYT A; 5A6V A; 1QB7 A; 3FLB A; 1AY0 A; 3OMX A; 1V9S A; 4YAL A; 4XLL A; 2R7M A; 2VQ6 A; 3QM1 A; 4J29 A; 4RK0 A; 3R5X A; 1XAJ A; 2DKF A; 1Z7W A; 4ORE X; 3NS7 A; 4LJL A; 4KNY A; 3QLU A; 4R81 A; 1W52 X; 3DNF A; 4UUQ A; 4D3Z A; 1GLG A; 3A9V A; 2PO5 A; 3I04 A; 3LYH A; 4MXD A; 5T7B A; 4XWG A; 3K70 C; 5JZO A; 1D6S A; 3DV0 A; 1XZG A; 1D1P A; 1XZK A; 3NVA A; 4Q9D A; 5K4H A; 3H6G A; 2H8G A; 2AB0 A; 2R8O A; 1T9D A; 1C4K A; 4JD5 A; 1I1Q B; 3VA7 A; 5LU8 A; 2GYU A; 1KWK A; 1Q6Z A; 4JJE A; 4D92 A; 5BST A; 3R6G A; 3ZDR A; 4WDR A; 3V6M A; 1I6W A; 1M2F A; 1ZPD A; 4D9M A; 3NGM A; 2L17 A; 5IUL C; 1ZDM A; 5K3A A; 1PBN A; 2XCS B; 1KU0 A; 3W0L B; 4QLA A; 1E1D A; 2FXI A; 1ITZ A; 4MWS A; 4LSZ A; 4KE8 A; 3NLL A; 4MCH A; 5IAR A; 3NHK A; 4IIJ F; 3DKI A; 3ABR B; 5CNK A; 4L5A A; 5A1J A; 1M2E A; 3ED1 A; 4DAB A; 1LTK A; 5BWB A; 3T38 A; 1XZA A; 1JDB C; 3PA8 A; 2AMJ A; 1Y1Q A; 1B6S A; 1VRV A; 2DHE A; 4PSU A; 4YK7 A; 3EY9 A; 4EMU A; 3NHR A; 2W6Z A; 2JK0 A; 5BMV F; 2QTH A; 2BFF B; 4NCB A; 4C7V A; 4E11 A; 1GSA A; 3W9B A; 3D8S A; 3FCA A; 1T8Y A; 3LCM A; 5CB4 F; 3T7A A; 2CB9 A; 3L1J A; 4CKB B; 1NXX A; 2WHQ A; 3EIS A; 5EWM B; 3AB7 A; 3Q1K A; 2ORY A; 3F2V A; 2O2J A; 3SIW A; 4EY5 A; 2OYS A; 3IAM 1; 2XQK A; 1CUF A; 5I9I A; 2Y6U A; 5CT5 A; 3FCE A; 4P54 A; 1U65 A; 3ECA A; 4CF6 A; 3CZC A; 4UBI A; 2DH5 A; 1FTG A; 3FCY A; 3F6E X; 3IQG X; 1UPX A; 3TEM A; 2OR3 A; 1CLE A; 4JOS A; 2HK6 A; 5EXD A; 3BBL A; 2FUG 1; 1F2D A; 5AHK A; 2XE7 A; 3DYV A; 3HUE A; 3ACB A; 1OFV A; 1K8Y B; 3CG0 A; 3ZY2 A; 3ONE A; 3AUK A; 5AXD A; 4QE3 A; 5K3B A; 3T9A A; 4JR0 A; 2HCR A; 4WD3 A; 1TKB A; 2HU5 A; 3V7N A; 1P0Q A; 3AEY A; 4LY0 A; 2VK4 A; 4J0D A; 2OAG A; 1USK A; 2HNA A; 3F7N A; 5FBH A; 1F0N A; 3OLZ A; 4H6P A; 1HX7 A; 2WU3 A; 1P81 A; 1C3X A; 2UZA A; 1M5U A; 4HN4 B; 4Q82 A; 2R7K A; 1UPU A; 3P0R A; 3R5J A; 3PF9 A; 4GEG A; 5EJA A; 4ENW A; 1A88 A; 1U9C A; 3BRS A; 3WYN A; 5ESD A; 1O8B A; 3OQM A; 3GH1 A; 2DY0 A; 3NWP A; 3PE3 A; 3LUC A; 3W9U A; 2RJN A; 3VKH A; 1V1M B; 1P0I A; 1U1C A; 5DW3 A; 1LJL A; 3QYA A; 1CY0 A; 1DU4 A; 4QA9 A; 4HZG A; 1H5N A; 4FQ5 A; 4AWA A; 1OLU B; 2B9V A; 1CUD A; 3BM5 A; 2OCD A; 4MQM A; 1AKN A; 5F7O A; 5HZG A; 1DJM A; 1UKC A; 3GLQ A; 3E9Y A; 4ENQ A; 3L1H A; 5AM2 A; 4GI5 A; 4BQO A; 1WIW A; 4AAY A; 3UK0 A; 2IQ5 A; 3QMM A; 5H3H A; 1SD2 A; 3VYT B; 4WZZ A; 4BAT A; 1D4Z A; 4JP5 A; 1XKT A; 4N6O A; 1Z7G A; 4BQN A; 1TJY A; 3WVN A; 1CUV A; 4Q7E A; 3UUG A; 3KSM A; 4AN0 A; 3F6C A; 3CUY A; 1MJG M; 1F8Y A; 1MTZ A; 1MX1 A; 4C22 A; 3VNQ A; 1VHJ A; 2HJ9 A; 1DQZ A; 2C2B A; 3QEM A; 5TQJ A; 3IR7 A; 3PBK A; 1GQF A; 1VME A; 2ACE A; 3WK7 A; 4EHD A; 3FWH A; 3OZC A; 3MT0 A; 1QTR A; 1UMD A; 2HFK A; 4AW9 A; 2JI7 A; 4TPK A; 1WEK A; 5TJ9 A; 4PO3 X; 4WFK A; 1PI3 A; 2EZU A; 1JHR A; 1KA9 H; 1VPV A; 1SY7 A; 3PC3 A; 2C3O A; 3QH4 A; 3UC9 A; 1CUE A; 4PS0 A; 4G25 A; 1GFW A; 3LII A; 1P5J A; 1PR2 A; 4XEU A; 1R1D A; 3CTP A; 1RXS A; 4QOI A; 3MJD A; 1DWU A; 3H5L A; 4P2G A; 4KMO A; 4LTN A; 2XI4 A; 4CHT A; 3QNW A; 4HS9 A; 1UPB A; 3G0B A; 4N6G A; 4IO0 A; 4BZZ A; 1WPX A; 1JT2 A; 1YCF A; 3HZH A; 5T8K C; 2JGD B; 4D8W A; 4K05 A; 1WHT A; 1VMK A; 4N82 A; 3T54 A; 2BUA A; 5JAD A; 4RLF A; 2XYH A; 1L4F A; 5F4Z A; 1GZ7 A; 4LOH A; 1QZT A; 4D6X A; 4CG1 A; 3RE1 A; 3BDV A; 1CUC A; 4ENV A; 2G7Z A; 2DKO A; 3TTU A; 1T9A A; 2HBY A; 1DP4 A; 5CYF A; 4CCW A; 3Q2O A; 5DNE A; 3ROT A; 5DNU A; 3L6U A; 1UMB B; 1HG1 A; 4WKB A; 1AUR A; 3O8N A; 5LZ9 A; 3NHM A; 4G9G A; 4GQI A; 3S81 A; 3KHS A; 3KXK A; 3NYR A; 3NUZ A; 4TXM A; 5DEI A; 4J3N A; 4BIM A; 4FU0 A; 1NVE A; 3ZXU B; 5AJU A; 5DTJ A; 1JDN A; 3IQI X; 5ELL A; 3D8R A; 2O1X A; 4PZ0 A; 1LZK A; 1NON A; 4HNS A; 3C6Z A; 1Z39 A; 5EAI A; 3GT7 A; 4N3A A; 4FHF A; 3PD1 A; 1SC3 A; 4ZPJ A; 2WJ6 A; 1GGJ A; 1PYD A; 2XE6 A; 3FYU A; 5JAR A; 1DTW B; 4WX2 B; 5FWX B; 2Q3B A; 4TWB A; 1I4O A; 4DG8 A; 4TLL B; 1Y1T A; 2PPL A; 4JWT A; 1FFA A; 1N3I A; 3SM9 A; 1S2L A; 4HX9 A; 1FX1 A; 3NND A; 4XX1 A; 3UW1 A; 2YRX A; 2H7C A; 4BAG A; 3O0D A; 1T5B A; 3C3W A; 4MRM B; 4UHF A; 3M7I A; 3UPT A; 1SCQ A; 5EA2 A; 4R7T A; 1LLF A; 2FVY A; 4D9J A; 5BT8 A; 3EXI A; 5I7W A; 3DLH A; 1ZI8 A; 5JZB A; 2JVI A; 3MIZ A; 5EWJ B; 1VL1 A; 3L6C A; 1VKH A; 3AHH A; 2WTM A; 3N3A C; 4P80 A; 4KEA A; 4EVQ A; 2O2Z A; 5ED4 A; 1GCG A; 1RWO A; 2UVP A; 4HPJ B; 1X7Y B; 1EWK A; 4R9N A; 1QS0 B; 4JR2 A; 2XWP A; 3LTN C; 1ULT A; 3OM8 A; 2E7Z A; 1WX1 A; 4F8Y A; 3EGW A; 1UM9 B; 3N0X A; 3S7Z A; 4X95 A; 3LCR A; 2JI9 A; 3QMV A; 2OCX A; 2VK2 A; 2H07 A; 2XCU A; 1QP0 A; 3OZD A; 1WTA A; 3MUN A; 2IPL A; 5KC8 A; 1XZ8 A; 1PK7 A; 1GGE A; 1AUV A; 2BGR A; 2YXP X; 1ISS A; 5EFZ A; 1T0B A; 1V4N A; 4MAM A; 2XUA A; 1B1C A; 4JUF A; 3DLB A; 3SBX A; 3HAC A; 4D3Y A; 1V41 E; 3FOW A; 1E4E B; 1K9S A; 2A9P A; 4KQC A; 4BRZ A; 3CKM A; 4C20 A; 5HYV A; 4LJK A; 3WZL A; 4DAR A; 4K9O A; 3W3A D; 2AR9 A; 4GS5 A; 4X84 A; 5EJ6 A; 2W6M A; 1XLW A; 1W75 A; 4UOQ A; 2QSU A; 5DIZ A; 2HDW A; 4EYG A; 1A7A A; 1UMB A; 1UJ6 A; 2AJL I; 1ACL A; 5ALT A; 4FE7 A; 1Y1L A; 4BQQ A; 3KXL A; 2V4U A; 3SZL A; 2IDM A; 1RWX A; 4GMK A; 3E15 A; 1OV6 A; 1XZJ A; 4U2E A; 4JJ8 A; 3UAZ A; 3GYB A; 2PDA A; 1KEZ A; 3G1W A; 2C2M A; 4OUK X; 4B9A A; 1QO0 A; 4D8V A; 1JBQ A; 4G37 A; 3PNP A; 2WFM A; 5HF5 A; 1XZI A; 1K6E A; 1SC9 A; 4OGL A; 2ROQ A; 3VU0 A; 4M46 A; 1FFB A; 3SC2 A; 2R96 A; 3L7K A; 5C1P A; 1ZCF A; 4LXQ A; 4PXS A; 3AFO A; 1J7J A; 3P2M A; 2FZV A; 3HAB A; 4XLT A; 3MGK A; 1V8Z A; 3ESZ A; 3NR2 A; 4GYW A; 2E3J A; 2FKA A; 2WMY A; 4HQ0 A; 3QHQ A; 3TGW A; 3RVM A; 2Z9C A; 3NM5 A; 4KPE C; 2JEZ A; 1A98 A; 4TGL A; 2FN3 A; 4LNH A; 4CG3 A; 4G0Q A; 3DOI A; 1P6U A; 3AO0 B; 5HQT A; 5JAL A; 4KRF A; 3H17 A; 2AG1 A; 1DJO A; 5ALG A; 2WC1 A; 2WG0 A; 4N3B A; 2RI1 A; 5FAK A; 5I51 A; 1AK1 A; 2PUH A; 2NUB A; 1JT9 A; 1SQS A; 5LOV F; 4EGC B; 4J4H A; 3H5I A; 1TKC A; 4X45 A; 4AMY A; 4C0W A; 3EJW A; 1K7X B; 3V48 A; 4MUY A; 3KLN A; 3OKF A; 3OJX A; 3FJO A; 5B36 A; 4EAB A; 3OX4 A; 3FSG A; 4F2W A; 3UUF A; 1BRT A; 1J42 A; 3LTE A; 1RR6 A; 4G4S O; 3K9F C; 1WSW A; 1L9X A; 4HXE B; 1LBS A; 5JNV A; 5K3D A; 3G9X A; 1SC1 A; 3VNR A; 3PNS A; 2E55 A; 4L50 A; 4EUS A; 2KLH A; 2GM4 A; 4F8E A; 4PNP A; 1ZD3 A; 2JIP A; 3FOF C; 1EXW A; 4C1B A; 1ECB A; 3TTV A; 4H4E A; 2CJY A; 4MS1 B; 3Q3I A; 2XOE A; 1AO0 A; 4JCC A; 3RV4 A; 3CHY A; 5EGN A; 3RVK A; 1H2Y A; 1OGY A; 2H06 A; 4Z3O A; 1KRX A; 3LMK A; 5A71 A; 4QEZ A; 3BXF A; 3VC3 A; 2R1T A; 4HDM B; 1T4M A; 2O7V A; 4FGU A; 1POX A; 4L9A A; 4I3F A; 3JVI A; 4QU8 A; 3L18 A; 3ITN A; 3B38 A; 4R1M A; 3TAX A; 2VDJ A; 3LGS A; 3LUP A; 2D1T A; 3AHF A; 3D8T A; 4JB2 A; 4XAR A; 2HHC A; 3OLW A; 4TVS A; 4WCV A; 4KR0 A; 4PV7 A; 2NZW A; 1ORW A; 1H2X A; 4W5Q A; 2QL5 A; 4LQ4 A; 2PLN A; 4RJJ A; 4F5Y A; 3HYS A; 5AO9 A; 3FAZ A; 2EKD A; 4YAS A; 2XLB A; 3FDF A; 2OC9 A; 4A23 A; 3OE1 A; 2R5N A; 4GZ6 A; 5LUB A; 1I7D A; 1Z5N A; 2OXF A; 4EBB A; 4FHC A; 4W8O A; 1BVY F; 2XED A; 3R5H A; 2WB4 A; 5JZS A; 2XUD A; 2W6O A; 3L7M A; 1K68 A; 3M3H A; 2QTC A; 3B6K A; 2FQX A; 4I1Y A; 4N03 A; 4Y7C A; 3OND A; 3HGQ A; 3KVR A; 1D6N A; 4O33 A; 5LLS A; 4J7A A; 5HF6 A; 4GE0 A; 5FNV F; 2PRZ A; 3VU3 A; 4A3S A; 1XRN A; 1CZN A; 2GAJ A; 1R5J A; 2Q3D A; 2VT6 A; 3A9T A; 3AIN A; 1CUX A; 4QOD A; 1L4K A; 1CJS A; 1N5M A; 4G41 A; 5IBC A; 1XQW A; 2JIR A; 1EHC A; 4YJK A; 3N58 A; 1J23 A; 4OSE A; 1Z68 A; 1CY6 A; 5KEI A; 2BGN A; 4BVL A; 3AIM A; 3BXH A; 4ZDK A; 4H3K B; 4DM7 A; 5CT9 A; 2QPL A; 4C21 A; 1KMI Y; 3LQ1 A; 3GUU A; 1BXR A; 4H60 A; 5T4H A; 4LJR A; 3ITE A; 1XKL A; 4RJK A; 2FX2 A; 2RG1 A; 5EY5 B; 2JIM A; 4P84 A; 4CKE B; 2C3P A; 4KAA A; 4L27 A; 1JA0 A; 4JJ7 A; 3ETH A; 1F6W A; 3KLO A; 1EZ1 A; 4Y1R B; 1QR2 A; 4GKA A; 4FNM A; 3GZT B; 2VJB A; 5LZ8 A; 4DIU A; 1RZR A; 1JU3 A; 4N3C A; 2OGT A; 5CFL A; 3D54 D; 3A3K A; 2XQG A; 2AJC A; 5I7R A; 1DWO A; 1M5T A; 4ARA A; 4ENT A; 4OP1 A; 4WFJ A; 3CE9 A; 4KAF A; 4D97 A; 1XTU A; 5BV0 A; 4PRY A; 2Z04 A; 4LXI A; 3KKL A; 4M99 A; 2EBF X; 3QUA A; 3FOE C; 3WO1 A; 1ECP A; 3A6Z A; 1HMP A; 1UXO A; 3I1Y A; 3H19 A; 4R2X A; 4C4Y A; 3UV6 A; 1PEA A; 4YR7 A; 1KU6 A; 4ZJZ A; 3EFE A; 5LVA A; 3PDC A; 3V33 A; 4FMP A; 2EC5 A; 2QTG A; 3RG2 A; 4QYW A; 1QIJ A; 3QBE A; 4XFK A; 5LYY A; 1N57 A; 1JSL A; 4BJ1 A; 4CU5 A; 5FB3 A; 1WPR A; 5CW2 A; 3CF4 A; 1IZY A; 2Q28 A; 1D2A A; 2QUB A; 3RAE C; 5CFM A; 1JR2 A; 1QCD A; 2V3V A; 4LGC A; 1LV8 A; 3FNI A; 3NHS A; 1OA0 A; 1KJQ A; 3O4I A; 3V46 A; 1V48 A; 2Z3W A; 1UT6 A; 4MEJ A; 3WL7 A; 1TTP B; 1SCI A; 3DNM A; 2BCE A; 5AI9 A; 1BDI A; 1ONS A; 4MYS A; 2V45 A; 4I2V A; 2J48 A; 5H0I A; 1KFC B; 4G8C A; 1R4Z A; 4J03 A; 1VMI A; 1HL7 A; 5LP1 A; 2QMZ A; 3VKG A; 5BSW A; 3E4D A; 4YO7 A; 2PSF A; 4ITV A; 4JB0 A; 2VBF A; 3EIU A; 3I0M A; 4QDO A; 3TTY A; 4O4L F; 3PGA 1; 2QJW A; 1BCS B; 4RJ2 A; 1M54 A; 3FOB A; 1Z7Y A; 3WKC A; 3SAJ A; 3F98 A; 3EXH C; 5CTA A; 3PKZ A; 3QOY A; 3U56 A; 4RGY A; 1PWH A; 3QX3 A; 1KFB B; 1Z5P A; 4F5D A; 5IAG A; 3HC7 A; 2BP7 B; 4LQL A; 1Z15 A; 4NVR A; 1VA5 A; 4QG3 A; 4G89 A; 1ZHH A; 3B36 A; 1C29 B; 1B8O A; 4R0V A; 4QWW A; 1XAH A; 4OP3 A; 2VKT A; 2WQZ A; 4M1E A; 5C1O A; 4MTC A; 4R9V A; 3JRX A; 5JFT A; 3AJA A; 3R40 A; 4Y0N A; 4YAF A; 4TLM B; 4CU2 A; 4BJ6 A; 3R3V A; 3IAE A; 5F52 A; 2FSU A; 5ALD A; 5LJL A; 3UWD A; 3F8W A; 1XQY A; 3PE4 A; 3NHZ A; 2IE8 A; 3T9C A; 1MDB A; 3HVU A; 3OWH A; 3EEQ A; 1C7E A; 5B3L A; 2RGR A; 2GQ0 A; 5EUC A; 2OHI A; 4X97 A; 1W9M A; 4X85 A; 1RWP A; 5ELM A; 4F1W A; 5I6D A; 2RSL A; 3DEI A; 3NWO A; 1QP7 A; 3D59 A; 1L8X A; 2XMD A; 1Z12 A; 1JSR A; 2JGJ A; 1LVU A; 2DHD A; 4R1O A; 4CDR A; 3GEL A; 3S70 A; 2Q5O A; 3EEI A; 2PNJ A; 4D6Y A; 4L5C A; 2OAE A; 2R8B A; 4QUA A; 2VDW B; 5BSR A; 2DQZ A; 1LH0 A; 4I2N A; 1AGX A; 3WKE A; 5K3F A; 4W63 A; 3TX2 A; 3TW6 A; 1W93 A; 2XT0 A; 5LZ2 A; 3L7I A; 1DTE A; 3NHL A; 3AZQ A; 1ZIT A; 4BFL A; 3EYA A; 5ESR A; 1CZR A; 5LBT A; 3CU5 A; 5C3U A; 5KB3 A; 2XVZ A; 5FDF A; 4D9G A; 3R0J A; 2O1S A; 4X6U A; 4B80 A; 1V26 A; 3QVK A; 3F5D A; 1NME A; 1IZ8 A; 4RZQ A; 3C6Q A; 4LI3 X; 2FLK A; 3H1G A; 4KQI A; 2W6Q A; 1OY1 A; 5TQ2 B; 4E14 A; 2PUA A; 4ESE A; 2QD4 A; 2XWS A; 4QRI A; 2IZ7 A; 3PFC A; 1L4G A; 3S44 A; 4KXX A; 3N6V A; 2DWC A; 4ZYL A; 3N0R A; 1X2E A; 1XRQ A; 1LPN A; 2HL6 A; 2G5G X; 2KLB A; 1V5F A; 3HB0 A; 2IOQ A; 1UZA A; 3I01 A; 4RH0 A; 2XUG A; 4D9K A; 3V1M A; 4YWH A; 4F5W A; 3WIO A; 3M9X A; 3QLT A; 1PNT A; 3OT1 B; 4ZV9 A; 4MQE B; 4QDX A; 1ULZ A; 2PSH A; 5ITS A; 2DRI A; 4FLF A; 1YHY A; 3WWC A; 5EHN A; 2CLH B; 1MN6 A; 3LUH A; 1XTV A; 3HI4 A; 2CZQ A; 4BBA A; 1UCF A; 3D7R A; 5BYB A; 1TRH A; 3B51 X; 3K6K A; 2PUF A; 2RBG A; 3HM9 A; 3KEF A; 4TXH A; 4DA0 A; 4DQD A; 4EHN A; 1WOM A; 1KFK B; 5I8P A; 3WWE A; 2J0E A; 4DLN A; 3H0E A; 5AIA A; 3IVM A; 4M0K A; 3OOS A; 4OGF A; 1XZE A; 5ULL A; 1ACJ A; 4EHA A; 2BFN A; 2JVK A; 3DGE C; 4KBY A; 5BRJ A; 3EUA A; 4LKO A; 3TEJ A; 3IMK A; 4L5I A; 4Q0M A; 3OJW A; 4AO8 A; 3DSR A; 4M6W A; 2BEV B; 3VBC A; 4YAW A; 4QGS A; 3UMJ A; 1Z35 A; 1XQX A; 1CUB A; 3C3C A; 1T2N A; 3D5E A; 3KB8 A; 3ES9 A; 5LUA A; 2KYR A; 2GZR A; 4XII A; 1JHM A; 4EA9 A; 2QTB A; 5ALV A; 4P35 A; 2R84 A; 2YV4 A; 3D8N A; 1G5F A; 1QIG A; 3MNF A; 1RCU A; 4QUD A; 5K4A A; 4W5R A; 1D1Q A; 4M8L A; 3DG9 A; 2GEB A; 3MJF A; 4OG2 A; 2FTK E; 1B73 A; 5FRD A; 1B8N A; 3EHD A; 1PV2 A; 2I7V A; 1TZK A; 4H18 A; 4QYT A; 2PU5 A; 3PQ2 A; 4NOJ A; 4BMX A; 3VSA A; 1JYF A; 4B40 A; 1SD5 A; 1WD5 A; 3GLK A; 1GRV A; 4ZQC B; 3I45 A; 1USG A; 4X7B A; 3TL6 A; 3OLV A; 2QF7 A; 1V4B A; 1W85 B; 4LOI A; 4M0C A; 5TPW A; 2GNP A; 2B20 A; 4QWM A; 1NVB A; 2Y0P A; 1C9E A; 1NNS A; 5D85 A; 4JJA A; 4DIL A; 2YHA A; 4LHE A; 2PSE A; 4MGH A; 3FKJ A; 5AD1 A; 3GAM A; 5J43 A; 2DMR A; 4FWB A; 4X6Y A; 3B5E A; 4FYK A; 4IX1 A; 2V03 A; 2HHA A; 1RWQ A; 1JHY A; 1UJ5 A; 3O2Q B; 3KOO A; 4PIC A; 4INZ A; 2OQR A; 3IB3 A; 1YNO A; 3JW8 A; 1W88 B; 4OD0 A; 4JNC A; 1YOB A; 4ZVR A; 5IVI A; 4QPZ A; 3Q3H A; 2XGO A; 1ZUW A; 3O1H B; 5K6T A; 4I4C A; 5IMS A; 1PEY A; 1RWK A; 3LY8 A; 4MS3 A; 3DEA A; 2OHJ A; 2OVB A; 2H4W A; 4FDM A; 4HDK A; 4EAR A; 4KMM A; 2TMY A; 4UHD A; 3OT1 A; 2GTH A; 3PQ8 A; 4Q1V A; 2XGS A; 5LU9 A; 1QNL A; 2G6T A; 1DBP A; 4HDN B; 4V39 A; 3R7B A; 2JRL A; 1ZOI A; 4G0O A; 4ZDJ A; 4M0F A; 4EUK A; 1CP3 A; 3UAV A; 1Y0B A; 5AH0 A; 3HUU A; 3STW A; 2JFN A; 4L95 C; 4EY4 A; 4JR1 A; 1EVQ A; 3M83 A; 1UMC A; 1EHI A; 1XQV A; 3P9Q A; 4K9Q A; 3R3Y A; 1QZ3 A; 2WSL A; 2WZC A; 2BFB B; 1O6G A; 2CJX A; 3OX3 A; 3AI7 A; 2FFJ A; 4OGG A; 2OKG A; 3SCZ A; 4C5A A; 4L85 A; 1A5B B; 4OJT A; 3W78 A; 2J30 A; 4ENR A; 4J0J A; 1CJB A; 2Z4T A; 2VAV A; 1LBI A; 1DRJ A; 2RDM A; 3OM0 A; 2GX6 A; 2GYV A; 2IHU A; 5DYN A; 1TD1 A; 1XWW A; 4RXM A; 2O1W A; 3R3Z A; 3IP6 A; 4RK7 A; 2XE4 A; 1JKM A; 2VK8 A; 4Z4E A; 4RK4 A; 1P8A A; 1C7F A; 5EWL B; 3PQ7 A; 5NUL A; 2E4Y A; 1N1M A; 3LIP A; 1J06 A; 4I3H A; 3T0G A; 3ESX A; 2GVN A; 4KGD A; 5KWJ A; 1MZP A; 3O8L A; 3F9C A; 1K8X B; 1OBV A; 3LOP A; 1QIH A; 5AK6 A; 3TGL A; 5D86 A; 3QLV A; 5J1T B; 5AOC A; 4PF1 A; 3WNZ A; 3KSA C; 4G0U A; 4YJ3 F; 4N41 A; 4QDZ A; 5HQ3 B; 3EIO A; 4O08 A; 3ILX A; 1VZ3 A; 1Z33 A; 3TJM A; 1QO0 D; 1LD3 A; 3KU4 A; 2BZ1 A; 4AJ9 A; 3IP7 A; 3IUJ A; 1MVO A; 1Z17 A; 4MS4 B; 4R2K A; 4WL0 A; 1QPB A; 3B53 X; 4UC0 A; 3ESA A; 4W9R A; 3WIB A; 1RCT E; 1TGY A; 4GE3 A; 3PHC A; 1L4H A; 1UK8 A; 4RAD A; 1OA1 A; 1UC9 A; 4NU8 X; 3V32 A; 3EUF A; 2JFV A; 1Y1R A; 4DY4 A; 1NMS A; 4NZO A; 3UHP A; 4MAA A; 4D9I A; 1T5D X; 1V5E A; 2XUO A; 3DF9 A; 1I0I A; 1W88 A; 4I19 A; 2JB9 A; 5KQP A; 1KXJ A; 1C7I A; 3A2N A; 1MU0 A; 2BFE B; 2QT9 A; 3CLH A; 1CUH A; 4P08 A; 3EDQ A; 2Y6V A; 1KFE B; 4W5O A; 1I1O A; 1I0L A; 1A0O A; 3FNQ A; 3LZD A; 1RE1 A; 3KWJ A; 5FWY A; 3BQ9 A; 3EGC A; 5C80 A; 1FY2 A; 3PUI A; 5FKJ A; 3D6M A; 3I2G A; 1JH9 A; 1ECL A; 4ZGB A; 3WJ2 A; 3BWE A; 3L4J A; 3WKB A; 5KKN B; 5F7Z A; 2IHZ A; 2W22 A; 4MPR A; 3LUD A; 5A62 A; 4K2A A; 3LPN A; 4G9E A; 4PZP A; 4KXU A; 1USV A; 2H65 A; 3FVW A; 2HJW A; 3N6R A; 1PFK A; 3NYQ A; 1GGH A; 1JVN A; 3DL7 A; 5D84 A; 2AC4 A; 3LKB A; 2FWN A; 1X70 A; 2MSW A; 3ZY4 A; 1U5B A; 1OLX A; 4D96 A; 4JBN A; 1SCK A; 5J5V A; 2FMH A; 3EDR A; 3ZY3 A; 3DP9 A; 4RQB A; 2L19 A; 1TK3 A; 3IP8 A; 3O75 A; 2QZP A; 3OTQ A; 4N12 A; 3GCD A; 4DDT A; 5IAN A; 4D74 A; 2WJ4 A; 4JOQ A; 3F90 A; 1CY1 A; 3V4S A; 4CIB A; 1ZY2 A; 5HQJ A; 4NOK A; 4YJ2 F; 4CCA A; 3MVE A; 1UIM A; 4L8F A; 5F64 A; 1WCI A; 3AHC A; 1OAS A; 1PW7 A; 3AEX A; 1V25 A; 3DKK A; 2PS1 A; 4LE2 A; 4OF4 A; 1ODL A; 4DNO A; 1F8X A; 5B7P A; 2XB6 A; 2WWV D; 4GVP A; 4NBN A; 5D3K A; 4PE5 B; 4RL4 A; 4UHK A; 1SHJ A; 3TTX A; 5I5G A; 5F51 A; 3R7N A; 4HEA 6; 3E61 A; 1QFM A; 1VHD A; 2O2H A; 4H4D A; 5K3O A; 1MNQ A; 1OJ7 A; 1QOP B; 2I8C A; 4G0Z A; 3B6I A; 3D7N A; 5ERX A; 5A7H A; 1THT A; 5IAK A; 1QE3 A; 1Y5L A; 1L7A A; 2OXE A; 2WID A; 3CRN A; 3QLU C; 4RJI A; 1UC8 A; 1UFO A; 5JNW A; 1E61 A; 5AXA A; 5LA6 F; 2V59 A; 1UOQ A; 1AZL A; 3I2F A; 3MF4 A; 4FKB A; 5KWI A; 5F78 A; 5F7J A; 1CVL A; 5EUB A; 5A5V A; 1TTQ B; 2ZUS A; 4Q3A A; 4HXG A; 4RZS A; 4OU5 A; 4LDN A; 4KKX B; 4Y2V A; 4TXJ A; 1NXW A; 1ZQ1 A; 4K5Q A; 3F96 A; 3I2I A; 4F12 A; 4FFS A; 1IZZ A; 3A70 A; 5HJE A; 4IGA A; 4N8D A; 4MQ5 A; 3VNS A; 1RWM A; 1IZ7 A; 4RY0 A; 2CMF A; 1YRY E; 1SIW A; 1NUL A; 4QYI A; 3RVJ A; 5K3E A; 4GPE A; 3OOE A; 4F2P A; 3OG9 A; 3OUR A; 5FBK A; 4MCA A; 4BXO A; 2QK4 A; 5HW0 A; 3LFT A; 1ZGZ A; 1L5N A; 2JGI A; 1U2P A; 3ZLT A; 5ALW A; 1J9G A; 5JS2 A; 1Z38 A; 3DEZ A; 3FWP A; 4OTZ A; 4KXN A; 2QMY A; 2GRU A; 4ML3 A; 3HFR A; 4RAC A; 1KY4 A; 4DAD A; 5AJH A; 2QX4 A; 1BN6 A; 1XAI A; 2NX2 A; 5GOT A; 3NOO A; 1F51 E; 1HOT A; 1E6M A; 3I28 A; 1K63 A; 5DCL A; 3KEG A; 1JFR A; 3C3K A; 5FPP A; 3W7A A; 1PFQ A; 3D7K A; 2V58 A; 3A9U A; 4YO8 A; 1P80 A; 1QON A; 2B8N A; 4PQE A; 1RWN A; 2PUJ A; 5NLL A; 1UOP A; 5BV7 A; 1L7R A; 1JAZ A; 1RZR D; 1AD2 A; 3JY6 A; 1V11 A; 1TGV A; 2EEP A; 1JDZ A; 3EXF B; 3JZD A; 2VPZ A; 3FOA A; 4OB8 A; 4E08 A; 2RH9 B; 4MYR A; 4WKC A; 5CH8 A; 3R3X A; 2ZDH A; 4KQF A; 1OI4 A; 2ZSI A; 3WLA A; 1OVG A; 4NJE B; 3TX6 A; 1JXQ A; 1M6V B; 4Y2T A; 3U7I A; 4II3 A; 1RQX A; 2ZIX B; 4PU0 A; 2JIQ A; 3NHF A; 4LYE A; 4PW0 A; 2RK4 A; 2X8B A; 1FLV A; 3NI2 A; 3UOA B; 2ZR8 A; 4V3B A; 2HSW A; 4P93 A; 1MAV A; 3HSS A; 1VCN A; 1CZK A; 1BZY A; 1CE8 B; 2QVX X; 2OZK A; 3VOT A; 1D4F A; 2JFO A; 2NYA A; 1C30 A; 4QSH A; 4EF5 A; 2RHT A; 1OTY A; 4G0P A; 2IYN A; 4P83 A; 3A2M A; 1A9S A; 4WKG A; 3U7R A; 1VHQ A; 4YRZ A; 2DHC A; 3FYT A; 4I36 A; 4D5G A; 2Y27 A; 3C5E A; 5BO5 A; 1I3C A; 3IEK A; 3G1U A; 5D5H A; 5KAM A; 4QLO A; 3KWF A; 4X9X A; 2BKX A; 1JDB B; 2IZ6 A; 4D1Y A; 3EXE A; 1EA5 A; 1ZN7 A; 1PYO A; 5DLP A; 3ORR A; 2HA6 A; 1XZC A; 2HSG A; 2ZYS A; 4FDL A; 4C4X A; 1A9X B; 2Q5C A; 5YAS A; 4K9L A; 5BRA A; 2VUI B; 5AK4 A; 3KE8 A; 1ZHO A; 4FYH A; 5I3C A; 3JRW A; 2WYI A; 5EKS A; 5EHX A; 3IR5 A; 3SSA A; 1V6S A; 4DMC A; 3S2Y A; 3CIS A; 3RAD C; 2NZX A; 3R76 A; 2LUO A; 4II2 A; 4DLK A; 4KOE C; 5IKX A; 1V16 B; 4XVC A; 4LRU A; 5HRA A; 1VZ2 A; 5AKG A; 1G8K A; 2HWU A; 2VA9 A; 4ROT A; 1U1D A; 3H2G A; 4U1R A; 4L72 A; 1JL3 A; 1XZL A; 3OWO A; 1LPA B; 4Y2Q A; 2YL2 A; 3O9V A; 3H11 B; 1ORO A; 1C4X A; 3OPY B; 3NHP A; 4KN6 A; 2AYX A; 3T5A A; 4B0P A; 3HMK A; 3L5O A; 2BEW B; 4CCY A; 3K1T A; 3WAG A; 4GDM A; 2A9R A; 3EWN A; 1URP A; 4DCO A; 1QJ4 A; 2GN0 A; 2AG0 A; 3FMG A; 3K2I A; 1I7Q B; 1BFD A; 3QW4 B; 3HXL A; 2P2C A; 4ZGG A; 5AP9 A; 3GBS A; 3IXM A; 4X44 A; 3DAY A; 1U1E A; 2H9A A; 4XDH A; 5GTD A; 2GBI A; 3HRH A; 2X13 A; 3QBJ A; 1CRL A; 4I1R A; 2WLT A; 3EXF A; 4ZVQ A; 5I4I A; 1CEX A; 1E9V A; 2AYZ A; 5A2G A; 1E2B A; 1OUM A; 5I47 A; 3BSS A; 4JC0 A; 1P19 A; 2RGY A; 4QOE A; 4IMJ B; 3NBQ A; 5KND D; 4DGQ A; 5I7H A; 4MV0 A; 4H7I A; 1ISR A; 3PC2 A; 4P92 A; 4W1P A; 1X7X B; 4TYM A; 3AIL A; 1L5L A; 3DOH A; 1HRK A; 2YS7 A; 5KNB D; 2CNK A; 1ABF A; 3IXL A; 3Q7S A; 4IWX A; 4B7Z A; 3NJ4 A; 3QK7 A; 3T9E A; 5D6O A; 4NZZ A; 5ERY A; 2LTA A; 3ZOK A; 2LVB A; 4MV5 A; 5DM6 0; 1MT3 A; 4JC5 A; 4OQP A; 1GUD A; 2EX0 A; 1CW2 B; 4UFP A; 1ODJ A; 4B2Y A; 5IX8 A; 5JAP A; 1ETH A; 1GPM A; 2JGK A; 2FQY A; 2XKK A; 3UO8 B; 1XRR A; 1ZOS A; 2FMI A; 3HV2 A; 1YDH A; 2XMZ A; 5CA1 F; 4K9K A; 5BR4 A; 4KRE A; 4ZXF A; 3AB8 A; 3L9W A; 2JH3 A; 3HN6 A; 3VYR B; 4YCS A; 4P81 A; 1V7C A; 3NTX A; 4D5E A; 1E89 A; 1A9X A; 1JJF A; 4H3H B; 3F73 A; 1J22 A; 5C8V A; 2CBG A; 1L8A A; 5K1Z A; 3OLQ A; 4HN2 A; 5BPF A; 4X94 A; 4IMI B; 4JC8 A; 1BA3 A; 4ZP1 A; 4KJX A; 2YH2 A; 3D2A A; 2MCF A; 4DG9 A; 1LCI A; 4ZI5 A; 3OUT A; 2WVH A; 4O4H F; 5HFN A; 3AZO A; 4S04 A; 1JA1 A; 1CF9 A; 1CD5 A; 1J0D A; 1U9Y A; 5F73 A; 1CIJ A; 3OQI A; 3HS3 A; 4FHE A; 5HF9 A; 4B85 A; 4E1B A; 1N0I A; 1XWF A; 1YVU A; 1J0E A; 2ZUT A; 4IO1 A; 5IWC A; 3WAK A; 2R87 A; 1C8V B; 3H9U A; 3OUZ A; 2WIF A; 5KAP A; 2JBW A; 2WU1 A; 2Y2U A; 2VBG A; 4MV9 A; 2WZB A; 4QU9 A; 3L4E A; 1OTX A; 1SOA A; 4FBL A; 2CLK B; 4XUG B; 3AJE A; 4FHD A; 2HA5 A; 3FYU C; 5IXJ A; 4ZVP A; 3I6Y A; 1K9V F; 1NMQ A; 4G23 A; 4OEH A; 3DBI A; 4AZ3 A; 3DTV A; 1SD1 A; 1PHP A; 1N0H A; 1BU5 A; 1DX4 A; 3LUG A; 2C61 A; 1A04 A; 1JJA A; 3PFB A; 2H1V A; 4D8X A; 3U17 A; 1IMJ A; 2F1O A; 3O4J A; 3OJC A; 1JDV A; 1LPB B; 5BUZ A; 3WMR A; 4IEF B; 2GPW A; 2G5P A; 4LE1 A; 1Z5O A; 1U63 A; 2J6C A; 1WNF A; 1JI3 A; 4RMN A; 3WRV A; 2IPA B; 1OBO A; 1S8N A; 5A6S B; 1G9T A; 1DOZ A; 1E8M A; 3VR2 D; 4Y7D A; 3OZA A; 3LBA A; 2HU8 A; 3GJR A; 2PRY A; 1PX2 A; 2AJT A; 3TO5 A; 2WAM A; 3ZKW B; 4H2D A; 1FUY B; 1W76 A; 4YX9 A; 2JBA A; 3KD2 A; 3BIW A; 5CXU A; 4KLA A; 4HDS B; 1ORV A; 1Y7L A; 4HTA A; 4X3C A; 2H1I A; 3P4U A; 1Y6Q A; 1W4L A; 3KKE A; 2B4A A; 3I12 A; 4MJ3 A; 1KWG A; 5B7N A; 1RU3 A; 4H7H A; 4PGA A; 3SIP A; 3H13 A; 2FMK A; 1HFJ A; 2XZT A; 1RYY A; 4AX4 A; 3DZD A; 2H4Y A; 1XTZ A; 1L1Q A; 4OQ4 A; 5C35 A; 1XRL A; 1T8R A; 5HM6 A; 1CR6 A; 4W1S A; 2X14 A; 2R25 B; 4GZ3 A; 4MV4 A; 2QMQ A; 4AUN A; 2ZVD A; 5DGI A; 4C7X A; 4G67 A; 2F48 A; 2ID7 A; 2V9C A; 2C7B A; 1KEE A; 4QU0 A; 3DEH A; 4KLR A; 1DQP A; 3RAF C; 4N8E A; 3KWL A; 3D3N A; 3HO1 A; 1YS6 A; 3ZHU A; 4MK4 A; 3LKV A; 3FFW A; 5ALN A; 3IUN A; 4Y9S A; 1RHU A; 2C4K A; 4IR7 A; 4Z9B A; 1NX9 A; 1AZW A; 4R8L A; 2DWU A; 2O20 A; 2PN1 A; 5E0K B; 1QFS A; 2H5J A; 5IAE A; 4NML A; 4UHB A; 4ZV7 A; 1AA6 A; 2Y0B A; 1C1H A; 1TC2 A; 4GLJ A; 4WY5 A; 3U9T A; 4WKN A; 4LC9 A; 5ESW A; 1Y37 A; 3ZHR A; 2QL9 A; 5JH7 F; 2F64 A; 1J2E A; 3IAU A; 4EY6 A; 3IEM A; 5JQG F; 3H1A A; 4QOJ A; 2DCM A; 4NZM A; 5T8K A; 4BP8 A; 5TQ0 B; 3CS3 A; 1DJP A; 4HEA 1; 1BDJ A; 1P4A A; 4ETM A; 5B1H A; 2R47 A; 1SU6 A; 4QTX A; 3O2S B; 1VE1 A; 3KSB C; 3KG2 A; 1D4A A; 4UCF A; 4N8S A; 5CT8 A; 1VST A; 1S2I A; 2ZIZ A; 3KUK A; 3R5K A; 4Y2J A; 5DYW A; 2JI8 A; 4B84 A; 1EH5 A; 2C0Q A; 3QEL A; 3GB9 A; 3HCR A; 3III A; 4L78 A; 4X00 A; 2VUH B; 3TQT A; 5LKR A; 4ETW A; 1PUX A; 3WK9 A; 4MB6 A; 2II6 A; 5JRO A; 1CUW A; 4JBL A; 1FXU A; 3RJM A; 4QQ8 A; 4MLD A; 2XVX A; 4FUT A; 3UTC A; 2XCQ A; 1EU1 A; 3QZU A; 3M3D A; 2BFC A; 1SQ6 A; 3G13 A; 3BSY A; 2GTI A; 3LUK A; 4N6N A; 5C5E A; 3W06 A; 1CZL A; 4MA5 A; 4EGS A; 1JDT A; 5JYC A; 2FAX A; 4UBJ A; 2QX9 A; 2VH1 A; 3E7X A; 2G4L A; 3NM4 A; 2HRC A; 4FBM A; 1SRR A; 3K2Q A; 3OZ7 A; 4NZN A; 3UHO A; 1GG9 A; 1H23 A; 1NVA A; 1QPG A; 2JPD A; 2VAX A; 1A8U A; 1XLV A; 4PNZ A; 5F77 A; 1FUE A; 5AI0 A; 1RCF A; 1RLJ A; 2H1W A; 2Q5L A; 2J9Z B; 4Q3K A; 5KYV A; 1LPP A; 2XUH A; 3UGJ A; 5FUQ A; 2C3Y A; 3IUR A; 4JP1 A; 5EIH A; 4Y2X A; 3GBP A; 2QZJ A; 3RVN A; 2LXN A; 3K8O E; 1A5A B; 1YD7 A; 3I2K A; 2FVX A; 2VJD A; 2ZSJ A; 4C1N A; 1POW A; 3ACD A; 4ZU1 X; 1OX4 B; 2DQY A; 2VEO A; 4F9T A; 2R97 A; 3E3A A; 3DDO A; 1MZV A; 2ISS D; 3DWG A; 3TDM A; 3DLP X; 5PNT A; 1JH8 A; 1SFR A; 5BSK A; 3OX2 A; 5AOA A; 3G8Y A; 2QJR A; 4B87 A; 4DMH A; 4F21 A; 4JUA A; 3AQI A; 2AC2 A; 1WB4 A; 1B41 A; 4Z4I A; 5BYA A; 1G8J A; 2H2W A; 2QTF A; 3LUF A; 4BRS A; 1T3T A; 2VYC A; 2CEK A; 1JDS A; 4TXL A; 2OGS A; 4TV8 F; 5DGC A; 3WV4 A; 3G02 A; 3H2H A; 1TQH A; 3X44 A; 2JBH A; 1F8U A; 1JE0 A; 3O73 A; 3OO1 A; 2UZ1 A; 1DKU A; 3TIG A; 1NI4 A; 1YCG A; 1TM2 A; 4P0P B; 3V34 A; 3CMM A; 1NRX A; 3X2F A; 2XCL A; 3FZN A; 4QGB A; 3HGT A; 3B2Q A; 1UPA A; 1OXK B; 2QWX A; 2HM7 A; 1PO7 A; 3WOV A; 5IBQ A; 3WV5 A; 1B6G A; 3VHX B; 3BF7 A; 4FFV A; 2YS6 A; 1DQN A; 5AKK A; 5K4G A; 1WBJ B; 3U7J A; 2P0A A; 1CZO A; 4NOL A; 1CVR A; 4AF8 A; 1Y9J A; 5KI6 A; 4G8J A; 2ZYI A; 2Z8Y A; 4L4Z A; 3PI6 A; 2YHB A; 3HUF A; 4RH1 A; 5CUR A; 2YYS A; 2WIJ A; 4HEQ A; 2PKY X; 4AUL A; 5BV1 A; 4PSD A; 4JQS A; 5JPC A; 3L6B A; 5E7Q A; 1TZM A; 3UAX A; 1Y5I A; 4TM8 A; 1ECJ A; 4NNJ A; 2B1J A; 2WJZ B; 4XI9 A; 3JS5 A; 4JA2 A; 4ZVO A; 1X80 A; 4ZVM A; 1QVZ A; 5ALH A; 4F9E A; 2J33 A; 4Y2Y A; 1I7S B; 3N75 A; 1RLI A; 2J9G A; 3DHV A; 3BRQ A; 3P9S A; 5SYM A; 1UFR A; 4Q3L A; 2XD4 A; 2WHR A; 3EFB A; 1I3O A; 2R1V B; 4RY8 A; 1UJ4 A; 3R5F A; 1MW8 X; 4CG2 A; 2HQO A; 2VPW A; 5ALS A; 4RKW A; 2WJA A; 2D5I A; 5AM1 A; 1LZL A; 5BUC A; 1IPH A; 2BHS A; 4DSA A; 4GLF A; 4EA6 A; 2Q62 A; 1JQK A; 3TTS A; 2E4Z A; 4IF4 A; 1PVD A; 3EQ6 A; 1CUI A; 1T36 B; 2K2Q B; 5ALL A; 1ZGG A; 3FFT A; 2V3W A; 5BNW A; 1IOW A; 1V1R A; 3T0F A; 5A4K A; 5LMC A; 4B0O A; 1Z16 A; 1FDO A; 3NZE A; 4N5I X; 2NT4 A; 1RXE A; 2XLC A; 4RV4 A; 5KNC D; 1CUS A; 2IUF A; 4Y9T A; 4LZL A; 1D07 A; 3D64 A; 2QXY A; 5KYT A; 3CNE A; 3NOV A; 4WFI A; 2W7T A; 5IBP A; 4CET A; 1JFV A; 3Q6M A; 1QTN A; 1M72 A; 5LJI A; 1FSS A; 2OG1 A; 4GMH A; 1YI1 A; 1VJ5 A; 4C0X A; 2GLT A; 3ICO A; 5C32 A; 1YAJ A; 5T6M A; 4ZVK A; 5AHO A; 5EJ4 A; 1AKU A; 3SY8 A; 3WK5 A; 3ZY6 A; 1KS2 A; 3H5O A; 4KQJ A; 5IC5 A; 1YQQ A; 13PK A; 2F8M A; 1C3O B; 4F5Z A; 1J25 A; 1SXI A; 3UT6 A; 1JHU A; 1ECG A; 4TMY A; 4Y9U A; 2IKS A; 1NR5 A; 4NAR A; 1GG5 A; 5CRI A; 4DAN A; 1RXY A; 4GNR A; 2HA0 A; 1W36 C; 5JNU A; 1K4Y A; 2QR2 A; 1QQA A; 2JIB A; 4MR8 A; 1DMS A; 2FQW A; 2V97 A; 4QYS A; 1D0V A; 4WY2 A; 3SF8 A; 3IPC A; 3HWW A; 1T8S A; 3T4U A; 4MR7 A; 4GWL A; 4GR5 A; 4S0X A; 3FB1 A; 4MRO A; 2YBE A; 4GPA A; 2CLF B; 4CBV A; 2R86 A; 4I50 F; 2WHP B; 4RS4 A; 3CV3 A; 5E3J A; 2IV2 X; 5IH7 A; 1X1Q A; 3ZJ5 A; 5HIA A; 2XE8 A; 4PFK A; 4MCJ A; 3D1V A; 4AMZ A; 3WL8 A; 5DNV A; 4IJQ A; 1WHS A; 2M98 A; 1E6L A; 1AUO A; 3WWD A; 3Q6N A; 1THG A; 4IH9 A; 2C3U A; 3ABS B; 1X7Z A; 4P53 A; 3DVA A; 3WK4 A; 4GXQ A; 1I13 A; 4I66 A; 2PSD A; 3MII A; 3BWX A; 1FQW A; 2RKB A; 4ZOL F; 2VFA A; 3T95 A; 3EEB A; 1CS0 A; 1Q1G A; 1HLG A; 1Y4Z A; 4DIK A; 5EJ7 A; 3Q7R A; 1RXC A; 1ODK A; 2FDX A; 4GHW A; 4IZ6 A; 4ZVL A; 3WDL A; 4YP9 A; 4KK3 A; 3R75 A; 1RWV A; 2BHT A; 1A9Q A; 3PUJ A; 2BGG A; 1DEA A; 3HK2 A; 5DLE A; 3GEB A; 1HDI A; 4AP5 A; 4D8U A; 1U1F A; 4F0J A; 3QX8 A; 4EAT A; 3CZ5 A; 2QU7 A; 3O4H A; 7ABP A; 1ZR2 A; 4QYM A; 3EZG A; 4XAQ A; 3NAG A; 4LYG A; 3I9V 6; 3VR6 D; 4JEI A; 5EI5 A; 4ES6 A; 2D7J A; 1OPR A; 5ALK A; 2FB9 A; 3KEM A; 2R7N A; 3EB9 A; 3Q3V A; 3DWI A; 1W96 A; 4K9M A; 1E66 A; 3EXH B; 2E4U A; 3KJN A; 5F76 A; 3H1F A; 2PUG A; 3LY9 A; 1A9P A; 5CHY A; 5AI8 A; 3V4O A; 4QTY A; 2ARK A; 3LWB A; 2CHE A; 5ESX A; 3DYI A; 1GKU B; 2ZAY A; 4I1P A; 1XCO A; 1TCC A; 1PKE A; 3EOD A; 2RJO A; 5DTE A; 1EK2 A; 3EN0 A; 1NAT A; 2W70 A; 2P9B A; 5I7A A; 1MCZ A; 3C97 A; 4YNB A; 2GBF A; 2CJP A; 1EAY A; 2HRQ A; 1BCR B; 4E46 A; 5BWC A; 2WVA A; 1GT6 A; 1W1I A; 3KVV A; 2ZDG A; 3HUT A; 1KBQ A; 2QR3 A; 4LKR A; 3C43 A; 3RLI A; 1T9C A; 3ABQ B; 4OHM A; 3L0Z A; 1LJU A; 4C4Z A; 3PUH A; 5K45 A; 3BJM A; 3GFQ A; 3U1T A; 1TC1 A; 1L5Y A; 1SG0 A; 2XQI A; 4P98 A; 1YAS A; 2VAT A; 4OY3 A; 4DAO A; 2HAD A; 1HO3 A; 4PFQ A; 5A5D A; 2WFL B; 4O2B F; 2QLF A; 4PX5 A; 3V4L A; 1AC5 A; 2BFF A; 3N2L A; 1P17 A; 1IK6 A; 1FFG A; 1ZJ4 A; 2FUN B; 4OLA A; 4B82 A; 5EWM A; 5KQL A; 5CYG A; 3OQN A; 1DC8 A; 3ML1 A; 3O3M B; 2ONC A; 4HQ6 A; 5C8Y F; 1EIN A; 1YS1 X; 2HIG A; 2OCG A; 3STU A; 1QIM A; 2P4S A; 4S0Z A; 3PIC A; 1ZWL A; 3X2E A; 2PJU A; 2B4K A; 1F4P A; 3ABO B; 3T4P A; 3PC4 A; 1EDE A; 5IEB A; 2ZGV A; 3NNS A; 3MBI A; 3FCC A; 4P5D A; 1CHN A; 3EQ7 A; 1DXO A; 4M0E A; 5JZ9 A; 1AGY A; 1K7F B; 3OOH A; 2F67 A; 4PR3 A; 4FUQ A; 3LT5 A; 2TSY B; 4OLH A; 2GBG A; 1ICE A; 3GQB B; 3LYA A; 3MMS A; 4D9N A; 4WT7 A; 3WAD A; 3OCC A; 5DGH A; 3CSS A; 3V6L A; 2VSN A; 4D9B A; 3QBD A; 2W6C X; 5FA6 A; 3ICW A; 3I04 M; 3CY6 A; 3GGC A; 3EXH A; 4OFX A; 1NE7 A; 5LD2 C; 1HFE L; 4I7E A; 3LX4 A; 1P7Z A; 4ENU A; 3N0T A; 2R0Q C; 3DS8 A; 4KRY A; 4YAU A; 4QAR A; 1V1M A; 4AB1 A; 4WJL A; 4D9E A; 3VR5 D; 2CLE B; 3H1B A; 1NW4 A; 3HBN A; 4Q3E A; 3ROF A; 1PWY E; 5D3Z A; 1TZJ A; 2DJI A; 5ALY A; 1G2O A; 3OD1 A; 5FT9 A; 1OLU A; 1D04 A; 1GN9 A; 4MYD A; 1Z18 A; 1WSA A; 2JGM A; 3VBE A; 5CFO A; 1SXG A; 2F2T A; 3VSC A; 5CFR A; 3STT A; 4O1I A; 2P8S A; 3C3M A; 3GV0 A; 2QTT A; 3ZHQ A; 3MUO A; 3G9U A; 2IPN A; 2YWB A; 4LXX A; 2C2K A; 3DTV D; 3T9B A; 4GXR A; 1JYE A; 1PJA A; 3O9M A; 3PE6 A; 4K9P A; 1E8D A; 1JHZ A; 2FEW B; 2DT8 A; 3OQV A; 3R0Z A; 1PHR A; 4L0D A; 2W6N A; 2ZKI A; 2PVS A; 2YZG A; 2OPH A; 1ZX1 A; 3MMN A; 5LIP A; 4X90 A; 1YMV A; 1JE1 A; 1ORD A; 4RAB A; 4MQF B; 2Z8X A; 1CUJ A; 2VJA A; 5K6R A; 4RM3 A; 1TCA A; 5I48 A; 1JDU A; 2QW0 X; 4IHJ F; 2VK1 A; 2BL4 A; 3NHY A; 1X77 A; 2CNN A; 3BFJ A; 4YP1 A; 2EZ4 A; 1ORE A; 1LPO A; 3RUP A; 5JD5 A; 5LBU A; 4MCI A; 1PK9 A; 1RHM A; 3Q8W A; 1ZN8 A; 3TIV A; 4IIL A; 3KVY A; 4MR9 B; 3TB6 A; 2FN9 A; 4GG1 A; 1QK3 A; 1C7J A; 3PFK A; 4HPX B; 4BP9 A; 2AI3 A; 3LUJ A; 1BU8 A; 1X2B A; 2D1S A; 2BFD B; 1KQ3 A; 1QBG A; 4P34 A; 3O83 A; 1DX6 A; 3FFX A; 1FLA A; 1GCA A; 3V3K A; 2PAF A; 2LIV A; 2JGD A; 2HW8 A; 5CNJ A; 4C88 A; 2L18 A; 4PEV A; 2PUE A; 1WL8 A; 1KJI A; 3NM6 B; 2WIK A; 3OHP A; 4QOG A; 4RE6 A; 2VF2 A; 4R84 A; 1XAL A; 1H9C A; 2C0P A; 2HY7 A; 1ZJ5 A; 4N7B A; 3ESB A; 2IHT A; 4RHY A; 1UWC A; 3L3B A; 4JQZ A; 1X7W B; 3VPB A; 3LHK A; 2VR1 A; 1FFW A; 3ENV A; 2HLH A; 2XMB A; 1TLF A; 5EVC A; 1QOQ B; 4ZVU A; 4J0H A; 1K0U A; 3U0V A; 3KJF A; 4N7J A; 4BTE A; 1DBW A; 1TDJ A; 2Q27 A; 4B9E A; 1P5F A; 2ZSH A; 4Q3B A; 2GAI A; 2DZD A; 2VPX A; 5B1I A; 2XT9 A; 2A0Y A; 5HHT A; 4MWT A; 5C31 A; 1G66 A; 4PX2 A; 2VLB A; 5LZ4 A; 5E38 A; 2XUQ A; 3BHN A; 3FNB A; 4TTA A; 2X0E A; 4LGY A; 5DVZ A; 2QD1 A; 4GBG A; 2C5G A; 1E2U A; 4N65 A; 3KCN A; 2XR1 A; 1QDL B; 2O5E A; 4GGM X; 3NAT A; 1TRK A; 1Z2E A; 2OEN G; 1BEE A; 2O54 A; 2ZWI A; 3MW8 A; 1ZIC A; 4EJF A; 2P1Z A; 3TRD A; 2ZPA A; 3HJC A; 4RS3 A; 1DTW A; 4G24 A; 3WRY A; 1Z25 A; 4FEA A; 3A10 A; 3AIO A; 1HEY A; 5FWX A; 2H48 A; 5KC9 A; 3E1G A; 2HIH A; 3HCN A; 1R9J A; 4RHT A; 4B09 A; 1XRO A; 1OAO A; 5B3J C; 5AHX A; 4FYM A; 1LBQ A; 3R7S A; 4Z53 A; 3EAF A; 3QX9 A; 1GKK A; 4MRM A; 2C4H A; 2P2N A; 2XYP A; 4QXQ A; 3JSX A; 5DV9 A; 3FLA A; 4LNA A; 4O2A F; 1GKL A; 5EWJ A; 3I5A A; 3PEE A; 1K3F A; 3WWO A; 3SWW A; 2EZT A; 5HKB A; 3ZLB A; 2K2W A; 2MYP A; 1KJ9 A; 1TCV A; 1V45 E; 3BIX A; 3B3A A; 2JLA A; 1QGO A; 4B6G A; 1X7Y A; 1PDV A; 1GNT A; 3MOS A; 4WML A; 2ZYR A; 1QS0 A; 2H7Y A; 3W05 A; 3WQH A; 3K5H A; 2BSX A; 1GXS B; 2NZU G; 4XB6 B; 1QHE A; 1YMU A; 3IBF A; 3ILY A; 4UDX X; 1UM9 A; 3HCW A; 3LUA A; 2X2P A; 1AKT A; 2QRU A; 2Z98 A; 4D8Y A; 3UTD A; 4OYY A; 5AKL A; 4N47 A; 2P2D A; 4QFB A; 2NAP A; 3QFR A; 5FX2 A; 5TPZ A; 4ETN A; 4ZI7 F; 3V4Z A; 1JFL A; 4TM7 A; 3UQZ A; 3O3O B; 5AI6 A; 2N0S A; 1XHE A; 3H2J A; 3TTT A; 1E4E A; 4YQE A; 1U0S Y; 9ABP A; 4C87 A; 1CY4 A; 1W30 A; 4A16 A; 1YB7 A; 1PDW A; 1I4E B; 3SOZ A; 1DT5 A; 4RA9 B; 3RVS A; 3PHB E; 1WDW B; 1DKR A; 3L1I A; 1N5R A; 2QM0 A; 3OLX A; 4LOK A; 1KMC A; 2VT7 A; 2Q4O A; 1OZG A; 3STX A; 2RAU A; 3R44 A; 5JNT A; 4K6G A; 2PUD A; 5CBK A; 4MQU A; 1E60 A; 2JC3 A; 4O5A A; 4ZRE A; 5IEJ A; 1XAG A; 1XZN A; 4RHX A; 1CYE A; 1ECF A; 4RH3 A; 3I6Z A; 4IZO A; 4NS1 A; 4G0M A; 2OQV A; 1L7Q A; 4ZHQ F; 2Z8Z A; 3DKR A; 3O7M A; 4U2C A; 5KQM A; 3I09 A; 4JH0 A; 5DND A; 5IUK C; 2J9F B; 3M34 A; 4L4Y A; 4LA4 A; 1Y9D A; 4D8T A; 4ZMS A; 3NOR A; 3Q9S A; 3RVP A; 3CE6 A; 4JAS B; 3I9V 1; 3R74 A; 2V5A A; 3OVM A; 1LK0 A; 2PBZ A; 2O19 A; 4FOL A; 2JFY A; 1ZGC A; 2GN1 A; 3RH0 A; 1CB0 A; 2C31 A; 3HJF A; 2OZL B; 2C2G A; 3HCO A; 3ZHT A; 3EQZ A; 5AKH A; 1JPV A; 2J6B A; 3PGK A; 3TMY A; 5BUQ A; 3ZHO A; 3ZJ4 A; 1FFE A; 2Q3J A; 3N0W A; 1W6R A; 1T34 A; 1A69 A; 5T4B A; 1HDE A; 4M9U A; 4RQA A; 4KLC A; 4KVF A; 1RYZ A; 2PMC A; 1NC1 A; 3GJS A; 5HKL A; 2QX6 A; 4I14 A; 3OUU A; 1I2A A; 3TBH A; 1JQA A; 4V38 A; 4QNN A; 4UHC A; 3C45 A; 1CV2 A; 3P0E A; 4RE5 A; 2G25 A; 3E3M A; 1PE0 A; 1MX9 A; 4HNU A; 3UV3 A; 4RGQ A; 3FSJ X; 1QPZ A; 3H6H A; 4Z1O A; 4G0V A; 4MNT A; 3QVJ A; 4AY5 A; 2YIC A; 4AXB A; 4KMR A; 1GQS A; 5JAS A; 3DV0 B; 1QII A; 2OHO A; 4G0Y A; 2PU7 A; 1WET A; 1WB5 A; 3M6M D; 3G85 A; 3L84 A; 4BMP B; 4HNR A; 4MS1 A; 3SS9 X; 3GL9 A; 5JVD F; 3P9P A; 4GR9 A; 5K3C A; 2I78 A; 1J0A A; 1NXO A; 5J1S B; 4JUB A; 2XMQ A; 1RTT A; 4G0W A; 3C8H A; 1TCU A; 5LZ7 A; 1J9Z A; 2PAA A; 16PK A; 1QF7 A; 2NZV G; 5CGQ B; 2EYQ A; 2XCI A; 3E35 A; 2BKL A; 4HDM A; 2BMV A; 3S5J A; 3ZHV A; 2YAS A; 2ON6 A; 1UIN A; 4EB8 A; 3LS2 A; 1YKG A; 5BSU A; 4OGK A; 2IGB A; 2G67 A; 3R0V A; 1L1R A; 3KLB A; 3GJT A; 1NTR A; 2FSP A; 4PS1 A; 3ANS A; 1MJG A; 1LK5 A; 2B2N A; 2PL1 A; 4Z4H A; 4FFW A; 4RNC A; 5KBY A; 4FGK A; 4G36 A; 4RXT A; 4OHP A; 1WB6 A; 1OXM A; 5BUR A; 4JYM A; 1WM1 A; 1E5D A; 4WKP A; 1USU A; 4G86 A; 1RP7 A; 4IWY A; 3N39 C; 2HA7 A; 2ZPU A; 1EWT A; 5JNS A; 2PKX A; 1FFS A; 2VBI A; 2GWR A; 3DL4 A; 3IAS 6; 2MHC A; 4O1H A; 2WRZ A; 4DJD C; 2VPQ A; 1SG6 A; 3IE1 A; 4K6H A; 1HGX A; 1I5E A; 5D8D A; 4G5X A; 2PPV A; 4TN5 A; 2PQM A; 1Z8N A; 4XJV A; 2AC7 A; 2QLB A; 1SHL A; 5CML A; 4U7G A; 4QDU A; 2JI4 A; 4Q3C A; 2TRS B; 3CN7 A; 4Q24 A; 4CIA A; 1ULA A; 4RVO A; 5LXT F; 2OZL A; 4F3C A; 1JMK C; 2PSJ A; 3OPM A; 1A96 A; 2YZK A; 4LTD A; 4BYM A; 4F3N A; 5IFK A; 1QID A; 4Q4D A; 4BJ5 A; 4M0P A; 2ID9 A; 1BN7 A; 3M3P A; 1XI2 A; 2H0A A; 2FMF A; 4U2B A; 1D0S A; 5IAJ A; 2MYU A; 4OYL A; 5I9B A; 2Q4D A; 2JCG A; 4NUL A; 1A5S B; 1K88 A; 3DD5 A; 2O7R A; 1MO2 A; 4BDT A; 3EF3 A; 4MV3 A; 4E7O A; 1PV1 A; 4Y1R A; 1XZF A; 3S8Y A; 1QK5 A; 3PQ3 A; 3GFR A; 4KRX A; 5FOQ A; 2ABW A; 3WL5 A; 3ILS A; 1XZM A; 4UBN A; 4JEN A; 4UZS A; 2MWM A; 3BDI A; 3FW1 A; 4N4A A; 2Y3I A; 1TIK A; 1TAH A; 1MAH A; 1BS9 A; 2YCB A; 2I8A A; 2K7Z A; 4P1Z A; 4WXY B; 4QZV A; 3P9R A; 5JJC A; 3TG8 A; 3UXE A; 4RXU A; 2RK3 A; 4HNV A; 3F8S A; 2CLO B; 1CFJ A; 3EXG 1; 1AX9 A; 5D87 A; 1Q7R A; 5HKA A; 4EHK A; 4FGJ A; 1K66 A; 2ODT X; 5B3A A; 2PGO A; 3RHT A; 3UXH A; 3KXI A; 4ZDI A; 2AD5 A; 1DCF A; 5JZM A; 1BEU B; 4W1R A; 1QVW A; 3U16 A; 2I7T A; 4MEA A; 4DDV A; 3QEM B; 3R0X A; 3KOM A; 3IST A; 3O5A A; 3L7L A; 4R7G A; 1UMD B; 4J3J A; 3W2T A; 4Z4F A; 1YCH A; 3KDA A; 5I9T A; 3FAK A; 2ZIV B; 3A2L A; 2YJN A; 1WCW A; 2YJG A; 3EUL A; 3SVL A; 2HBQ A; 2V98 A; 1YI0 A; 4LDZ A; 4QUL A; 2I03 A; 4HRX A; 3BG5 A; 5LIR A; 4TXN A; 3HLK A; 4N9Q A; 4W5N A; 4V2U A; 1B00 A; 3OQO A; 3GRO A; 5JPI A; 5C34 B; 1JFS A; 5I3Z A; 4C76 A; 2H08 A; 3CNB A; 5HSG A; 1U2Q A; 3FBW A; 3EYW A; 5BV2 P; 1ZGB A; 4WXM A; 1ZN9 A; 4EAA A; 1VLQ A; 4XXH A; 3V1K A; 2JFZ A; 2IIQ A; 4DTC A; 2EQA A; 4DDU A; 1BCS A; 1JHO A; 1T35 A; 3T99 A; 1Y4S A; 3VKF A; 1S2G A; 4ZVN A; 1WHT B; 1PZM A; 4P36 A; 2I3D A; 3NKF A; 4RKR A; 3K8Q A; 3D6F A; 3K1Y A; 4D4G A; 2R5F A; 4UHE A; 2BP7 A; 2L2Q A; 1SXH A; 3GUV A; 4UHJ A; 3UAY A; 1DV1 A; 2X5X A; 1JHQ A; 2RHB A; 2JFP A; 5H80 A; 1KFJ B; 2FZ5 A; 4C6H A; 3IOM A; 3V1L A; 4PFJ A; 4D9F A; 4X91 A; 3NOX A; 4H7J A; 3N3B C; 4DNQ A; 2IPM A; 2FEX A; 2H3H A; 1FS6 A; 5KIN B; 4J49 A; 3WRX A; 3CG4 A; 5AM5 A; 2QV0 A; 2WDP A; 2IIV A; 4M9C A; 4N49 A; 2XUP A; 3GRC A; 3ESC A; 2EX1 A; 5CXS A; 1JX6 A; 1L4N A; 1FSP A; 3JZI A; 2OVF A; 4U2K A; 3EFH A; 2YZN A; 4NPA A; 3CW9 A; 3AF6 A; 1K3U B; 2E4X A; 2C83 A; 3FYU B; 2CUT A; 2A0W A; 5JXW A; 6PFK A; 5F12 A; 5KX5 F; 2IEA A; 1X74 A; 1BJT A; 2QXU A; 3SNK A; 1JF8 A; 1L5M A; 3HYN A; 1LGY A; 1IU9 A; 1IIB A; 2FQQ A; 3EXI B; 1Q0R A; 5BR1 A; 4OB6 A; 4LY3 A; 1CQZ A; 1P2F A; 4QXP A; 2A5L A; 1NGS A; 4DA8 A; 2WJ3 A; 4JK3 A; 4HS4 A; 5EJ9 A; 3I6M A; 4W5T A; 5TQ2 A; 5K8C A; 3W5S A; 2LP4 Y; 4M88 A; 4H7E A; 5BN4 B; 3H04 A; 2ZYH A; 2C5F A; 3KTO A; 4PXY A; 1EFA A; 1MPX A; 3H5V A; 2UVP B; 4LZN A; 1LK7 A; 1QCC A; 4KE7 A; 4LDP A; 3G7S A; 2RHW A; 4MB0 A; 4MZX A; 4XAS A; 1P0P A; 3U9S A; 3AFI A; 4G4S P; 4MB2 A; 1NY5 A; 2XGM A; 2OHG A; 3G0I A; 4FEG A; 4GZ5 A; 4QUE A; 2KM1 A; 1NW9 B; 4MQE A; 4HYE A; 3RKS A; 3YAS A; 2P9Q A; 4KXM A; 1AMO A; 3ONF A; 3F67 A; 4N5H X; 4IYR A; 5SYN A; 5AI5 A; 3ZI7 A; 1JHA A; 3Q7T A; 1JHV A; 2CWD A; 1ABE A; 2XWQ A; 4ZVS A; 4AN1 A; 1ZIY A; 3OQJ A; 1CQW A; 4PCU A; 3KJX A; 3HYL A; 5I4B A; 3G8C A; 5BOV A; 2J4F A; 2J32 A; 1Y6Z A; 1RHQ A; 3QFC A; 2XMG A; 3DMR A; 3H2K A; 4Z9A A; 1JFT A; 5A7G A; 4ZJP A; 2IY8 A; 4MV6 A; 4TTJ A; 4HVT A; 4D9H A; 5JAU A; 4EYK A; 1Z26 A; 2X0F A; 1G2Q A; 4EYO A; 1HG0 A; 4HNT A; 4IQ4 A; 4JEL A; 3DJY A; 2BEV A; 1QWS A; 1TMY A; 3GEP A; 3WZM A; 2WZD A; 2DSN A; 5CT6 A; 3MGB A; 1GPL A; 4G4J A; 4PX3 A; 2HIM A; 4IP0 A; 4GP9 A; 3QPD A; 2QTA A; 4QXR A; 2O2G A; 4AQD A; 2JFW A; 2LBP A; 2M6R A; 1BRO A; 1R8J A; 1JBE A; 1CY2 A; 4Z49 A; 1Y4U A; 4JIT A; 2ZJF A; 3N53 A; 5KNL A; 4EZI A; 3OPV A; 3BSW A; 4OHR A; 2VHE A; 4BC0 A; 1JLS A; 4TVK A; 4WWH A; 3SC2 B; 3P9Y A; 1I7N A; 4P5P A; 5B53 A; 4BC1 A; 2OQI A; 1DIN A; 5I7O A; 2VJY A; 4EGJ A; 3ONV A; 1QP4 A; 4Y6G B; 1K27 A; 4UOZ A; 4LOJ A; 1H69 A; 5FV4 A; 3C6X A; 1Y89 A; 4PAW A; 3NBM A; 2BI4 A; 1VKR A; 5J3J A; 2DX7 A; 4P9N A; 1A8S A; 3IUM A; 5FLE X; 1B0P A; 1W85 A; 2YW2 A; 5T4E A; 3JTE A; 5AKE A; 1U04 A; 4BX9 A; 4MAR A; 2ECO A; 3L4K A; 5AHR A; 2YZM A; 4E7P A; 4EHJ A; 3LP5 A; 3C5V A; 3E0Q A; 2XDY A; 5AOB A; 5TE9 A; 5HVN A; 1XHF A; 5HJ7 A; 3EMV A; 3VPC A; 4FM9 A; 1XRP A; 1FYE A; 1TJP B; 4NOM A; 2AJD A; 2YWC A; 5HM8 A; 3BJE A; 4EHH A; 3NNN A; 4OAT A; 4AO6 A; 3BL6 A; 4U2G A; 2EZ9 A; 5KRE A; 3TIN A; 1UJN A; 3P0F A; 2IHK A; 2A1I A; 2CUN A; 5HKO A; 4QIC A; 1Y1S A; 3D8U A; 5BN5 B; 2HRR A; 2Q9U A; 4EYQ A; 1JDP A; 1AUX A; 1XYV A; 4Y1G A; 1KY5 A; 4K6K A; 3I01 M; 4OXI A; 3Q41 A; 5HK9 A; 3RVL A; 5CXQ A; 5BQO A; 4D98 A; 3E7F A; 4D02 A; 1QRD A; 3S7T A; 4DIM A; 3WYD A; 3SK0 A; 2XCR B; 3W77 A; 5DKF A; 2CHU A; 3BXE A; 1SC4 A; 4NBK A; 2GSW A; 2XT6 A; 3KAP A; 1XZD A; 3GA7 A; 1G9S A; 2NT3 A; 2ZIW B; 4HDN A; 1LPM A; 4Z4C A; 1J9E A; 4NBL A; 2BF4 A; 3O8O B; 4NG4 A; 1HFK A; 3QE2 A; 3VR4 D; 3G9T A; 4DA6 A; 1VLZ A; 3RIX A; 4YAO A; 1ZVV A; 4DN6 A; 4RA6 B; 4TUY F; 5ADW A; 2WUF A; 1B3R A; 1OZH A; 2JF0 A; 4GM0 A; 3MA0 A; 3P9Z A; 1UOO A; 2MSL A; 1TA9 A; 2I3Z A; 2R1T B; 1JLK A; 2D1R A; 5BSM A; 5C8X A; 2BFB A; 2WIL A; 2V5V A; 4UPD A; 5CT4 A; 5ALE A; 4L1K A; 2YWJ A; 4C14 A; 1MJ5 A; 1RRM A; 4Y2U A; 2W6P A; 2ECQ A; 4ZFN A; 1VDM A; 3RVO A; 3OY2 A; 3HA2 A; 4I55 F; 4RU1 A; 3IAS 1; 5ETJ A; 5TPZ D; 3QYJ A; 3ANT A; 3HJH A; 1YAH A; 4JEU A; 4P3K A; 4EHF A; 3VJL A; 4N5D A; 4UBO A; 2R11 A; 5EWL A; 3ZWQ A; 2VQD A; 2O5C A; 4MPJ A; 2JI6 A; 4NCA A; 2C1E A; 5FLK A; 1SU8 A; 2WG1 A; 4U2F A; 2XDW A; 3D2B A; 2WYH A; 3ZR4 B; 1PBT A; 2Y1K A; 5JNR A; 3CN9 A; 2ZAI A; 1L4M A; 3C5W P; 1B6R A; 4P0Q B; 3FX2 A; 1CZU A; 4J0C A; 3NCT A; 4ICI A; 1A97 A; 2NXW A; 4Q3D A; 2JLC A; 4LVC A; 5HQ3 A; 3QEK A; 5D7B A; 4L8W B; 1VE5 A; 5BSV A; 4QOF A; 3M4Z A; 4F8J A; 5BQ3 A; 3ENZ A; 5A7F A; 4MS4 A; 4RHU A; 3PCX A; 1N8S A; 3RV3 A; 3ACC A; 1VJC A; 4HDR B; 1HFW A; 5CA0 F; 4BZW A; 4QUI A; 1LBH A; 1S3F A; 4FE4 A; 3CFY A; 1KJJ A; 5C1D A; 3D6H A; 4JUD X; 3UMM A; 1ISP A; 4JUC A; 3OC0 A; 1ZH2 A; 5HBG A; 3R6L A; 3HL0 A; 4JQY A; 4B83 A; 5JAT A; 4OJY A; 1FLN A; 1BXR B; 2WD9 A; 5DMX A; 4JUI A; 1D03 A; 4N39 A; 4EG0 A; 1DCM A; 5CFP A; 3A4Y A; 4R1P A; 3R3W A; 4Y1F A; 2XQJ A; 3OM1 A; 4YML A; 5AIC A; 3JPY A; 2BFE A; 1DWP A; 2ZJ1 A; 2FOX A; 1FCJ A; 1MW9 X; 1E5T A; 4XDG A; 2F8T A; 4N0P A; 1EB9 A; 2H7X A; 4QAT A; 5CFN A; 3DUF B; 1USI A; 3QPC A; 4D4H A; 4FNG A; 3LA8 A; 1K5P A; 3AW8 A; 4COO A; 4B31 A; 3JYH A; 3M82 A; 5DM7 0; 5EHQ A; 5C5C A; 4RDC A; 3O8O A; 1YK0 A; 2CZG A; 2YRW A; 4EHB A; 1DT3 A; 1IBS A; 1J56 A; 5K51 A; 4CI9 A; 1YTU A; 2FX5 A; 2NV2 B; 3SZU A; 5JQW A; 2WUD A; 3K4H A; 2Y1L A; 2OCL A; 3LY7 A; 2Z9B A; 3EJ6 A; 3ER6 A; 2OEC A; 2JGL A; 3K5I A; 3DDU A; 2Y4O A; 3I2H A; 3KXP A; 2C8J A; 2O59 A; 5ALM A; 1CX9 B; 4L0C A; 4MX8 A; 3W1W A; 5IJW A; 4FLE A; 4B7A A; 2VSY A; 3PD0 A; 2WT4 A; 2VPI A; 4DA7 A; 2BJH A; 3T5C A; 4G8B A; 4BCB A; 3E53 A; 1QIE A; 3WO0 A; 2EZ8 A; 1EDD A; 5IUJ C; 1L4B A; 2O2E A; 3SPX A; 4KYV A; 4MV8 A; 4DMK A; 3DEK A; 1UBS B; 4YVF A; 2M9M A; 1QO7 A; 5IPF A; 3BVP A; 1GUB A; 2J3Q A; 4LOL A; 1T9B A; 1K8Q A; 3HWX 1; 3H2I A; 1VHW A; 1Y7I A; 1DRK A; 5CCD A; 1QMP A; 3HDG A; 3RVQ A; 3DF7 A; 3QPA A; 1VJD A; 3WJ1 A; 3FGZ A; 3HHE A; 1E3Q A; 3RM3 A; 1I14 A; 4MPP A; 2CLL B; 2W5U A; 5J2U F; 1QGD A; 2BEU B; 2W48 A; 2HA2 A; 2WVG A; 1NXV A; 4S2U A; 1Q84 A; 2JKZ A; 4C89 A; 1M0S A; 1LI4 A; 6NUL A; 2ECF A; 4ZU6 X; 3QIT A; 5ALP A; 3G0G A; 2QD2 A; 4J0I A; 4DNP A; 1TLL A; 3C6Y A; 3N8D A; 1OXB B; 4ADS G; 3O2J A; 4OPM A; 1I80 A; 3GJQ A; 1TZ2 A; 5HCU A; 1KL7 A; 5TLS A; 3ZPX A; 4J6E A; 1NXT A; 2A9Q A; 1BEZ A; 4HDR A; 4FB4 A; 2A9O A; 1U8E A; 3UWM A; 1KEK A; 3IAF A; 4L6I A; 1QTI A; 2UZ0 A; 3V1N A; 1O1Y A; 4WN3 A; 1QLW A; 4L3W A; 1IUO A; 5J2T F; 2MT9 A; 4JB8 A; 5K2M A; 3O2N A; 2BAG A; 2M1Z A; 1YS2 X; 2G5T A; 1MGP A; 3WL6 A; 5EPU A; 3L7N A; 2XHE A; 3LPL A; 3VYU B; 4Y1E A; 5SYA A; 2HNB A; 3V9A A; 5EFO A; 2BN4 A; 2DR0 A; 4OXX A; 1W25 A; 1LBT A; 2J31 A; 3AIK A; 4MSU A; 2N1M A; 2W42 A; 4MUX A; 4F4D A; 2AF3 C; 2N3Z A; 2Z5G A; 1RSZ A; 3IEL A; 3DUF A; 2FLW A; 4LIP D; 1FRZ A; 3U40 A; 3H5T A; 2X0D A; 3E5N A; 3UHJ A; 2I7X A; 3NYI A; 4DYH A; 4U2D A; 3IEX A; 3Q15 C; 2EDA A; 4TTI A; 3VXK A; 1L4L A; 1ODT C; 3OZF A; 1V8B A; 5AK5 A; 1EK1 A; 2PBL A; 1GLV A; 5DWV A; 2X7J A; 1EB8 A; 3VYS B; 3BIL A; 2YIV X; 5JV5 A; 3UV7 A; 2WU4 A; 3JVD A; 1KGS A; 2XKJ E; 4QUJ A; 2PGA A; 2FJP A; 2DE0 X; 2FLV A; 4FHZ A; 3KWM A; 4NMW A; 4NSN A; 1PR5 A; 4KQH A; 2EDC A; 4GYY A; 4WXE A; 4B1F A; 1ZH4 A; 4DSZ A; 1BNC A; 4LXG A; 5CNI A; 5DNW A; 2IIT A; 4RAQ A; 1H0H A; 3KS9 A; 1J24 A; 3SS7 X; 2E4V A; 5K3F B; 1M6V A; 1E5V A; 4FHG A; 3VDX A; 2WP0 A; 2O0M A; 2ZIX A; 1U8T A; 3G0D A; 3IR6 A; 4L5J A; 1UKA A; 5ALQ A; 2LRH A; 2A0K A; 1M33 A; 4O3F A; 4COK A; 5LLD A; 1GDT A; 1CE8 A; 4KXV A; 2IOY A; 4D99 A; 3I42 A; 4DJE C; 2BEU A; 2AI2 A; 1HPL A; 3H0C A; 3FIJ A; 1YB6 A; 4J4B A; 1P18 A; 4LY9 A; 4WBN F; 3LWD A; 4Z99 A; 4YLE A; 2EQ5 A; 2Q2O A; 4E5V A; 5K5E A; 4OHK A; 1Y7H A; 1E5X A; 1GPK A; 3VNB A; 3HR4 A; 3K7E A; 2I4T A; 4G4I A; 3WK8 A; 1TIB A; 3BE8 A; 4LZA A; 4UBK A; 5EZY F; 3RFC A; 4BE4 A; 4NY1 A; 3GID A; 5IAB A; 4NS4 A; 1APB A; 2D0D A; 2LV8 A; 4BMY A; 3SIX A; 4LXT A; 2K87 A; 5CNM A; 4UFN A; 1UK7 A; 4PSC A; 3OMW A; 2AXE A; 2JKY A; 2R4Q A; 4IHA A; 1BKS B; 5BUS A; 1V16 A; 6CHY A; 4IL5 A; 4RGV A; 2XQF A; 2L69 A; 5DGD A; 2MSK A; 5E9A A; 3IDO A; 1DQS A; 2BUC A; 3HO6 A; 4LE0 A; 3BRW A; 1M73 E; 4O4I F; 4GEC A; 4F60 A; 1SJ9 A; 3M6L A; 3H11 A; 3OPY A; 4C5B A; 1FDI A; 1MB0 A; 4HVA A; 4M6W B; 5AKI A; 2BEW A; 1DXQ A; 1OVM A; 1VPW A; 3PUK A; 3A0R B; 3UKJ A; 1V3Q E; 1NVD A; 4RAO A; 4D57 A; 2WG2 A; 5CH3 A; 1HBJ A; 5CVC A; 1Z13 A; 3R23 A; 5LNK 6; 1DX9 A; 3IV7 A; 6YAS A; 5ALU A; 1WCX A; 4ISB A; 1A95 A; 3GFS A; 5B7G A; 3RVR A; 1ODI A; 1BE0 A; 5I4C A; 2GYW A; 5JIB A; 3R6W A; 1PR4 A; 1J1I A; 1V5G A; 5E4J A; 4ETI A; 1R88 A; 3ENQ A; 2LCI A; 4OQQ A; 4HNQ A; 4I4I A; 3WZ2 A; 3LHI A; 2EBH X; 2G28 A; 3ESY A; 7YAS A; 3GZJ A; 3H5W A; 3I2J A; 5TPW B; 3FLE A; 1ZI9 A; 1FVH A; 3H83 A; 2Q7D A; 3KE9 A; 3SQN A; 4M0M A; 1F0P A; 3B7W A; 1X7X A; 4BE9 A; 2XMS A; 3WDM A; 3G7N A; 3OLY A; 3B3Q A; 1I51 A; 2J4C A; 3OZE A; 3EQ9 A; 4H1T A; 4WY8 A; 3KYJ B; 5DTI A; 3DQZ A; 2ISQ A; 1DG9 A; 5IP5 A; 1GHT A; 4BN0 A; 1Z37 A; 5EIA A; 4E15 A; 4MQL A; 1A8Q A; 1I7L A; 4MS3 B; 3T6K A; 1P7Y A; 3MAJ A; 3S99 A; 4HDK B; 3FMV A; 1YS7 A; 4UNI A; 2JFQ A; 2H54 A; 5LWH A; 3KSR A; 2C2Z A; 4LMA A; 5E2I A; 4GLB A; 5FUM A; 2RHG B; 3WAJ A; 4F9G A; 4BBZ A; 1NU6 A; 5LYJ F; 2FP3 A; 3VIS A; 3URM A; 4J8P A; 1PWE A; 2JK1 A; 2QVB A; 3G5M A; 3QI7 A; 3LP8 A; 4QUB A; 4BVJ A; 1UMC B; 4LAF A; 1STO A; 3OX1 A; 2R1U A; 2XUK A; 4H4C A; 2TYS B; 4DMR A; 4EMT A; 1XRM A; 3CW8 X; 5AI4 A; 2CD7 A; 5LP6 F; 4UFO A; 4KSY A; 2WM2 A; 4RAN A; 4LEI A; 3WI7 A; 1O6F A; 5JD4 A; 3TDN A; 2YIJ A; 4M0U A; 4Z4Q A; 1E8N A; 2V9Z A; 3M49 A; 2R8P A; 4KC0 A; 3K3P A; 4Y2S A; 2GZS A; 4QEK A; 4OB7 A; 3BIJ A; 3O3N B; 5HK8 A; 4FEE A; 4ZVT A; 2F62 A; 3PQ5 A; 4HCJ A; 4MGE A; 5B3K A; 1ZI6 A; 3EDO A; 3H75 A; 4HRY A; 3U39 A; 2VJC A; 1J0B A; 3MTG A; 5H9W A; 1R3D A; 2XF2 A; 2X4I A; 1TD9 A; 4LX8 A; 1E6K A; 3UK1 A; 4EA7 A; 4M3N A; 5LWQ A; 1JJB A; 2GUW A; 3NON A; 2WFZ A; 1SRQ A; 4QVI A; 1GSH A; 4G8D A; 1J07 A; 4OCZ A; 3T9F A; 4RV5 A; 1G42 A; 1O2D A; 2DJ5 A; 2ZUW A; 3ODG A; 3GVP A; 1DC7 A; 1AKV A; 4FXO A; 1T0I A; 1YR3 A; 1OAO C; 2CLM B; 2R85 A; 1FQO A; 1QVV A; 3HEB A; 2HBZ A; 4U7F A; 1PF7 E; 5IW8 A; 1PR1 A; 3D0K A; 1FSG A; 5JAN A; 1GL9 B; 5HRC A; 1Q2U A; 2XYG A; 2ILV A; 2A8Y A; 8ABP A; 3HCP A; 2GK3 A; 2HQB A; 1O5O A; 2C84 A; 1BGW A; 1V71 A; 3GFV A; 4K2H A; 3HNO A; 2HRD A; 3FLM A; 1W9H A; 1R9N A; 5E4T A; 5A6S A; 4PTY A; 2CNO A; 1UK6 A; 4H7F A; 2B3D A; 4DDW A; 1TKA A; 3JPW A; 3C6B A; 1JYS A; 5ALZ A; 2ZIU B; 2F8S A; 3KAQ A; 6ABP A; 3ZKW A; 2AI1 A; 1A9T A; 3IE2 A; 2HPV A; 4QXO A; 1XZH A; 3RF7 A; 4GI1 A; 3IPL A; 1PK8 A; 4HDS A; 5EJM A; 3GRA A; 2Q2N A; 3UMY A; 1JLR A; 3G0C A; 4K6O A; 3QVM A; 4LTM A; 3HSY A; 1CY7 A; 1O58 A; 4H35 A; 3L9X A; 1ZES A; 3GPC A; 2JID A; 4G26 A; 3D4L A; 4B81 A; 1GSO A; 2L82 A; 5FWY B; 1YSC A; 4X6X A; 4RUL A; 3O95 A; 1O7J A; 4G1F A; 2ZJ6 A; 2CNL A; 4RK1 A; 2LR0 A; 4OHC A; 3MB8 A; 5I7U A; 2NN3 C; 4DNF A; 4BVK A; 1RHR A; 4YPV A; 1KZH A; 1EVE A; 1BD4 A; 1UK9 A; 4Q6W A; 4ECA A; 4EGQ A; 1VCO A; 1IVY A; 3ZLV A; 1WSB A; 4N48 A; 1U2E A; 1JWL A; 3UAW A; 3FYS A; 3I0N A; 1ZXX A; 1DMR A; 1XLU A; 3VPD A; 3FVR A; 1S2D A; 2W71 A; 2EGU A; 3AHJ A; 2XUF A; 2N9U A; 1C9D B; 2GI4 A; 4V3C A; 2RGU A; 1U5B B; 1FFC A; 2MR6 A; 3ICV A; 1OLX B; 3ESD A; 1OZF A; 3ZLU A; 3L49 A; 4MV7 A; 2ZAG A; 2DH6 A; 3VJM A; 2PX6 A; 4LZW A; 1F9E A; 1IBC A; 2IHJ A; 3IBT A; 4NLL A; 2X7X A; 3OD5 A; 4HQR A; 2V96 A; 5IVK A; 3I1I A; 5GAJ A; 5CB0 A; 3U4G A; 4HXF B; 3RAR A; 3EXG 2; 3M81 A; 1H66 A; 4UBL A; 5TQ0 A; 1TMO A; 1MIH A; 5AKY A; 4S05 A; 3O74 A; 4DCP A; 3OZB A; 1OIL A; 1OCE A; 4H0C A; 1WCI B; 3E9Z A; 2Y4N A; 1UKB A; 2IZ5 A; 5IVD A; 1EX9 A; 4BDS A; 1MNA A; 2IHV A; 4BMO B; 2XUI A; 1T5H X; 5ISM A; 3F7T A; 4X71 A; 2VH2 A; 1XTT A; 1RT9 A; 3DCN A;
#similar chains in the Genus database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unknotted ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...